Open Access Article
Open Access ArticlePeptide-based nanomaterials for gene therapy
Wei
Zhang
a,
Qing
Chen
a,
Feng
Wu
a,
Jun
Dai
 b,
Defang
Ding
*a,
Jun
Wu
b,
Defang
Ding
*a,
Jun
Wu
 *a,
Xiaoding
Lou
*a,
Xiaoding
Lou
 a and
Fan
Xia
a and
Fan
Xia
 a
a
aEngineering Research Center of Nano-Geomaterials of Ministry of Education, Faculty of Materials Science and Chemistry, China University of Geosciences, Wuhan 430074, China. E-mail: dingdf@cug.edu.cn; junwu@cug.edu.cn
bDepartment of Obstetrics and Gynecology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430074, China
First published on 8th December 2020
Abstract
Gene therapy is a novel therapeutic method and widely used for treating numerous diseases. However, the utilization of suitable materials for specifically and effectively delivering therapeutic oligonucleotides (ODNs) into a targeted site remains a great challenge. Regarding this, by possessing the capabilities of high loading rate, good stability, targeting ability and biological barrier penetration, peptide-based nanomaterials are developing rapidly and becoming a new trend in the application of gene therapy. In this review, a variety of peptide-based nanomaterials designed for gene therapy are briefly demonstrated, focusing on their main constituents in terms of three aspects: (1) small molecules, (2) nanoparticles and (3) polymers. Using peptide-based nanomaterials, a controlled gene delivery process can be achieved and directly monitored in a real-time mode. In future work, different designs of peptide-based nanomaterials need to be explored for higher gene transfection efficiency and a better therapeutic effect. In the end, some challenges and deficiencies are mentioned for bringing more attention to accelerate the research of peptide-based nanomaterials.
1 Introduction
In recent decades, gene therapy enjoyed growing interest in the field of clinical disease treatment.1–6 However, because of being easily subjected to degradation and poor cellular targeting and endosome escaping abilities, direct utilization of naked therapeutic ODNs as drugs was gradually avoided.5,7,8 Hence, establishing a system of selectively delivering therapeutic ODNs into targeted cells in pursuit of an effective disease therapy has drawn extensive attention.9–11Peptides, composed of amino acid monomers with amide bonds,12,13 are recognized as one group of the fundamental units to construct numerous bio-nanomaterials for applications including gene therapy.14–22 By possessing different R groups, amino acids have their individual structure and function.23,24 Currently, there are twenty natural amino acids and a large number of nonstandard amino acids. The combination and arrangement of different amino acids are highly diversified and varied, making the peptides rich in a variety of capabilities,25 such as tissue targeting,26–28 membrane penetration,29–33 endosome escaping34–37 and nuclear localization.38–40 Moreover, functional components including luminogens and polymers are widely used to conjugate with peptides, resulting in a variety of peptide-based nanomaterials.41–47 Compared with peptides without functional components, peptide-based nanomaterials can provide more therapeutic applications.48–52 For example, to achieve a high loading rate of drugs and better stability in vivo, the effective method is to introduce biopolymers. Herein, peptide-based nanomaterials have been largely enriched by linking with different components, which ensured specificity and selectivity and conferred satisfactory functions of gene therapy.53–58
Numerous significant studies of peptide-based nanomaterials for the purpose of gene therapy have been published recently. In this vein, we review several recent studies on peptide-based nanomaterials (Fig. 1) consisting of three different functional components including (1) small molecules, (2) nanoparticles, and (3) polymers, and focus on the design strategy for gene therapy. Finally, the difficulties and challenges of using peptide-based nanomaterials for gene therapy would be discussed, and a brief perspective of future tasks in this field is pointed out.
2 Function of peptides
We have summarized some functional peptides as shown in Table 1.| Type | Sequence | Function | Ref. |
|---|---|---|---|
| Targeting peptides | ASSLNIA | Treating heart and skeletal muscle | 59 |
| DDDDDD | Targeting hard tissue (bone and teeth) | 60 | |
| RGD | Targeting multiple integrin overexpression tumor cells | 61–63 | |
| KCCYSL | Targeting HER2-positive tumor cells | 64 and 65 | |
| (KLAKLAK)2 | Targeted destruction of the mitochondrial membrane | 66–69 | |
| Cell-penetrating peptides (CPPs) | RKKRQRRR | Penetrating the cell membrane | 29 and 70 |
| R9 | Penetrating the cell membrane | 71 and 72 | |
| CRGDKGPDC | Penetrating the cell membrane | 72 | |
| Endosomal escape peptides | GLFGAIAGFIENGWEGMIDG | Destructing the membrane of endosomes | 73 |
| WEAALAEAALEAALEAALEAALEAALAALA | Destructing the membrane of endosomes | 74 | |
| Nuclear localization signal (NLS) peptides | PKKKRKV | Providing intranuclear transport | 75 and 76 |
| GYGPKKKRKVGGC | Providing intranuclear transport | 77 | |
| KRRRR | Nucleocytoplasmic trafficking | 78 and 79 | |
| Responsive peptides | PLGVR, LGLAG | Responsive to metalloproteinase-2 (MMP-2) | 67 and 80–82 |
| DEVD | Responsive to caspase-3 | 83 and 84 | |
| GFLG | Responsive to cathepsin B (CB) | 85 |
3 Peptide-based nanomaterials combined with different molecules for gene therapy
3.1 Small molecules
Small molecules can easily diffuse through the cell membrane and reach the target site. And some small molecules such as fluorescent molecules86–91 and positively charged peptides92–94 are able to combine with negatively charged nucleic acids stably, thus improving the loading capacity, the targeting ability and the transfection efficiency of peptide-based nanomaterials and enabling the materials to be imaged in real-time. In recent decades, traditional fluorescent molecules have been studied widely, but their emissions could be weakened at high concentrations because of the aggregation-caused quenching (ACQ) effect.95,96 In 2001, Tang and his colleagues proposed the concept of aggregation-induced emission (AIE) to illustrate a series of fluorescent molecules that emit strong fluorescence in the highly aggregated state.97 Different from the case of fluorescent molecules affected by the ACQ effect, for AIE luminogens (AIEgens) there is no need to control their concentration during use and they can emit fluorescence in the aggregated state, and some AIEgens have photosensitivity, so they are widely used in bioimaging,98–101 biosensing102,103 and the field of biological therapy.104–109 Our group have reviewed some applications of AIE-based materials.110–112The covalent binding of AIEgens and peptides allows not only the biological functions of peptides but also the aggregation luminescence properties of AIEgens, which helps deliver gene therapy drugs. Our group developed self-assembled nanoparticles named TNCP/ASO-NPs (Fig. 2a), which could efficiently deliver antisense single-stranded DNA oligonucleotides (ASO) to the targeted cells.113 TNCP/ASO-NPs self-assembled from a peptide-conjugated AIEgen (TNCP) and ASO. The amphiphilic TNCP consisted of the hydrophobic AIEgen PyTPE and a peptide sequence which contained three parts: (1) a peptide (DGR or RGD) that helped TNCP/ASO-NPs to target integrin αvβ3; (2) a nuclear localization sequence (KRRRR) that assisted ASO to enter the nucleus through nuclear pore complexes; and (3) a CPP (RRRR) that assisted nanoparticles to enter the cell and escape from endosomes. As the hydrophobic core, the PyTPE promoted the self-assembly of TNCP/ASO-NPs into nanoparticles, mainly distributed between 76 and 198 nm (Fig. 2b). TNCP/ASO-NPs selectively bound to integrin αvβ3 and rapidly entered the cytoplasm, then partially released ASO in the cytoplasm to produce intracytoplasmic interference. What's more, TNCP/ASO-NPs selectively combined with importin and transported into the nucleus, then released most of the ASO in the nucleus to produce endonuclear interference. Within 5 minutes, PyTPE showed yellow fluorescence on the cell membrane, lasting more than 150 minutes, showing good real-time imaging characteristics. The correlation coefficient of TNCP and ASO-Cy5 and the confocal laser scanning microscopy (CLSM) and the biological electron microscopy (bio-TEM) cell images suggested that TNCP/ASO-NPs were delivered into overexpressed integrin αvβ3 cells effectively and disassembled effectively. In vivo experiments of tumor-bearing mice and western blot assays of tumor tissue showed that ASO was delivered into tumor tissues and it inhibited tumor growth by inhibiting Bcl-2 expression (Fig. 2c). By combining AIE with peptides and self-assembling with ASO, TNCP/ASO-NPs have significant specific nuclear imaging functions and tumor treatment effects, confirming the important role of small molecules in the imaging and structural self-assembly of peptide-based nanomaterials used for gene therapy.
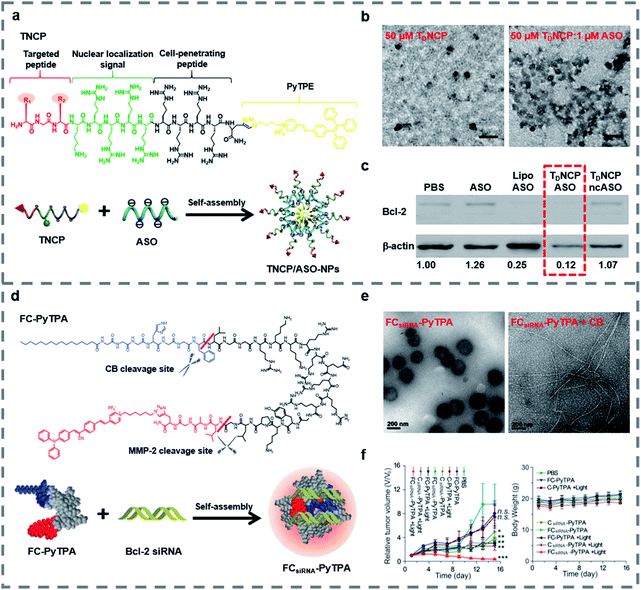 | ||
| Fig. 2 (a) Structure and function of TNCP and TNCP/ASO-NPs. (b) Transmission electron microscopy (TEM) images of TDNCP and TDNCP/ASO-NPs, scale bars: 200 nm. (c) Western blot of MDA-MB-231 cells incubated with different gene therapeutic agents. Reproduced with permission.113 Copyright 2019 Wiley-VCH. (d) Self-assembly of the FCsiRNA–PyTPA complex and schematic of cellular uptake and inhibition of Bcl-2 expression. (e) TEM images of FCsiRNA–PyTPA and FCsiRNA–PyTPA incubated with CB. (f) Relative volume changes (V/V0) and body weight change of tumors in each group during treatment. Reproduced with permission.114 Copyright 2020 Wiley-VCH. | ||
Recently, in order to further study peptide-conjugated AIEgen nanomaterials for gene therapy, our group reported a multiple-agent-therapy probe (MATP) named FCsiRNA–PyTPA (Fig. 2d).114 FCsiRNA–PyTPA was self-assembled from peptide-based molecular FC-PyTPA and siRNA, where FC-PyTPA was synthesized by conjugating small molecules C16 and AIEgen PyTPA with a short peptide. The short peptide was used to transport siRNA and respond to the tumor microenvironment, the amphiphilic structure with C16 and GGGH could self-assemble to form nanofibers and kill cancer cells and the AIEgen PyTPA was used for image-guided photodynamic therapy (PDT). When FCsiRNA–PyTPA reached the tumor site, MMP-2 divided FCsiRNA–PyTPA into FCsiRNA and PyTPA, which were internalized by macropinocytosis and caveolae-mediated endocytosis, respectively. Then, FCsiRNA entered the cells and self-assembled into nanofibers to destroy the structure of lysosomes through the reaction of CB, which was highly expressed in lysosomes. siRNA escaped from the lysosomes and silenced the gene. At the same time, we applied light at the right time to give the PDT effect of PyTPA full play. According to TEM (Fig. 2e), FCsiRNA self-assembled into nanoparticles, then transformed into nanofibers under the action of CB to destroy lysosomes as shown in bio-TEM. The CLSM and absorption spectra suggested good imaging characteristics and PDT activity of PyTPA. The co-localization experiment and western blot assay indicated that siRNA was carried into the cell, where it successfully escaped from lysosomes and down-regulated the expression of Bcl-2 to promote cancer cell death. Finally, the triple combination therapy successfully down-regulated the expression of anti-apoptotic proteins and effectively inhibited tumor growth (Fig. 2f). This work showed us that by combining small molecules with functional peptides, siRNA can be effectively transported, and also proved the important role of peptide-based nanomaterials in gene therapy and multiple therapy.
There are also other small molecules used in peptide-based nanomaterials to promote structural self-assembly and for gene therapy. For example, Kostarelos and his co-workers53 designed self-assembled peptide nanofibers (PNFs) composed of palmitoyl and a peptide (GGGAAAKRK), which could deliver siRNA and silence the target Bcl-2 in specific loci of the brain. Liu and co-workers115 combined a stearyl segment that self-assembled inside the structure in an aqueous environment with peptide to construct an α-helical structure and promote hydrophobic interactions with the cell membrane, which led to the improvement of gene transfection efficiency.
3.2 Nanoparticles
Nanoparticles such as gold nanoparticles (AuNPs), nanodiamonds (NDs), and porous silicon nanoparticles are widely studied because of their special physical and chemical properties, such as easy surface functionalization, good stability and easy size control, so they are often used in the field of gene therapy.116–118 When a peptide is combined with nanoparticles, it can improve the targeting ability of nanomaterials, reduce the biological toxicity and improve the therapeutic effect.AuNPs are some of the promising nanoparticles as they are nontoxic and can be easily modified on the surface or a good single nanoparticle carrier.119–121 As gold has been approved by the FDA for preclinical trials, gene modification of rat mesenchymal stem cells (rMSC) with AuNPs is expected to generate therapeutic cell lines. Strouse and co-workers122 designed a solid gold nanoparticle AuNP-complex for delivering genes into mesenchymal stem cells (MSC) (Fig. 3a). The surface of AuNPs was modified with a BDNF/mCherry fusion gene (6.6 kbp), which could express the BDNF/mCherry fusion protein, and a Ku70 peptide whose N-terminal was modified with cysteine through thiol linkage chemistry to form the AuNP-complex of about 130 nm (Fig. 3b). The Ku70 peptide is an intriguing pentapeptide from Ku70 which is a DNA repair binding sequence and plays a role in antiapoptosis. What's more, the N-terminal cysteine modified Ku70 peptide is zwitterionic and reduces electrostatic interactions between the BDNF/mCherry fusion gene and AuNP peptide surface which is effective for improving nanoparticle's gene transfection. According to the calculated loading level of the peptide per AuNP, a value of 3.6 ligands per nm2 was obtained. The optical images showed that more than 80% of rMSC cells expressed after transfection with the AuNP-complex within 4 days, and a Cedex Cell Counter and analyzer system showed that 98.3% of rMSC cells survived after single transfection, which indicated that the gene transfection of MSC population was successfully achieved by using the AuNP-complex with no significant effects on downstream of the MSCs. And the western blot analysis showed that the expression of BDNF protein was time-dependent in the media after gene transfection (Fig. 3c). This study established a new method to transfect the BDNF/mCherry fusion gene and proved that AuNPs modified by a gene and Ku70 peptide can effectively change the protein expression of MSCs and induce one-step transformation of rMSCs, while avoiding an apoptotic response.
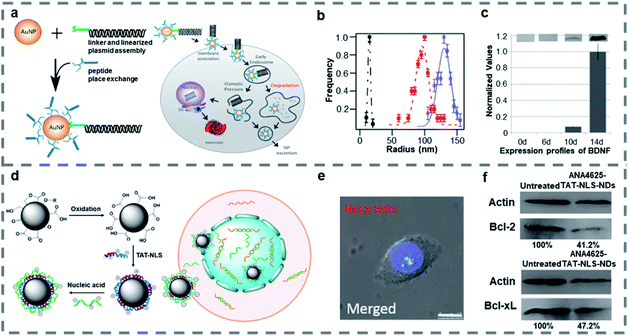 | ||
| Fig. 3 (a) Assembly of the AuNP-complex, the fusion gene and schematic of cellular uptake of the AuNP-complex and subsequent exogenous BDNF/mCherry protein expression. (b) Dynamic light scattering data for the AuNP-gene (blue, 130 nm), the linearized gene (red, 97 nm), and 6.6 nm water-soluble BSPP-AuNPs (black, 15 nm). (c) Time-dependent secretion levels for BDNF expression determined using western blot analysis. Reproduced with permission.122 Copyright 2014 American Chemical Society. (d) Synthetic scheme of TAT-NLS-NDs and cargo-loaded TAT-NLS-NDs and schematic of cargo-loaded TAT-NLS-NDs entering cells and nucleus. (e) CLSM of ANA4625-Cy3-TAT-NLS-NDs (red) in HeLa cells stained with Hoechst (blue), scale bar: 10 μm. (f) Western blot analysis of Bcl-2 and Bcl-xL expressions in MCF-7 cells after treatment with or without ANA4625-TAT-NLS NDs. Reproduced with permission.126 Copyright 2018 American Chemical Society. | ||
The NDs are attractive because of their long-term particle tracking in vivo and fluorescence cell labelling/imaging in vitro with no inflammatory indications and no immune response in animal models.123–125 Lo and co-workers126 reported cationic peptide-functionalized NDs to enhance cellular uptake and deliver loaded antisense ODNs to the nucleus (Fig. 3d). The 30 nm NDs were modified with dual-function, cationic TAT-NLS peptides in aqueous media, then the nucleic acid ANA4625 was loaded by electrostatic interaction. The TAT-NLS was formed from HIV TAT protein and SV40 large T protein NLS, which could improve the efficiency of gene transfection and expression. And the ANA4625 could reduce Bcl-2 and Bcl-xL gene expression in lung carcinoma cells. Wide-field optical imaging showed that the ANA4625 was delivered into the cells by TAT-NLS-NDs more effectively than by carboxyl-NDs, and CLSM showed that only TAT-NLS-NDs with smaller size could enter the nucleus without aggregation (Fig. 3e). Thiazolyl blue tetrazolium bromide (MTT) assay and western blot analysis (Fig. 3f) indicated that ANA4625 carried by TAT-NLS-NDs exhibited high cytotoxicity in MCF-7 cells by inhibiting the Bcl-2 and Bcl-xL gene expression. Compared to uncoated NDs, the TAT-NLS-NDs were confirmed as a more efficient carrier, which promoted the cellular uptake and delivered the nucleic acid into the nucleus with low cytotoxicity.
There are many other nanoparticles used in gene therapy. For example, Sailor and co-workers127 reported porous silicon nanoparticles combined with fusogenic liposomes and the outer sheath of homing peptides to selectively target macrophages and transfect an ODN into the cytosol. Langel and co-workers128 developed a new platform for transfecting plasmid (pGL3), SCO and siRNA by using magnetic nanoparticles (MNPs, Fe3O4) and CPPs.
3.3 Polymers
Polymers can form a stable complex with a gene and have the characteristics of high safety, low immunogenicity and easy structure modification.129,130 In gene therapy, cationic polymers and hydrophilic polymers are widely used. Cationic polymers such as PEI and chitosan can compress DNA and avoid degradation by lysosomes.131,132 Hydrophilic polymers such as PEO and PEG can improve the water solubility and stability of the materials.133,134 When the polymers are combined with peptides, the targeting property, stability and gene transfection efficiency of the materials can be improved.Some peptides are hydrophobic and easily precipitate and aggregate in vivo. When peptides combine with hydrophilic polymers, they can form a self-assembly structure, which increases the hydrophilicity, the in vivo stability of the materials and contributes to the uptake by target cells.135–138 Leong and co-workers139 reported PEGylated nanoparticles (P-HNPs) (Fig. 4a) self-assembled from the cationic α-helical peptide named PPABLG for delivering Cas9 expression plasmid with sgRNA. The traditional CPPs either were too small to penetrate the cell membrane or lacked an adequate cationic charge density, which made CPPs unable to be used as an independent gene vector. However, PPABLG could bind and concentrate short siRNA and plasmid DNA while maintaining high membrane-penetrating ability for enhancing the ability of cellular internalization and endosomal escape. In order to enhance the extracellular stability and the potential applications in vivo, the PPABLG-Cas9 expression plasmid and sgRNA complexes (HNPs) were modified using PEG-polythymine40 (PEG-T40) in the formulation without changing the size of the nanoparticles (Fig. 4b). By studying the internalization pathway of P-HNPs, it was found that the internalization of P-HNPs was related to the clathrin-mediated pathway, macropinocytosis and caveolae-mediated uptake. The colloidal stable-HNPs achieved up to 60% Cas9 transfection efficiency and 67.4% sgRNA uptake efficiency. In vitro, P-HNPs were used to deliver Cas9 expression plasmids and Plk1-targeting sgRNAs into HeLa cells, resulting in obvious inhibition at the target site and causing cell apoptosis by 46.2%. Experiments in vivo and western blot analysis (Fig. 4c) confirmed that P-HNPPCas9+sgPlk1 led to a tumor suppression of >71% and decreased the expression level of Plk1 protein by 67%.
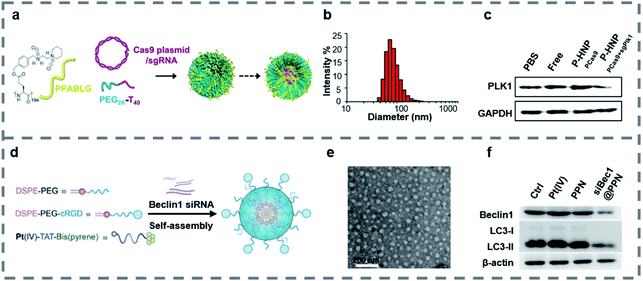 | ||
Fig. 4 (a) Schematic illustration showing the formation of P-HNPs and the intracellular activity of Cas9 expression plasmid/sgRNA in performing genome editing or gene activation. (b) Size of P-HNPs formed from PPABLG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) px458 px458![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG2K-T40 at a ratio of 30 PEG2K-T40 at a ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. (c) The expression level of Plk1 protein in tumor tissues. Reproduced with permission.139 Copyright 2018 the author(s) published by PNAS. (d) Illustration of the self-assembled nanoprodrug system. (e) TEM images of siBec1@PPN. (f) Western blot of Beclin1 and LC3-II in Pt-resistant A549 tumor cells treated with PBS (Ctrl), Pt(IV), PPN, and siBec1@PPN. Reproduced with permission.145 Copyright 2019 American Chemical Society. 1. (c) The expression level of Plk1 protein in tumor tissues. Reproduced with permission.139 Copyright 2018 the author(s) published by PNAS. (d) Illustration of the self-assembled nanoprodrug system. (e) TEM images of siBec1@PPN. (f) Western blot of Beclin1 and LC3-II in Pt-resistant A549 tumor cells treated with PBS (Ctrl), Pt(IV), PPN, and siBec1@PPN. Reproduced with permission.145 Copyright 2019 American Chemical Society. | ||
Platinum is a widely used chemotherapy drug to treat a variety of cancers.140,141 And various polymeric nanomaterials were designed to deliver cisplatin, which improved the effects of the circulation in vivo of cisplatin.142,143 What's more, inhibition of autophagy may reduce the resistance of tumor cells to cisplatin.144 Wang and co-workers145 developed a self-assembled nanoprodrug platform named siBec1@PPN containing cisplatin and Beclin1 RNAi (siBec1 is an autophagy initiation factor), which could inhibit the growth of the cisplatin-resistant tumor. The nanoprodrug platform was self-assembled using three parts including Pt(IV)-peptide-bis(pyrene), DSPE-PEG and cRGD-modified DSPE-PEG (Fig. 4d). The Pt(IV)-peptide-bis(pyrene) loaded Pt(IV) with a load rate up to 95%, released Pt(II) in the presence of glutathione (GSH) and delivered siBec1 into the cytoplasm, which inhibited autophagy and increased the cell apoptosis. The DSPE-PEG increased the stability and biocompatibility of the nanoparticles and the cRGD helped nanoparticles to target the tumor cells. As TEM images (Fig. 4e) show, the siBec1@PPN is a spherical structure with a size of 54.44 ± 11.13 nm. What's more, the western blot (Fig. 4f) showed that the siBec1 was transfected successfully and it inhibited the Beclin1 expression on A549 tumor cells. Animal study results showed that the siBec1@PPN reversed Pt resistance by siBec1-mediated autophagy inhibition and the tumor cells were apoptotic. Overall, siBec1@PPN showed us the ability of peptide–polymer complexes to transfer genes effectively and inhibit the growth of tumor cells.
Due to the excellent properties of polymers, a large number of studies have been carried out. For example, Rosenecker and co-workers146 developed compact and monodisperse peptide-poloxamine nanoparticles, which could long-term repair the cystic fibrosis transmembrane conductance regulator defect in vitro and in vivo with a safe profile. Wang and co-workers147 reported that polyurethane nanomicelles modified by an osteoblast-targeting peptide could selectively target osteoblasts to deliver siRNA/microRNA without overt toxicity or causing an immune response in vivo.
4 Conclusions
In this review, several peptide-based nanomaterials self-assembled from peptides and three individual functional components are introduced for the application of gene therapy. Compared with pure peptide sequences, peptide-based nanomaterials with other functional components appear to be more ideal gene delivery materials by taking advantage of multifunctionality, good biocompatibility, high load rate etc. In order to improve the specific targeting and transfection efficiency for gene therapy and add additional features to the design, more details but not limited to the following issues need to be considered: (1) the design of small molecule–peptide nanomaterials requires rational selection of suitable functional polypeptides and small molecules, in terms of the length, electrical properties, hydrophilicity and hydrophobicity of peptides and structures of small molecules, to avoid the interference between peptides and small molecules; (2) there is room for improvement of preparation of nanoparticle–peptide nanomaterials to enhance the homogeneity of products and reduce the side effects in vivo; (3) the non-specific adsorption of polymers could be further eliminated, and accelerating the targeting of polymer–peptide nanomaterials to tumor cells would be more favored; (4) the design strategy of peptide-based materials such as the load part of ODNs, biological functional peptides, the ratio of peptides and other components and the self-assembly structure should be considered. However, peptide-based nanomaterials used in gene therapy still have some problems such as poor structural stability in serum, being easily degraded in vivo and low gene loading rate. What's more, the combination of peptides with other components without affecting the function of each part is still a problem to be solved. In the future, it is necessary to study the secondary structure of peptides in depth and to improve the circulation time in vivo and the accumulation of peptide-based nanomaterials in the target tissue for clinical use. And the applications of peptide-based nanomaterials in other fields, such as cancer treatment, bioimaging, and tissue biomaterial engineering, need to be further studied.On the basis of notable achievements, peptide-based nanomaterials will be developed with more novel and efficient designs in the near future. In order to obtain advanced functional peptide-based nanomaterials, for example, it is necessary to explore the complex structure of peptides containing unnatural amino acids and optimize multifunctional components to ensure their precise functions. Because of the space limitation, this review is confined to three components (small molecules, nanoparticles, and polymers) which construct peptide-based nanomaterials. However, we hope that it will be able to promote the research of peptide-based nanomaterials for gene therapy and accelerate their development in the fields of nanomaterials, biology and chemistry.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Key R&D Program of China (2020YFA0211200), the National Natural Science Foundation of China (21722507, 21525523, 21974128, 21874121 and 21802130), the Natural Science Foundation of Hubei Province (2019CFA043), the project funded by the China Postdoctoral Science Foundation (2020M672436), the Open Research Fund of State Key Laboratory of Bioelectronics, Southeast University, and the Hubei Postdoctoral Innovative Research Foundation (to Jun Wu).Notes and references
- O. Khorkova and C. Wahlestedt, Nat. Biotechnol., 2017, 35, 249–263 CrossRef CAS.
- M. Matsui and D. R. Corey, Nat. Rev. Drug Discovery, 2017, 16, 167–179 CrossRef CAS.
- A. Wittrup and J. Lieberman, Nat. Rev. Genet., 2015, 16, 543–552 CrossRef CAS.
- C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359, eaan4672 CrossRef.
- S. F. Dowdy, Nat. Biotechnol., 2017, 35, 222–229 CrossRef CAS.
- M. Wang, Z. A. Glass and Q. Xu, Gene Ther., 2017, 24, 144–150 CrossRef CAS.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS.
- J. Chen, L. Lin, Z. P. Guo, C. N. Xu, H. Y. Tian, K. Park and X. S. Chen, J. Drug Deliv. Sci. Technol., 2015, 30, 417–423 CrossRef CAS.
- D. Ibraheem, A. Elaissari and H. Fessi, Int. J. Pharm., 2014, 459, 70–83 CrossRef CAS.
- M. R. Molla and P. A. Levkin, Adv. Mater., 2016, 28, 1159–1175 CrossRef CAS.
- H. J. Kim, A. Kim, K. Miyata and K. Kataoka, Adv. Drug Delivery Rev., 2016, 104, 61–77 CrossRef CAS.
- G. Qi, Y. Gao, L. Wang and H. Wang, Adv. Mater., 2018, 30, 1703444 CrossRef.
- C. Zhang, W. Wu, R. Li, W. Qiu, Z. Zhuang, S. Cheng and X. Zhang, Adv. Funct. Mater., 2018, 28, 1804492 CrossRef.
- C. D. Fjell, J. A. Hiss, R. E. Hancock and G. Schneider, Nat. Rev. Drug Discovery, 2012, 11, 37 CrossRef CAS.
- M. Shadidi and M. Sioud, Drug Resist. Updates, 2003, 6, 363 CrossRef CAS.
- P. Vlieghe, V. Lisowski, J. Martinez and M. Khrestchatisky, Drug Discovery Today, 2010, 15, 40 CrossRef CAS.
- M. Abbas, Q. Zou, S. Li and X. Yan, Adv. Mater., 2017, 29, 201605021 CrossRef.
- K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82 CrossRef CAS.
- P. Midoux, C. Pichon, J. Yaouanc and P. Jaffrès, Br. J. Pharmacol., 2009, 157, 166–178 CrossRef CAS.
- M. E. Martin and K. G. Rice, AAPS J., 2007, 9, E18–E29 CrossRef CAS.
- H. Wang, Z. Feng and B. Xu, Theranostics, 2019, 9, 3212–3222 Search PubMed.
- J. Hoyer and I. Neundorf, Acc. Chem. Res., 2012, 45, 1048–1056 CrossRef CAS.
- R. V. Ulijn and A. M. Smith, Chem. Soc. Rev., 2008, 37, 664–675 RSC.
- A. Kidera, Y. Konishi, M. Oka, T. Ooi and H. A. Scheraga, J. Protein Chem., 1985, 4, 23–55 CrossRef CAS.
- Z. Kang, Q. Meng and K. Liu, J. Mater. Chem. B, 2019, 7, 1824–1841 RSC.
- W. Wang and Z. Hu, Adv. Mater., 2019, 31, 1804827 CrossRef CAS.
- S. Lee, J. Xie and X. Chen, Biochemistry, 2010, 49, 1364–1376 CrossRef CAS.
- F. Araste, K. Abnous, M. Hashemi, S. M. Taghdisi, M. Ramezani and M. Alibolandi, J. Controlled Release, 2018, 292, 141–162 CrossRef CAS.
- M. Green, M. Ishino and P. M. Loewenstein, Cell, 1989, 58, 215–223 CrossRef CAS.
- P. Lundberg and Ü. Langel, J. Mol. Recognit., 2003, 16, 227–233 CrossRef CAS.
- J. Temsamani and P. Vidal, Drug Discovery Today, 2004, 9, 1012–1019 CrossRef CAS.
- T. Lehto, K. Ezzat, M. J. Wood and S. E. Andaloussi, Adv. Drug Delivery Rev., 2016, 106, 172–182 CrossRef CAS.
- N. Nakayama, S. Takaoka, M. Ota, K. Takagaki and K. I. Sano, Langmuir, 2018, 34, 14286–14293 CrossRef CAS.
- P. Midoux and M. Monsigny, Bioconjugate Chem., 1999, 10, 406–411 CrossRef CAS.
- S. L. Lo and S. Wang, Biomaterials, 2008, 29, 2408–2414 CrossRef CAS.
- K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef CAS.
- J. G. Schellinger, J. A. Pahang, R. N. Johnson, D. S. H. Chu, D. L. Sellers, D. O. Maris, A. J. Convertine, P. S. Stayton, P. J. Horner and S. H. Pun, Biomaterials, 2013, 34, 2318–2326 CrossRef CAS.
- M. van der Aa, E. Mastrobattista, R. S. Oosting, W. E. Hennink, G. A. Koning and D. J. A. Crommelin, Pharm. Res., 2006, 23, 447–459 CrossRef CAS.
- J. Robbins, S. M. Dilworth, R. A. Laskey and C. Dingwall, Cell, 1991, 64, 615–623 CrossRef CAS.
- Y. Y. Xu, W. L. Liang, Y. S. Qiu, M. Cespi, G. F. Palmieri, A. J. Mason and J. K. W. Lam, Mol. Pharm., 2016, 13, 3141–3152 CrossRef CAS.
- T. Jiang, T. A. Meyer, C. Modlin, X. Zuo, V. P. Conticello and Y. Ke, J. Am. Chem. Soc., 2017, 139, 14025–14028 CrossRef CAS.
- A. T. Preslar, G. Parigi, M. T. McClendon, S. S. Sefick, T. J. Moyer, C. R. Haney, E. A. Waters, K. W. MacRenaris, C. Luchinat and S. I. Stupp, ACS Nano, 2014, 8, 7325 CrossRef CAS.
- S. Silva, A. J. Almeida and N. Vale, Biomolecules, 2019, 9, 22 CrossRef.
- G. Wei, Y. Wang, X. Huang, H. Hou and S. Zhou, Small Methods, 2018, 2, 1700358 CrossRef.
- J. Y. Shu, B. Panganiban and T. Xu, Annu. Rev. Phys. Chem., 2013, 64, 631–657 CrossRef CAS.
- J. Couet, J. D. J. S. Samuel, A. Kopyshev, S. Santer and M. Biesalski, Angew. Chem., Int. Ed., 2005, 44, 3297–3301 CrossRef CAS.
- D. Zaramella, P. Scrimin and L. J. Prins, J. Am. Chem. Soc., 2012, 134, 8396–8399 CrossRef CAS.
- L. Zhang, D. Jing, N. Jiang, T. Rojalin, C. M. Baehr, D. Zhang, W. Xiao, Y. Wu, Z. Cong, J. J. Li, Y. Li, L. Wang and K. S. Lam, Nat. Nanotechnol., 2020, 15, 145–153 CrossRef CAS.
- Q. Luo, Y. Lin, P. Yang, Y. Wang, G. Qi, Z. Qiao, B. Li, K. Zhang, J. Zhang, L. Wang and H. Wang, Nat. Commun., 2018, 9, 1802 CrossRef.
- K. Han, W. Zhang, J. Zhang, Q. Lei, S. Wang, J. Liu, X. Zhang and H. Han, Adv. Funct. Mater., 2016, 26, 4351–4361 CrossRef CAS.
- H. Wang, Z. Feng, W. Tan and B. Xu, Bioconjugate Chem., 2019, 30, 2528–2532 CrossRef CAS.
- G. Qi, F. Hu, Kenry, L. Shi, M. Wu and B. Liu, Angew. Chem., Int. Ed., 2019, 58, 16229–16235 CrossRef CAS.
- M. Mazza, M. Hadjidemetriou, I. de Lázaro, C. Bussy and K. Kostarelos, ACS Nano, 2015, 9, 1137–1149 CrossRef CAS.
- Y. Ruff, T. Moyer, C. J. Newcomb, B. Demeler and S. I. Stupp, J. Am. Chem. Soc., 2013, 135, 6211–6219 CrossRef CAS.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS.
- T. Fan, X. Yu, B. Shen and L. Sun, J. Nanomater., 2017, 2017, 4562474 Search PubMed.
- R. Kedmi, N. Veiga, S. Ramishetti, M. Goldsmith, D. Rosenblum, N. Dammes, I. Hazan-Halevy, L. Nahary, S. Leviatan-Ben-Arye, M. Harlev, M. Behlke, I. Benhar, J. Lieberman and D. Peer, Nat. Nanotechnol., 2018, 13, 214–219 CrossRef CAS.
- Li. Liu, Z. Li, L. Rong, S. Qin, Q. Lei, H. Cheng, X. Zhou, R. Zhuo and X. Zhang, ACS Macro Lett., 2014, 3, 467–471 CrossRef CAS.
- C. Y. Yu, Z. Yuan, Z. Cao, B. Wang, C. Qiao, J. Li and X. Xiao, Gene Ther., 2009, 16, 953–962 CrossRef CAS.
- S. Kasugai, R. Fujisawa, Y. Waki, K. I. Miyamoto and K. Ohya, J. Bone Miner. Res., 2000, 15, 936–943 CrossRef CAS.
- Y. Dong, T. Yu, L. Ding, E. Laurini, Y. Huang, M. Zhang, Y. Weng, S. Lin, P. Chen, D. Marson, Y. Jiang, S. Giorgio, S. Pricl, X. Liu, P. Rocchi and L. Peng, J. Am. Chem. Soc., 2018, 140, 16264–16274 CrossRef CAS.
- Y. Liu, L. Mei, C. Xu, Q. Yu, K. Shi, L. Zhang, Y. Wang, Q. Zhang, H. Gao, Z. Zhang and Q. He, Theranostics, 2016, 6, 177–191 CrossRef CAS.
- C. Mas-Moruno, R. Fraioli, F. Rechenmacher, S. Neubauer, T. G. Kapp and H. Kessler, Angew. Chem., Int. Ed., 2016, 55, 7048–7067 CrossRef CAS.
- S. R. Kumar, F. A. Gallazzi, R. Ferdani, C. J. Anderson, T. P. Quinn and S. L. Deutscher, Cancer Biother.Radiopharm., 2010, 25, 693–703 CrossRef CAS.
- S. R. Kumar, T. P. Quinn and S. L. Deutscher, Clin. Cancer Res., 2007, 13, 6070–6079 CrossRef CAS.
- H. M. Ellerby, W. Arap, L. M. Ellerby, R. Kain, R. Andrusiak, G. D. Rio, S. Krajewski, C. R. Lombardo, R. Rao, E. Ruoslahti, D. E. Bredesen and R. Pasqualini, Nat. Med., 1999, 5, 1032–1038 CrossRef CAS.
- K. Han, Q. Lei, S. B. Wang, J. J. Hu, W. X. Qiu, J. Y. Zhu, W. N. Yin, X. Luo and X. Z. Zhang, Adv. Funct. Mater., 2015, 25, 2961–2971 CrossRef CAS.
- L. Agemy, D. Friedmann-Morvinski, V. R. Kotamraju, L. Roth, K. N. Sugahara, O. M. Girard, R. F. Mattrey, I. M. Verma and E. Ruoslahti, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 17450–17455 CrossRef CAS.
- Z. Y. Qiao, Y. X. Lin, W. J. Lai, C. Y. Hou, Y. Wang, S. L. Qiao, D. Zhang, Q. J. Fang and H. Wang, Adv. Mater., 2016, 28, 1859–1867 CrossRef CAS.
- A. D. Frankel and C. O. Pabo, Cell, 1988, 55, 1189–1193 CrossRef CAS.
- S. Y. Li, H. Cheng, W. X. Qiu, L. H. Liu, S. Chen, Y. Hu, B. R. Xie, B. Li and X. Z. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 28319–28329 CrossRef CAS.
- Y. Cheng, F. Huang, X. Min, P. Gao, T. Zhang, X. Li, B. Liu, Y. Hong, X. Lou and F. Xia, Anal. Chem., 2016, 88, 8913–8919 CrossRef CAS.
- K. N. Sugahara, P. Scodeller, G. B. Braun, T. H. de Mendoza, C. M. Yamazaki, M. D. Kluger, J. Kitayama, E. Alvarez, S. B. Howell, T. Teesalu, E. Ruoslahti and A. M. Lowy, J. Controlled Release, 2015, 212, 59–69 CrossRef CAS.
- S. F. Ye, M. M. Tian, T. X. Wang, L. Ren, D. Wang, L. H. Shen and T. Shang, Nanomed. Nanotechnol. Biol. Med., 2012, 8, 833–841 CrossRef CAS.
- N. K. Subbarao, R. A. Parente, F. C. Szoka Jr, L. Nadasdi and K. Pongracz, Biochemistry, 1987, 26, 2964–2972 CrossRef CAS.
- D. Kalderon, W. D. Richardson, A. F. Markham and A. E. Smith, Nature, 1984, 311, 33–38 CrossRef CAS.
- D. Kalderon and A. E. Smith, Virology, 1984, 139, 109–137 CrossRef CAS.
- B. K. Kim, H. Kang, K. O. Doh, S. H. Lee, J. W. Park, S. J. Lee and T. J. Lee, Bioorg. Med. Chem. Lett., 2012, 22, 5415–5418 CrossRef CAS.
- A. Leonidova, V. Pierroz, R. Rubbiani, Y. Lan, A. G. Schmitz, A. Kaech, R. K. O. Sigel, S. Ferrari and G. Gasser, Chem. Sci., 2014, 5, 4044–4056 RSC.
- Y. Liu, D. Zhang, Z. Y. Qiao, G. B. Qi, X. J. Liang, X. G. Chen and H. Wang, Adv. Mater., 2015, 27, 5034–5042 CrossRef CAS.
- P. D. Nguyen, V. T. Cong, C. Baek and J. Min, Biosens. Bioelectron., 2017, 89, 666–672 CrossRef CAS.
- L. I. Fessler, K. G. Duncan, J. H. Fessler, T. Salo and K. Tryggvason, J. Biol. Chem., 1984, 259, 9783–9789 CAS.
- Y. Lu, W. Sun and Z. Gu, J. Controlled Release, 2014, 194, 1–19 CrossRef CAS.
- W. Song, J. Kuang, C. X. Li, M. K. Zhang, D. W. Zheng, X. Zeng, C. J. Liu and X. Z. Zhang, ACS Nano, 2018, 12, 1978–1989 CrossRef CAS.
- Y. Yuan, C. J. Zhang, M. Gao, R. Zhang, B. Z. Tang and B. Liu, Angew. Chem., Int. Ed., 2015, 54, 1780–1786 CrossRef CAS.
- M. Gao and B. Z. Tang, Coord. Chem. Rev., 2020, 402, UNSP 213076 CrossRef.
- Z. Zhao, H. Zhang, J. W. Y. Lam and B. Z. Tang, Angew. Chem., Int. Ed., 2020, 59, 9888–9907 CrossRef CAS.
- D. Wang and B. Z. Tang, Acc. Chem. Res., 2019, 52, 2559–2570 CrossRef CAS.
- H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC.
- X. Wu, W. Shi, X. Li and H. Ma, Acc. Chem. Res., 2019, 52, 1892–1904 CrossRef CAS.
- S. Ding and Y. Hong, Chem. Soc. Rev., 2020, 49, 8354–8389 RSC.
- R. A. Petros and J. M. DeSimone, Nat. Rev. Drug Discovery, 2010, 9, 615–627 CrossRef CAS.
- A. Nel, T. Xia, L. Mdler and N. Li, Science, 2006, 311, 622–627 CrossRef CAS.
- I. W. Hamley, Chem. Rev., 2017, 117, 14015–14041 CrossRef CAS.
- D. Ding, K. Li, B. Liu and B. Z. Tang, Acc. Chem. Res., 2013, 46, 2441–2453 CrossRef CAS.
- J. Mei, Y. Huang and H. Tian, ACS Appl. Mater. Interfaces, 2018, 10, 12217–12261 CrossRef CAS.
- J. Luo, Z. Xie, J. W. Y. Lam, L. Cheng, H. Chen, C. Qiu, H. S. Kwok, X. Zhan, Y. Liu, D. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740–1741 RSC.
- X. Wang, J. Dai, X. Min, Z. Yu, Y. Cheng, K. Huang, J. Yang, X. Yi, X. Lou and F. Xia, Anal. Chem., 2018, 90, 8162–8169 CrossRef CAS.
- Y. Cheng, J. Dai, C. Sun, R. Liu, T. Zhai, X. Lou and F. Xia, Angew. Chem., Int. Ed., 2018, 57, 3123–3127 CrossRef CAS.
- R. Hu, B. Chen, Z. Wang, A. Qin, Z. Zhao, X. Lou and B. Z. Tang, Biomaterials, 2019, 203, 43–51 CrossRef CAS.
- X. Ni, X. Zhang, X. Duan, H. Zheng, X. Xue and D. Ding, Nano Lett., 2019, 19, 318–330 CrossRef CAS.
- X. Xu, W. Zhao, P. Gao, H. Li, G. Feng and X. Lou, NPG Asia Mater., 2016, 8, e234 CrossRef CAS.
- Z. Long, S. Zhan, P. Gao, Y. Wang, X. Lou and F. Xia, Anal. Chem., 2018, 90, 577–588 CrossRef CAS.
- J. Dai, Y. Li, Z. Long, R. Jiang, Z. Zhuang, Z. Wang, Z. Zhao, X. Lou, F. Xia and B. Z. Tang, ACS Nano, 2020, 14, 854–866 CrossRef CAS.
- X. Yi, J. Dai, Y. Han, M. Xu, X. Zhang, S. Zhen, Z. Zhao, X. Lou and F. Xia, Commun. Biol., 2018, 1, 202 CrossRef.
- C. Chen, X. Ni, S. Jia, Y. Liang, X. Wu, D. Kong and D. Ding, Adv. Mater., 2019, 31, 1904914 CrossRef CAS.
- C. Chen, H. Ou, R. Liu and D. Ding, Adv. Mater., 2020, 32, 1806331 CrossRef CAS.
- C. Chen, X. Ni, H. Tian, Q. Liu, D. Guo and D. Ding, Angew. Chem., Int. Ed., 2020, 59, 10008–10012 CrossRef CAS.
- J. Dai, Y. Cheng, J. Wu, Q. Wang, W. Wang, J. Yang, Z. Zhao, X. Lou, F. Xia, S. Wang and B. Z. Tang, ACS Nano, 2020, 14, 14698–14714 CrossRef.
- F. Xia, J. Wu, X. Wu, Q. Hu, J. Dai and X. Lou, Acc. Chem. Res., 2019, 52, 3064–3074 CrossRef CAS.
- F. Wu, X. Wu, Z. Duan, Y. Huang, X. Lou and F. Xia, Small, 2019, 15, 1804839 CrossRef.
- J. Yang, J. Wei, F. Luo, J. Dai, J. J. Hu, X. Lou and F. Xia, Top. Curr. Chem., 2020, 378, 47 CrossRef CAS.
- Y. Cheng, C. Sun, R. Liu, J. Yang, J. Dai, T. Zhai, X. Lou and F. Xia, Angew. Chem., Int. Ed., 2019, 58, 5049–5053 CrossRef CAS.
- J. Yang, J. Dai, Q. Wang, Y. Cheng, J. Guo, Z. Zhao, Y. Hong, X. Lou and F. Xia, Angew. Chem., Int. Ed., 2020, 59, 20405–20410 CrossRef CAS.
- Z. Meng, Z. Kang, C. Sun, S. Yang, B. Zhao, S. Feng, Q. Meng and K. Liu, Nanoscale, 2018, 10, 1215–1227 RSC.
- N. Li, H. Yang, Z. Yu, Y. Li, W. Pan, H. Wang and B. Tang, Chem. Sci., 2017, 8, 2816–2822 RSC.
- A. K. Oyelere, P. C. Chen, X. Huang, I. H. El-Sayed and M. A. El-Sayed, Bioconjugate Chem., 2007, 18, 1490–1497 CrossRef CAS.
- Y. Wang, J. Chen and J. Irudayaraj, ACS Nano, 2011, 5, 9718–9725 CrossRef CAS.
- M. E. Muroski, J. M. Kogot and G. F. Strouse, J. Am. Chem. Soc., 2012, 134, 19722–19730 CrossRef CAS.
- L. Q. Chen, S. J. Xiao, P. P. Hu, L. Peng, J. Ma, L. F. Luo, Y. F. Li and C. Z. Huang, Anal. Chem., 2012, 84, 3099–3110 CrossRef CAS.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027–1030 CrossRef CAS.
- M. E. Muroski, T. J. Morgan Jr, C. W. Levenson and G. F. Strouse, J. Am. Chem. Soc., 2014, 136, 14763–14771 CrossRef CAS.
- D. Wang, Y. Tong, Y. Li, Z. Tian, R. Cao and B. Yang, Diamond Relat. Mater., 2013, 36, 26–34 CrossRef CAS.
- A. Bumb, S. K. Sarkar, N. Billington, M. W. Brechbiel and K. C. Neuman, J. Am. Chem. Soc., 2013, 135, 7815–7818 CrossRef CAS.
- S. J. Yu, M. W. Kang, H. C. Chang, K. M. Chen and Y. C. Yu, J. Am. Chem. Soc., 2005, 127, 17604–17605 CrossRef CAS.
- H. M. Leung, M. S. Chan, L. S. Liu, S. W. Wong, T. W. Lo, C. H. Lau, C. Tin and P. K. Lo, ACS Sustainable Chem. Eng., 2018, 6, 9671–9681 CrossRef CAS.
- B. Kim, H. B. Pang, J. Kang, J. H. Park, E. Ruoslahti and M. J. Sailor, Nat. Commun., 2018, 9, 1969 CrossRef.
- M. Dowaidar, H. N. Abdelhamid, M. Hällbrink, K. Freimann, K. Kurrikoff, X. Zou and Ü. Langel, Sci. Rep., 2017, 7, 9159 CrossRef.
- A. Bolhassani, S. Javanzad, T. Saleh, M. Hashemi, M. R. Aghasadeghi and S. M. Sadat, Hum. Vaccines Immunother., 2014, 10, 321–332 CrossRef CAS.
- G. Im II, J. Biomed. Mater. Res., Part A, 2013, 101, 3009–3018 CrossRef.
- W. T. Godbey, K. K. Wu and A. G. Mikos, J. Controlled Release, 1999, 60, 149–160 CrossRef CAS.
- G. Borchard, Adv. Drug Delivery Rev., 2001, 52, 145–150 CrossRef CAS.
- S. Roy, K. Zhang, T. Roth, S. Vinogradov, R. S. Kao and A. Kabanov, Nat. Biotechnol., 1999, 17, 476–479 CrossRef CAS.
- J. Dai, M. Xu, Q. Wang, J. Yang, J. Zhang, P. Cui, W. Wang, X. Lou, F. Xia and S. Wang, Theranostics, 2020, 10, 2385–2398 CrossRef CAS.
- V. Toncheva, M. A. Wolfert, P. R. Dash, D. Oupicky, K. Ulbrich, L. W. Seymour and E. H. Schacht, Biochim. Biophys. Acta, 1998, 1380, 354–368 CrossRef CAS.
- D. Cheng, X. Zhang, Y. Gao, L. Ji, D. Hou, Z. Wang, W. Xu, Z. Qiao and H. Wang, J. Am. Chem. Soc., 2019, 141, 7235–7239 CrossRef CAS.
- L. Liu, W. Qiu, Y. Zhang, B. Li, C. Zhang, F. Gao, L. Zhang and X. Zhang, Adv. Funct. Mater., 2017, 27, 1700220 CrossRef.
- H. Hosseinkhani, F. Abedini, K. Ou and A. J. Domb, Polym. Adv. Technol., 2015, 26, 198–211 CrossRef CAS.
- H. X. Wang, Z. Song, Y. H. Lao, X. Xu, J. Gong, D. Cheng, S. Chakraborty, J. S. Park, M. Li, D. Huang, L. Yin, J. Cheng and K. W. Leong, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4903–4908 CrossRef CAS.
- L. Kelland, Nat. Rev. Cancer, 2007, 7, 573–584 CrossRef CAS.
- D. Wang and S. J. Lippard, Nat. Rev. Drug Discovery, 2005, 4, 307–320 CrossRef CAS.
- S. Dhar, F. X. Gu, R. Langer, O. C. Farokhzad and S. J. Lippard, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17356–17361 CrossRef CAS.
- S. He, C. Li, Q. i Zhang, J. Ding, X. J. Liang, X. Chen, H. Xiao, X. Chen, D. Zhou and Y. Huang, ACS Nano, 2018, 12, 7272–7281 CrossRef CAS.
- J. M. Farrow, J. C. Yang and C. P. Evans, Nat. Rev. Urol., 2014, 11, 508–516 CrossRef CAS.
- Y. Lin, Y. Wang, H. An, B. Qi, J. Wang, L. Wang, J. Shi, L. Mei and H. Wang, Nano Lett., 2019, 19, 2968–2978 CrossRef CAS.
- S. Guan, A. Munder, S. Hedtfeld, P. Braubach, S. Glage, L. Zhang, S. Lienenklaus, A. Schultze, G. Hasenpusch, W. Garrels, F. Stanke, C. Miskey, S. M. Johler, Y. Kumar, B. Tümmler, C. Rudolph, Z. Ivics and J. Rosenecker, Nat. Nanotechnol., 2019, 14, 287–297 CrossRef CAS.
- Y. Sun, X. Ye, M. Cai, X. Liu, J. Xiao, C. Zhang, Y. Wang, L. Yang, J. Liu, S. Li, C. Kang, B. Zhang, Q. Zhang, Z. Wang, A. Hong and X. Wang, ACS Nano, 2016, 10, 5759–5768 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |