 Open Access Article
Open Access ArticleA plant-mediated synthesis of nanostructured hydroxyapatite for biomedical applications: a review
Kingdom Alorku ,
M. Manoj
,
M. Manoj * and
Aihua Yuan
* and
Aihua Yuan *
*
School of Environmental and Chemical Engineering, Jiangsu University of Science and Technology, Zhenjiang 212003, Jiangsu Province, PR China. E-mail: aihua.yuan@just.edu.cn; manoajkrr@gmail.com; Tel: +86-511-85639001
First published on 10th November 2020
Abstract
The engineering of calcium-based phosphate materials at the nanoscale gains several unique properties compared to the bulky state. The effort to scale down, e.g., from bulky state to nanoscale in order to control the morphology and improve structural properties requires the use of varying reagents that can be detrimental to the environment. A typical example of these materials is hydroxyapatite (HAp), one of the well-known calcium phosphate materials, which has a close resemblance to human bone tissue. HAp has valuable applications in catalysis, drug delivery, bone and dental implant formation, and adsorption. Hydroxyapatite-based nanomaterials synthesized through conventional routes make use of reagents that are not environmental friendly and are very costly. Since the current research trends are geared towards producing/synthesizing nanomaterials through an eco-friendly approach, there is the need to consider the techniques and reagents involved in the synthesis of HAp. This review touches on the possible replacement of such synthetic chemical reagents, synthesis routes, and toxic capping agents with plant extracts for synthesizing HAp-based nanomaterials for multi-functional applications. The influence of biomolecules from plants on synthesized HAps and the attainable mechanism during these green approaches are discussed. Viable future modifications of the methods used to obtain extracts from plants are also studied.
1 Introduction
Apatite is a known constituent of igneous, sedimentary, and metamorphic rocks. The general formula of apatite is A10(BO4)6X2; where A is Ca or rare metals (e.g., Sr, Pb, Cd); BO4 represents anionic groups such as PO43−, VO43−, AsO43−, CO32−, and SiO43−; and X in the general formula stands for monovalent anions (OH−, F−, Cl−) or bivalent anions (CO32−, O2−).1 The well-known form of apatite is the calcium phosphate ceramic material with an OH− group, and is referred to as hydroxyapatite (HAp). HAp is represented by the molecular formula of Ca10(PO4)6(OH)2, instead of Ca5(PO4)3(OH). This is because one crystal structure of HAp contains two units.2 The unit cell dimensions of HAp have been calculated as a = b = 9.432, c = 6.881, and a space group of P63/m.3This ceramic material (HAp) occurs in nature, and it has been known to be an essential material used for making dental fillings due to its splendid biocompatibility.4,5 Some biomedical applications include bone regeneration for mammals (osteoclast and osteoblast cells),6 bio imaging, cancer treatment, and drug loading.7–9 Aside from the biomedical applications of HAp, the usefulness of this ceramic material has also been found in catalysis. The HAp nanomaterial has been identified as an exceptional material when coupled with other metals or metal oxides for applications in the field of catalysis. Epoxidation reactions, oxidation of alcohols, Knoevenagel reaction, Michael addition, photocatalysis, and the hydrogen evolution reaction are just a few catalytic applications of nano-HAp.1,10–15
For the diverse applications of HAp, there is the need to always scale down the crystal size of HAp to the nanoscale. This is well-explained by the fact that materials that have smaller size (crystallite size) are more reactive and exhibit enhanced physicochemical properties due to the larger exposed surface area.16–18 This positions the nano-synthesized materials as better options compared to their bulky counterparts. Nanomaterials are thus defined as particles with at least one dimension on the nanoscale (1–100 nm), which are purposely structured on this scale so that their properties can be exploited according to their dimensions.19 Recent research trends are more aligned towards manipulating synthesis routes in order to keep materials within the nanoscale. This is because of the benefits that come with the applications of nanomaterials and the economic importance.20
To ensure synthesized HAp is within the nanoscale or to control their agglomeration and morphology, researchers employ specific reagents as chelating agents.21–24 Precursor materials, capping agents, ligands, structure directing agents, stabilizing agents, templates, and surfactants are several names used to describe the chemicals that are used in nanoparticle synthesis to stabilize the size, control the morphology, and vary some tunable properties of the synthesized materials.25–31 Some capping agents are even used to manipulate the spectral, structural, photo catalytic, and photo luminescence properties of nanomaterials.32–34
Unfortunately, not all of the capping agents used in nanomaterial preparation easily degrade after they have been expended. They leave long-term effects on the environment since it takes a very long time for these reagents to degrade into less toxic or completely acceptable forms in the environment.35–37 Although reducing or capping agents are essential owing to the various important roles they play in nanoparticle synthesis, there is a need to find greener alternatives for these reagents. Greener routes of nanoparticle synthesis are so imperative in this modern era driven by concerns about the adverse effects of chemicals used in the manufacturing processes. In some instances, the plant-derived materials served as alternatives to the traditional reagents to make the preparation (nanomaterials) procedure become a greener route.38–40 These routes are referred to as Plant-Mediated Synthesis (PMS). The important reason to synthesize pure HAp without using toxic capping agents has to do with the biomedical applications of HAp for drug delivery,41,42 antimicrobial purposes,43 antiviral applications,44 gene vector applications,45 and in vitro cell viability studies,46,47 all of which require nanomaterials of very high purity and very low toxicity (good bio compatibility).
Hydroxyapatite can be synthesized through different kinds of routes (methods). Solid-state reactions, microemulsions, hydrothermal method, sol–gel method, template-guiding method, are some of the main routes/methods.10,48–52 The above-mentioned processes encounter the common problem of agglomeration. Aggregation or agglomeration does not only occur during the synthesis of HAp, but also during the synthesis of other nanomaterials. Agglomeration suppresses the mechanical properties of nanoparticles due to the restriction of the interfacial area.53,54 To achieve a good dispersion of particles by preventing the attractive forces between the nanoparticles, the problem of agglomeration is solved by incorporating capping agents or surfactants.55
Examples of capping agents/chelates that have been used to prevent agglomeration include EDTA (ethylenediaminetetraacetic acid), polyvinylpyrrolidone (PVP), glycine, glycine–acrylic acid (GLY–AA), pentaethyleneglycoldodecylether, 2-hydroxyacetophenone, and dipalmitoylphosphatidylcholine (DPPC).49,56–60 These chelating agents require a higher temperature for synthesis, and are sometimes not environmentally friendly because they take a long time to disintegrate into the acceptable forms after handling.61 EDTA is a chemical compound that is used extensively in industrial and household processes. It has been under scrutiny for some time now due to environmental concerns, and also has known toxic effects on mammals.62 It has also been labeled as one of the anthropogenic compounds with the highest concentrations in inland European waters.63 Another reason for considering HAp prepared with readily available natural precursors is the costly nature (expensive) of the final material. Traditionally prepared HAps require the use of high purity commercial reagents, which are expensive.64–66
The green alternative to traditional reagents used to prevent agglomeration is called plant extract. The plant extracts are known to contain phytochemicals that have reducing and antioxidant effects. Therefore, they are used to control the morphology, and to overcome the setback of agglomeration during the preparation of a wide range of nanoparticles.67,68 There are a few reviews that focus on obtaining hydroxyapatite from natural sources. However, no work has been published yet that categorically focuses on the plant-mediated synthesis (PMS) of hydroxyapatite. Most of these reviews also gave insight into obtaining calcium precursors from nature, but failed to discuss the importance of phosphate precursors from natural sources such as plants. The purpose of the present review is to discuss the preparation approaches of hydroxyapatite using plant-derived materials (biomolecules) (such as chelating agents, surfactants, templates, ligands, and structure-directing agents), and the use of phosphate sources from nature, deriving phosphate sources from vegetable wastes, and the synthesis of hydroxyapatite directly from plants without using any conventional calcium or phosphate precursor materials.
2 Plant-mediated synthesis (PMS) of HAp
2.1 Processing of plants to get extracts for PMS
The preparation of capping agents is one of the vital steps in the synthesis procedure. Among the sample preparation procedures, such as Soxhlet extraction, distillation, adsorption chromatography, and HPLC, some traditional basic separation techniques are also employed. Boiling and filtration are the basic traditional techniques that are suitable for obtaining natural capping agents from plants in the crude or unrefined form. Plant-derived biochemicals, such as pectin, malic acid, tartaric acid, piperine, and sugar (sucrose), are all obtained from plant sources by separation processes, of which extraction is one of the most important processes.69,70In some synthesis procedures (methods), the plant extracts are directly used after the treatment techniques, such as boiling and filtration. The resultant plant extract was used as the medium for preparing HAp nanoparticles, as the plant extracts contain certain phytochemicals that could initiate and control the morphology to achieve the desired nanoparticles or nanostructures. These methods did not include chromatographic separation techniques (methods), such as HPLC, to carefully obtain specific extracts (such as alkaloids or plant organic acids) for further usage as capping agents. A generic schematic diagram is provided in Fig. 1 that describes the formation of HAp using plant extracts. The methods of obtaining plant extracts (crude and sophisticated) and simple reaction procedures are outlined.
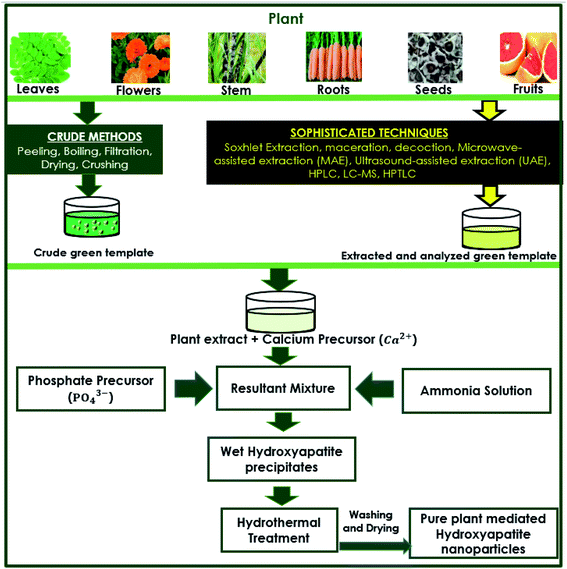 | ||
| Fig. 1 Generic schematic flow diagram of plant-mediated synthesis of HAp and extraction techniques of plant materials. | ||
Kalaiselvi et al., prepared Moringa oleifera flower extract-capped hydroxyapatite (MOFE:HAp) powders by directly employing the flower extract. In their method, 50 g of Moringa oleifera flower was dried at room temperature, mixed with 250 mL de-ionized water, and then boiled for 2 hours. The obtained plant extract was cooled down to room temperature, filtered, and used directly as a capping agent for HAp nanoparticle synthesis. The probable reason behind their choice of not using extraction techniques to obtain specific phytochemicals for their procedure is because the Moringa oleifera plant is known to contain polyphenols, alkaloids, tannins, flavonoids, vitamins, minerals, and carotenoids.71 All of these antioxidants provide a favorable reaction medium because of their reduction abilities. Furthermore, they also control the morphology during the nanoparticle synthesis process. Several studies took advantage of these polyphenols by using Moringa oleifera plant parts during the green synthesis of nanoparticles.72–76
Similarly, Gopi et al. used three kinds of fruit for preparing direct green templates for HAp synthesis. The extracts were prepared from tamarind, grape, and banana. The peels and seeds of the individual fruits were discarded, and 1 kg of the pulp was crushed into 100 mL sterile double distilled water. The resultant mixture was filtered using Whatman No. 1 filter paper and dried at 40 °C. The resultant powder was directly used as a green template.77 Their quest was to take advantage of the tartaric acid content of these three fruits for a good templating effect. Grapes, for example, are known to contain excellent phenolic molecules that can serve this purpose.78
The widely used, well known extraction technique to obtain certain desired capping or reducing agents for hydroxyapatite synthesis is high-performance liquid chromatography (HPLC). HPLC is an instrumental form of liquid chromatography that uses stationary phases comprising small particles, thereby achieving more efficient separations than those used in conventional liquid chromatography.79 HPLC can be used to accurately extract the desired materials, and identify the main constituents of plants and its extracts. Sometimes, HPLC is coupled with other techniques such as gas chromatography (GC), and modified or optimized to obtain chemical fingerprints of plants.80–86 Chemical fingerprint is the constituents of a plant that determine the active ingredients and identifies the available impurities.80 HPLC is one of the important techniques for alkaloid metabolite extraction and analysis.87 Green leafy plants are also one of the richest sources of phenolics with unique antioxidants. Sometimes, mass spectrometry (MS) is coupled with liquid chromatography (LC) for profiling plant metabolites. Typical among these substances are flavonoids and phenolic acids.88 Plants are known to contain biomolecules that may be more useful for the green synthesis of nanoparticles. Modifications of HPLC, such as reversed phase-high performance liquid chromatography (RP-HPLC), are used as a sample preparation method for obtaining specific plant extracts.89
Gopi et al. used LC techniques for the sample preparation and extraction of malic acid from apple fruits. In a typical experimental setup for obtaining the fruit extract, slices of nearly 20–40 g of the edible parts of the fruits were blended with 100% ethanol to a final concentration of 80% ethanol. The slurry formed was refluxed while stirring for 2 h with a condenser. Whatman No. 54 paper was used to filter the slurry, and 80% ethanol was used to wash the residue and the flask. The resultant extract was reduced to less than 25 mL using a rotary vacuum evaporator. Distilled water was used to make a 25 mL of fruit extract after filtering using Whatman No. 42 paper.90 For the extraction part, HPLC columns (200 × 7 mm i.d. fitted with a cation exchange resin in the hydrogen ion form, a 250 × 4 mm i.d. 5 μm Spherisorb NH2, and a 250 × 4 mm i.d. Spherisorb ODS-2 (C 18)) using the LKB Model 2140 rapid spectral detector and an IBM data station were used. For the column monitoring purposes, LKB (Bromma, Sweden) liquid chromatography was employed. Effluents were analyzed at a wavelength of 206 nm. Isocratic analysis was performed at a volumetric flow rate of 0.5 mL min−1 and 25 °C.91–93 The extracted malic acid from apple juice was dried and crushed, and the powdered malic acid was used as a green template for synthesizing HAp. Results indicated that the HAp obtained from PMS is less toxic, and has improved antibacterial property compared to the HAp synthesized using commercially purchased malic acid. Using LC, the antibacterial/antioxidant properties of malic acid extracted from apples have been retained, whereas commercial processes led to a reduction in such properties. However, it was found that the plant extracted malic acid based HAp particles have low crystallinity, which is a problem associated with most of the HAps synthesized (prepared) via PMS. In a different work of Gopi et al., sucrose was extracted from the stems of sugarcane (Saccharum officinarum), carrot (Daucus carota), and pineapple (Ananas comosus). Liquid chromatography in conjunction with mass spectrometry (LC-MS) was used for analyzing sucrose in the three fruits used in the experiment. The process of extraction was carried out by HPLC (Varian Inc., USA) equipped with a model 410 Prostar Binary LC with a photodiode array (PDA) detector. The column in this process was amino-bonded carbohydrate (10 μm, 300 × 4.1 mm) together with an acetonitrile–water (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mobile phase. The column was operated under isocratic conditions at a volumetric flow rate of 1.4 mL min−1. The obtained sucrose in the form of crystals was ground and used as a green templating agent.56,94 The three fruits, however, produced HAp with lower crystallinity compared with commercially purchased sucrose.
1) mobile phase. The column was operated under isocratic conditions at a volumetric flow rate of 1.4 mL min−1. The obtained sucrose in the form of crystals was ground and used as a green templating agent.56,94 The three fruits, however, produced HAp with lower crystallinity compared with commercially purchased sucrose.
Piperine is an important alkaloid obtained from black pepper (Piper nigrum), and it is studied for so many pharmaceutical properties due to its phytochemical attributes.95,96 One technique used to extract piperine from plants is high-performance thin-layer chromatography (HPTLC)97 because of its useful and robust nature.98 Another recent and novel technique for acquiring or finding piperine from black pepper is a voltammetric sensor based on glassy carbon (GC) electrode with ethanol for extraction. This method is efficient, less time-consuming, and makes use of only one step.99 Subramanian et al. also extracted piperine from commercially available black pepper powder, and it was used as a green template for HAp synthesis. A methanolic extract was used to isolate piperine, and it was purified afterwards using column chromatography.100 In a related article, they went further by comparing Soxhlet extraction with double bypass sidearm Soxhlet apparatus extraction in terms of the extraction time, extraction cycle, yield of crude piperine, and volume of solvent used. The double bypass sidearm method was found to be more effective than the Soxhlet extraction method.101
Green synthesis, especially those that make use of plant extracts, is becoming common not only for nano-HAp synthesis, but for a broad range of nanoparticle synthesis. Some of these nanoparticles are even applied in the biomedical and pharmaceutical fields. Therefore, the use of crude green plant extracts for synthesis should be looked into. The potential solution of the incorporation of extraction techniques is the use of less solvent volume, less time consuming, less expensive, and can be optimized for extracting phytochemicals from plants. For example, a comprehensive work by Castro-López et al. investigated the impact of extraction techniques on antioxidant capacities, and the phytochemical composition of polyphenol-rich extracts of five different plants. The techniques considered were maceration, decoction (an extraction technique used exclusively for water-soluble and thermostable constituents),102 microwave-assisted extraction (MAE), and ultrasound-assisted extraction (UAE).81,103–107 After the analysis using liquid chromatography coupled with tandem mass spectrometry (LC-MS2), which offers a high-throughput and rich information,108 there was an indication that the extraction methods have significant influence on the phytochemical profile. Thus, microwave-assisted extraction (MAE) was found to be better compared to the other methods.109 MAE is one of the green extraction techniques that can be automated and has many advantages, such as a reduction of the extraction time, lower solvent consumption, and the potential of simultaneously extracting multiple (up to 40) samples, which enhances the sample throughput. The greenness of this technique is in tandem with the calls for environmentally friendly techniques for most synthesis procedures, including the extraction of biochemicals for green HAp synthesis.110 Ultrasound-assisted extraction (UAE) method has more advantages over the Soxhlet method used by Subramanian et al. during their extraction process for green templates, and other works that made use of Soxhlet extraction.100,111 Some of these advantages are a shorter operating time compared with Soxhlet extraction. UAE allows the addition of a co-extractant to increase the polarity of the liquid phase, possibly increasing the extraction efficiency, and permits the leaching of thermolabile analytes that are altered during Soxhlet extraction.112 These methods may be far more suitable for biomolecule analysis (extraction from plants) for green synthesis compared to conventional ones.
2.2 Mechanism of PMS
The plant-mediated synthesis of HAp falls under the wet process (synthesis) route. The wet process (synthesis) refers to the preparation of HAp from an aqueous reaction medium, such as chemical precipitation, hydrolysis, and hydrothermal routes, which all fall under the wet process (synthesis) route. It is easier to control the morphology and the size of HAp during the wet process (synthesis) route. Hence, this method is suitable for making/preparing plant-mediated HAps.The mechanism of PMS of HAp highly depends on the formation of chelates/complexes with Ca2+. Precursors for complexation come from reactive groups (functional groups) in plant polyphenols. Terpenoids, flavonoids, amino groups (R–CH(NH2)–COOH) of plants bear (–COOH), (–OH), carboxyl derivatives, or an organic acid can provide a chelating effect. During PMS, the higher electrostatic force between the carboxyl derivatives/reactive groups and Ca2+ aid in the Ca-chelate formation. The resultant Ca-complex then reacts with the phosphate precursor (PO43−) to start the nucleation process. HAp formation is also reliant on the pH of the reaction medium. An ammonia solution (liq. NH3) is used to provide a suitable basic medium (pH 9–10) for the reaction to proceed. At higher pH (9–10), excess OH− strips off H+ from H2PO4− to form PO43−. The supersaturation of PO43− in the reaction solution leads to the precipitation of HAp, rather than other calcium phosphate phases. The Ca/P ratio is therefore maintained at 1.67.56,113,114 However, in some reports, there was no record of the use of ammonia solution, revealing only the precursors used and the plant extract function (role) for the chelating effect under the already provided suitable pH conditions.24,40,115 The formed precipitates are treated hydrothermally at temperatures around (100 °C), and then washed with water or ethanol to obtain the pure HAp nanoparticles. A probable mechanism that describes the role of biomolecules in hydroxyapatite synthesis is the work of Zhou et al. described in Fig. 2. Eqn (1) describes the stoichiometric reaction that occurred. They used the wet process (synthesis) route (chemical precipitation), and maintained pH at 9 using aqueous ammonia. They proposed that grape seed polyphenol (GSP) formed a chelate with Ca2+ ions under favorable pH. The positively charged Ca–GSP chelate reacts with the negatively charged PO43− to affect the co-precipitation, dispersion, and nucleation. The polyphenols from grape seed easily get attached to the HAp surface to enhance steric hindrance and electrostatic repulsion, which in turn prevents the aggregation of particles.116
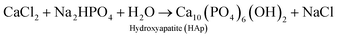 | (1) |
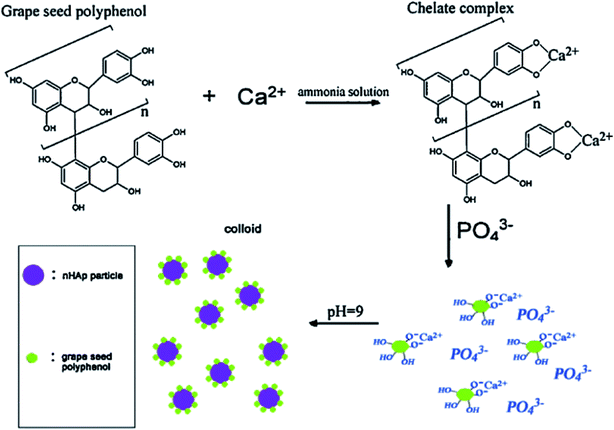 | ||
| Fig. 2 Schematic flow diagram of polyphenol extracted from grape seed used as a capping agent in nano-HAp synthesis (adapted from ref. 116, with permission from Elsevier). | ||
The synthesis of HAp using plant parts as capping agents has received significant attention over the years, owing to their eco-friendly nature, reliability, and ease of reaction. Earlier synthesis routes incorporated green capping agents from plants for efficient HAp synthesis.116–119 There are also a lot of reports on the upsurge of using green methods or plant extracts to synthesize metallic nanoparticles. Some examples include the use of Clinacanthus nutans Lindau (C. Lindau) to synthesize gold nanoparticles,120 Saccharum spontaneum (Ss) to synthesize silver nanoparticles,121 pomegranate (Punica granatum L.) fruit peels to synthesize platinum nanoparticles,122 and Aloe vera plant extract solution to synthesize In2O3.123 For hydroxyapatite, the earliest sole PMS of hydroxyapatite was carried out by Klinkaewnarong et al., where they used the Aloe vera plant extract as the main solution for dissolving calcium and phosphate precursors (Ca(NO3)2·4H2O and (NH4)2HPO4). The precursor materials were mixed at a molar ratio of 1.67 (Ca/P), which is very important for the formation of pure HAp without other calcium phosphate phases. The pH of the reaction medium was not changed, which may be due to the suitable resulting mixture of calcium-phosphate precursors and the Aloe vera extract for HAp formation. The formed precipitate was calcined above 550 °C to obtain nanocrystalline HAp powders.117 In 2003, Bose et al. also synthesized HAp and controlled the morphology of the nanoparticles using sucrose. However, the sucrose employed in this procedure was commercially purchased, but not directly extracted from plants.124 In one of our previous work, HAp nanosheets were synthesized from natural sources, and waste eggshells were used as a calcium precursor. We replaced commercial phosphate with extracts of ginger. Remarkably, we did not use liquid ammonia to alter the pH of the resultant mixture. The reason behind our procedure is the favorable basic pH range already provided by Ca(OH)2. After heating the calcium precursor (eggshells) at a higher temperature (900 °C), the calcium carbonate (CaCO3) in the eggshells was thermally decomposed to form calcium oxide (CaO). In an aqueous medium, CaO was then converted to Ca(OH)2. The excess OH− released in the solution helps to attain a suitable basic reaction medium for nano-HAp formation.40
The works of Gopi et al. showed that the plant extracts could function as a directing agent during HAp synthesis. They used dried powders from banana (Musa acuminta), grape (Vitis) and tamarind (Tamarindus indica), which are sources of tartaric acid as a green templating agent to synthesize HAp.77 Pectin extracted from banana peel powder,125 sucrose extracted from different fruits (pineapple, carrot, and sugarcane),56 malic acid extracted from apples,91 pectin from prickly pear peel (Opuntia ficus indica), and pectin from bitter orange peel (Citrus aurantium) were used as a directing agent to prepare nano-HAp and other calcium phosphate phases, like beta-tricalcium phosphate (β-TCP).126 In most works of Gopi et al., they postulated that the chelating effect of –COO or the carboxylic acid derivatives of plant extracts were the major driving forces behind PMS of HAp. This observation has also been made by other researchers.127 It should be noted that several transformations of plant extracts occur before chelation with Ca2+ begins. Plant active ingredients are first converted into sugars, organic acids with hydroxyl groups, carbonyl groups, or organic functional groups that can initiate a conjugation effect with Ca2+. This transformation then leads to complex formation. One drawback of PMS of HAp is the low complexation effect of some plant extracts. For example, the complexation effect of the –OH groups is low compared to –COO groups. Therefore, it will not produce HAp with the desired morphology.56 A summary of the research works that made use of plant extracts to synthesize HAp has been given in Table 1 below.
| Plant | Part | Extract/templating agent | Processing/extraction method | Type of HAp | Crystallite size (nm) | Morphology | References |
|---|---|---|---|---|---|---|---|
| Natural rubber latex (NRL) (Hevea brasiliensis) | — | Latex | Dilution with DI, preservation with ammonia solution | Nano-HAP | X = 0, (30.81 to 66.98) | Nanorods | 24 |
| X = 1, (21.10 to 33.49) | |||||||
| Ginger | Roots | Aqueous filtrate | Distillation and filtration | HAp nanosheets | 47 nm | Nanosheets | 40 |
| Tamarind (Tamarindus indica) | Fruit | Aqueous extract (tartaric acid) | Crushing, filtration, boiling, drying and crushing into powder | Nano-HAP | 49 | Uniform nanorods | 77 |
| Grape (Vitis) | Fruit | Aqueous extract (tartaric acid) | Crushing, filtration, boiling, drying, and crushing into powder | Nano-HAP | 53 | Nanorods (rod-like structure) | 77 |
| Banana (Musa acuminata) | Fruit | Aqueous extract (tartaric acid) | Crushing, filtration, boiling, drying and crushing into powder | Nano-HAP | 57 | Nanorods in the form of ice cubes | 77 |
| Banana (Musa paradisiaca) | Peel | Pectin | Fractional extraction | HAp, TCP | 25.17–47.20 | Discrete spherical HAP | 125 |
| Apple (Malus domestica) | Fruit (juice) | Malic acid | High-performance liquid chromatography (HPLC) | Pure nano-HAp | 40 | Ball-like structure | 91 |
| Prickly pear fruit (Opentia ficus-indica) | Peel | Pectin | Drying and grinding | HAp and beta tricalcium phosphate (TCP) | 24 | Cluster-like/granular-like | 126 |
| Bitter orange (Citrus aurantium) | Peel | Pectin | Drying and grinding | HAp and tricalcium phosphate (TCP) | — | Bead-like structure | 126 |
| Pineapple | Fruit (juice) | Sucrose | Water extraction method by using HPLC | Nano-HAp | — | Cubic-like | 126 |
| Sugar cane | Stem | Sucrose | Water extraction method by using HPLC | Nano-HAp | — | Spherical shape | 126 |
| Carrot | Root | Sucrose | Water extraction method by using HPLC | Nano-HAp | — | Capsule-like | 126 |
| Moringa oleifera | Flower | Aqueous flower extract | Drying and boiling | Nano-HAp | 41 | Nanorods | 71 |
| Moringa oleifera | Flower | Flower extract | Soxhlet extraction | HAp, TCP | 200 | Crystalline plate like structure | 128 |
| Azadirachta indica | Leaves | — | Boiling and filtration | Nano-HAP | 53 | Hexagonal structure | 129 |
| Coccinia grandis | Leaves | — | Boiling and filtration | Nano-HAp | 64 | Hexagonal structure | 129 |
| Potato | Skin/peel | 18-Hydroxyoctadec-9-enoic acid, octadec-9ene-1,18dioc acid | Boiling and filtration | — | 250–500 | — | 130 |
| Papaya | Leaf | Beta-carotene, calcium, papain, and vitamins | Boiling and filtration | — | — | — | 130 |
| Orange | Orange peel | Oil 90–95% limonene content | Boiling and filtration | — | ∼50 | Rectangular and elongated | 130 |
| Calendula | Calendula flowers | 19 different carotenoids | Boiling and filtration | — | — | — | 130 |
| Native pine wood | — | Carbon templates | Pyrolysis of ligneous raw materials | Biomimetic hydroxyapatite scaffolds | — | — | 131 |
| Native rattan wood | — | — | Pyrolysis of ligneous raw materials | Biomimetic hydroxyapatite scaffolds | — | Spherical form | 131 |
| Carrot | Peel | — | Boil, distillation, filtration | Nano-HAP | 50 | Spherical | 132 |
| Rice | Seeds | Glutinous rice | Boiling | HAp, β-Ca3(PO4)2, CaO, and β-NaCa(PO4) | — | Rod-shaped | 133 |
| Black pepper | Black pepper powder | Piperine (isolated from the methanolic extract) | Column chromatography | Nano-HAP | 29.55–30.80 | Well defined nanorods | 100 |
| Parkia biglobosa (African locust bean) | Pods | Pectin | Acidification, boiling, and filtration | Nano-HAP | 17.5–25.3 | Flake-like, spherical | 127 |
2.3 Characterization of HAp obtained via PMS
Research has shown that the majority of HAPs obtained via PMS are applied in the biomedical field.134,135 Materials applied to the biomedical field need careful analysis to describe their composition, structure, morphology, and more importantly, the type of chemical bonds presented. Useful material testing techniques, such as powder X-ray diffraction crystallography (XRD), X-ray photoelectron spectroscopy (XPS), Fourier transform infrared (FTIR) spectroscopy, nuclear magnetic resonance spectroscopy (NMR), Raman spectroscopy, electron microscopy (EM) coupled with energy-dispersive X-ray spectroscopy (EDS), are used to confirm the formation of the calcium phosphate materials. Prior to PMS of HAp, it is necessary to subject the green templates (plant extracts) to FTIR and NMR to analyze the type of chemical bonds that exist therein. The major rationale behind these two techniques is that the plant organics (biomolecules), which restore specific properties (e.g., antimicrobial, antibacterial) to the final HAp powder, could harbor harmful chemical components. FTIR provides fine details about the type of chemical bonds and the composition of any compound available in the materials tested. In this sense, it reveals the type of covalent bonds available in the plant extract. This test goes further to uncover the specific functional groups present in the plant extracts meant for PMS.56,91,125,127 Undesirable functional groups can therefore be verified and isolated. Although NMR analysis provides data on the types of chemical bonds in crystalline HAp,136–138 it can also be employed to reveal vital information on the organic compounds available in plant extracts used for PMS. Proton-1 nuclear magnetic resonance spectroscopy (1H-NMR) helps in the determination of different hydrogen bonds in plant biomolecules, whereas carbon-13 nuclear magnetic resonance (13C-NMR) spectroscopy determines the different types of carbon environment available. 1H-NMR and 13C-NMR collectively confirm the types of polyphenols used for preparing HAp for biomedical purposes by analyzing the correlation between the carbon atoms and protons.2.4 Role of phytochemicals in PMS of nano-HAp
Phytochemicals from plants are bioactive compounds that may be vitamins, thiols, terpenoids, or phenolics, such as tannins, lignins, flavonoids, most of which have antioxidant properties.139 The rich nutrient contents of some plants result in their diverse applications, ranging from biomedicals to nanosciences. For example, the unique antibacterial and antioxidation activity of plants has been studied, in addition to the possible medicinal uses and food preservation potential indicated from known research works.140–144 In some instances, plant extracts used as capping agents are known to have a great impact on nanoparticles during preparation.33 Capping agents (biomolecules) derived from plants have also played a significant role during metallic nanoparticle synthesis.67,145–148Traditional capping agents, such as triethanolamine (TEA), ethylenediaminetetraacetic acid (EDTA), diethanolamine (DEA), ethylene glycol (EG),149 citric acid, sodium citrate,150,151 polyethylene glycol,152 sodium dodecyl sulphate, and sodium dodecylbenzene sulphonate153 were used to control the particle nucleation and growth to produce nanosized HAp. These conventional organic molecules used during HAp nanoparticle synthesis help in the transformation of elements involved in the crystal structure, thereby changing the morphology and ensuring that the synthesized calcium phosphate is within the nanoscale. The transformation mechanism of these traditional agents is similar to chemicals in plants called phytochemicals. Phytochemicals have the ability to capture active ions during synthesis, in other words, helping in the reduction process. Biomolecules such as proteins and glucose help in controlling the size and the morphology of HAp nanoparticles. Amide groups of proteins (extracted from flowers or other plant parts) serve as the capping ligand during nano-HAp transformation.71,154
Enoic acid (from potato peels) was able to form a chelate with calcium ions and help control the morphology.86 The particles formed have unique shapes and are nanosized. The leaves of papaya were also noted to contain beta-carotenes that control the size and shape of the HAp nanoparticles.86 Carotenoids from Calendula helped in the formation of HAp with elongated structure. A control experiment was set up to compare the role of carotenoids in the HAp synthesis using HAp prepared without biomolecules, and yielded results with no change in size or shape, which confirmed the essence of the biomolecules.130
The polyphenols of Moringa leaves contain tannins. Sundrarajan et al. proposed that the hydroxyl groups of tannins resulted in a p-track conjugation effect (the binding of a hydroxyl group with metal ions) to form a chelate with Ca2+ during PMS. The Ca–tanninate complex formed in the presence of 1-butyl-3-methylimidazolium tetrafluoroborate ([BMIM]BF4) controls the morphology and crystallinity of HAp. The formation of hierarchical structures was due to the interactions of tannins from the Moringa oleifera plant.128,157 From this study, there is an indication that the HAp formation was not entirely due to the extracts from the Moringa leaves (tannins), but also due to ([BMIM]BF4). Also, the major chelating effect of the plant source (Moringa leaves) in this study will be due to the chlorogenic acid content of tannins. There is a –COOH group of chlorogenic acid that favored the formation of the Ca–tanninate complex. Although some other acids (such as L-ascorbic acid and quercetin) are present in tannins,76 the –OH groups of these acids can have a tendency to form a complex. However, the complexation effect of –COOH is higher than –OH, which indicates that chlorogenic acid is the key component for chelate formation in this study.56
The effect of plant-derived sucrose used as a chelating agent was obvious after comparing the sizes of nano-HAps calculated from the Debye–Scherrer's formula with nano-HAp synthesized from commercially purchased sucrose. HAps derived from PMS were found to have smaller crystallite sizes. Less agglomeration, uniform particle sizes, and well-defined morphology were also reported than HAp prepared with commercially purchased sucrose.56 This could probably be due to the excessive purification of commercially purchased sucrose, which could have led to a reduction in the chelating abilities. Biomolecules of Gum acacia or Gum arabia have good emulsifying potential, stabilizing tendency, and are very good at dispersing formed nanoparticles. The Gum acacia solution greatly controls the morphology of the synthesized HAps, as the TEM images of the control experiment showed no result. Between the use of 0.5% and 1% of Gum acacia for preparing the nanoparticles, the one with 1% Gum acacia changes the morphology into leaf-like structures. The observed report indicates that the changes were due to an interaction of the carboxyl functional group of Gum acacia with Ca cations.158,159
In Fig. 3a, the SEM image of HAp synthesized by commercially purchased malic acid as the template shows particles that are larger, agglomerated, and look like balls. The present elements (Ca, P, and O) were confirmed from EDX (Fig. 3a1) spectra. In contrast, the SEM image of HAp synthesized by malic acid extract from apple fruit (Fig. 3b) showed a smaller particle size, lower agglomeration, and reduced crystallite size (Table 2, a). Aside from the better antibacterial activity of the apple extract mediated-HAp, the extracted malic acid from the apples resulted in additional elements (Mg, Na, Zn), as confirmed by EDS (Fig. 3b1). This enhanced antibacterial activity of natural malic acid is attributed to the presence of Mg2+, Na+, and Zn2+.133 The presence of these cations in apples is known to cause antioxidant effects.160 Although the presence of the detected elements was explained as the entrapment of minerals resulting from the fruit extract used,91 the elements may have been substituted or co-substituted with Ca2+ of HAp from the apple extract-mediated synthesis. Metal ions from plant extracts are known to substitute Ca2+ in HAp during PMS.133 Substituted HAps are currently being investigated for use as orthopedic prostheses, dental implants, and other applications.161–164 Therefore, the substituted HAp obtained via PMS would provide a greener alternative to the ones prepared via traditional methods/feedstocks. In that case, HAps of this nature could be improved by providing suitable sintering conditions to enhance their mechanical strength since the mechanical strength of such HAps are sometimes low.165
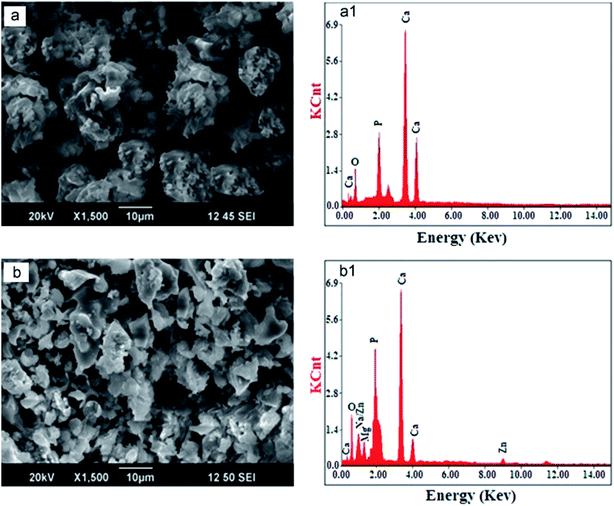 | ||
| Fig. 3 SEM micrographs and EDS spectra of HAp synthesized by commercial malic acid (a and a1) and malic acid extracted from natural apple fruit (b and b1) (adapted from ref. 91 with permission from Elsevier). | ||
| S. No. | Templating/capping agent | Degree of crystallinity | Average crystallite size (nm) | References |
|---|---|---|---|---|
| a | Commercial tartaric acid | 4.8 | 62 | 77 |
| Tartaric acid from banana extract | 3.8 | 57 | ||
| Tartaric acid from grape extract | 3.0 | 53 | ||
| Tartaric acid from tamarind | 2.4 | 49 | ||
| b | Commercial malic acid | 1.70 | 44 | 91 |
| Malic acid extracted from natural apple fruit | 1.49 | 40 | ||
| c | Piperine (20 mg) | 2.23 | 29.55 ± 05 | 100 |
| Piperine (10 mg) | 2.73 | 30.80 ± 04 | ||
| Piperine (0 mg) | 3.00 | 24.87 ± 02 |
Fulmer et al. compared the dissolution rates of the bone source, Norian cranial repair system (types of HAp used for repairing cranial defects, bone fracture, and others), and a sintered hydroxyapatite under known physiological conditions (tris buffer solution, pH 7.4, 37 °C) with 7.5, 7.4, and 1.4 ppm, respectively. Dissolution rates of 0.0465, 0.1589, and 0 mg min−1 were achieved for the bone source, CRS, and calcite (sintered hydroxyapatite), respectively, within 10 min. The higher dissolution rates of the bone source and Norian CRS materials were assignable to their low degree of crystallinity. In contrast, the highly crystalline calcite stayed in solution longer without easily dissolving.171 This includes the fact that highly crystalline HAps are less soluble, while those with poor crystallinity are more (easily) soluble under the same dissolution parameters. Other researchers also studied the trend in the dissolution rate of crystalline HAps versus amorphous HAps. Poor crystalline (amorphous) HAps have been found to have higher dissolution rates.172 However, there are applications for poor crystalline HAps. Low crystalline HAps from PMS could be applied in biological cell imaging (the study of cell behaviors under controlled conditions), ink-based printing for security information storage,173 and drug delivery.174
Taking the methods of preparation of HAp into consideration, capping agents used in the preparation of HAps may affect the crystallinity. For example, Table 2 summarizes the degree of crystallinity of HAp prepared with commercial capping agents and plant-derived capping agents. Although the average crystallite size of HAps decreases with plant-derived extracts, the decreasing degree of crystallinity also indicates that these HAps may highly soluble compared to their counterparts that were synthesized with or without commercial capping agents. The highly soluble HAps may not be suitable for biomedical applications, such as dental and orthopedic implants engineering. This points to the fact that not all of the plant-mediated HAps can be used for applications requiring a very excellent crystalline nature, unless specific modifications are made in the unit lattice structure.
3 Nature of templating agents and possible applications of resultant nano-HAp in biomedicine
Since phytochemicals serve as the driving force in shaping the type of HAp nanoparticles produced with respect to the morphology, factors affecting their reaction in the presence of other solvents in the reaction media are quite imperative. Research has shown that the nature of these organic molecules plays a vital role in the applications of nanoparticles. For example, hydrophilic organic molecules are biocompatible and can be exploited for use in drug delivery applications. Natural polysaccharide gums (derived from tamarind seed) and guar gum functionalized nano-HAp showed that the viability in cells is more than 95%. This result confirms the excellent biocompatibility of these natural plant-mediated HAp nanoparticles. Guar gum-mediated HAp has an outstanding drug-loading capacity of 236.18 mg g−1 and drug loading efficiency of up to 74.79%.175The treatment of cancer cells may be viable by using biosynthesized hydroxyapatite nanoparticles. The pectin-based nano-HAp reveals a dose-dependent cytotoxicity towards prostate cancer cells (PC3 cells).176 Modified pectin has anticancer potentials177,178 and therefore, can be used in conjunction with HAp for cancer treatment applications since some research works indicated the desirable drug loading potential of HAp-based materials for anticancer activity.179–181 This paves the way for research into cancer therapy developed by using plant-mediated HAp.
Another less explored topic of plant extracts used in HAp synthesis is the antibacterial activity of these nanoparticles. Hydroxyapatite synthesized using plant extracts has the ability to suppress the growth (activity) of bacteria. HAp is one of the calcium phosphate ceramic materials widely explored for biomedical purposes. The antibacterial property will further help the cause of research in this area. HAp synthesized using Azadirachta indica and Coccinia grandis leaf extract was very effective against Escherichia coli and Staphylococcus aureus, as the nanoparticles showed significant inhibition zones against both bacteria.129 Comparably, the lavender oil-mediated HAp showed higher inhibition potential against Escherichia coli and Staphylococcus aureus than pure HAp (without plant extract). The explanation given was that the excellent carrier properties of nano-HAp even with low concentrations of lavender oil enabled this antimicrobial effect.182 Hence, the HAp prepared via PMS has acquired the ability to suppress the bacterial growth. In addition, the purity of the nanomaterials plays a vital role against microorganisms such as bacteria. Commercially produced HAp requires thorough purification due to the nature of the chemical reagents used. These kinds of HAps might not have antibacterial properties unless they are inoculated with purified constituents afterwards. PMS thus provides a comprehensive one-step approach to producing HAp with biomedical properties compared with HAp produced from commercial chemical reagents. Therefore, the biomedical application of nanomaterials requires materials that are not just within the nano-range, but of higher purity and capability (structural modifications due to PMS). Conventional or synthetic chemical reagents used during nanosynthesis requires multiple purification steps to get rid of the excess and unreacted chemicals or templates. In the case of using plant-derived materials, templates help in increasing the biocompatibility and bioactivity of the resultant nanomaterials.183,184 HAp is of no exception; therefore, the risk of toxicity is not of high concern.117,185 Excess or unreacted plant extracts can be easily washed off. Even if they get into the environment, they degrade easily under environmental factors. This gives plant extracts an edge over their synthetic substitutes as templates for biomedical HAp preparation. A summary of HAp synthesized using plant parts and their respective biomedical applications have been given in Table 3.
| Type of HAp | Plant used for preparing extract/template | Biomedical application of nanostructured HAp | References |
|---|---|---|---|
| HAp nanosheets | Ginger (Zingiber officinale Roscoe) | Bone tissue regenerative medicine | 40 |
| Nano-HAp | Azadirachta indica, Coccinia grandis | Antibacterial activity | 129 |
| Nano-HAP | Parkia biglobosa | Antibacterial activity | 127 |
| Nano-HAp | Tamarind seed, guar gum | Drug loading and in vitro drug release | 175 |
| Nano-HAp | Basil, lavender | Antimicrobial activity | 182 |
| Nano-HAp | Peppermint plant (M. piperita) | Antimicrobial activity | 186 |
| Honeycomb-like poly(lactic acid) nano-HAp | Bitter gourd fruits | Bone tissue regeneration | 187 |
| Cocoon shaped nano AgHAp | Azadirachta indica gum | Antibacterial activity | 188 |
| Strontium-hydroxyapatite (SrHA) | Hippocampus kuda Bleeler | Drug delivery system for bone tissue repair | 189 |
| Reduced graphene oxide/hydroxyapatite composite (RGO–HA) | Melissa officinalis | Cell viability | 190 |
4 HAp directly obtained from plants, other plant-based calcium and phosphate sources, and feasible future directions
Most research and review papers published on the greener sources of HAp stressed on obtaining calcium to phosphate precursors [Ca2+]/[P] from nature (e.g., seashells, bone of animals).191–194 However, little attention was given to obtaining [Ca2+]/[P] directly from plants or methods that combined plant extracts and other natural sources. The leaves of Moringa oleifera are known to contain certain percentages of calcium and phosphate. Selected works indicated the presence of 68.5% of Ca and 1.6% of phosphorus for the elemental composition of Moringa oleifera leaf powder. The rest of the composition included other elements, such as Fe, K, Mo, and Ba.195 Susanto et al. also pegged the contents at 68.5% for calcium from the conventional method and 73.0% from the unconventional method, and 1.65% for phosphorus from the conventional method and 1.4% from the non-conventional method, when the leaves were analyzed for potential advanced bioengineering applications. This was done after trying two methods of processing for the leaves.196 This finding indicates a higher percentage of calcium and lower percentage of phosphate precursors if the leaves are considered for direct extraction of nano-hydroxyapatite. A greener and more feasible way to produce a quantifiable amount of HAp from Moringa leaves would be by supplementing the extracts with wastes from potatoes, almonds, and other vegetables with high phosphate content.197–199Shaltout et al. analyzed the content of Catha edulis (C. edulis) by getting rid of the organic contents of the plants through a higher calcination process. The inorganic products of the leaves and stalks were identified to contain a quantifiable amount of calcium hydroxyapatite after employing characterization techniques, such as FTIR, XRD, and thermal analysis (TGA, DTG, DSC). Hydroxyapatite with formula Ca10(PO4)6(OH)2 was also identified in plants such as basil, mint, green tea, and trifolium by using the same procedure for C. edulis. The amount of HAp was low in these plants compared with C. edulis. Their work is one of the few that discusses the synthesis of HAp solely from plants, thus not making use of any calcium precursor, phosphate precursor, or binding materials from conventional reagents.200 Although the burning of plants to produce HAp is not economically viable, their method paves the way for research into producing HAp from plants. An alternative to calcining plants should be considered in order to make the process more practical.
In the work of Monballiu et al., calcium phosphate (hydroxyapatite) was recovered from the diluted industrial effluent of a Belgian potato processing plant using the upflow anaerobic sludge blanket (UASB) technique. UASB is a robust and widely used technology for treating sewage through a high rate anaerobic process.201–207 Laboratory Continuous Stirred Tank Reactors (CSTRs) were used to perform this experiment. In the typical experimental setup illustrated in Fig. 4, two CSTRs were connected in series. The first reactor was fed with the effluent (nitrifying sludge from the wastewater treatment of potato processing), aerated, and used for nitrification. The first reactor containing the potato sludge was operated at room temperature. The second reactor connected with the first receives effluent from the nitrification reactor, which is then used for forming calcium phosphate precipitates. The pH of the reaction medium was adjusted using calculated amounts of NaOH (0.5 mol L−1). For the experiment conducted by using only one tank, the pH was kept in the range of 8.0 to 8.5. In contrast, the pH for CSTRs connected in series was pegged at 10.5. CaCl2 solution was added to the second tank for the connected CSTRs to manipulate the [Ca2+]/[P] ratio of the formed precipitates.208 This ratio is more important during the HAp synthesis because it determines the phase composition of the final product.209–211 Although their method is a little greener due to the use of vegetable wastes, incorporating calcium precursors from seashells, eggshells, and other sources would be more environmentally friendly compared to the commercial CaCl2.
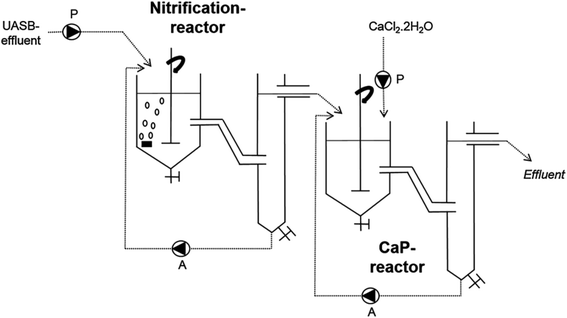 | ||
| Fig. 4 UASB set-up used to convert potato waste to hydroxyapatite (adapted from ref. 208 with permission from Elsevier). | ||
Likewise, Wu et al. used fruit wastes as the source of phosphates to hydrothermally produce HAp. The wastes of grapes, sweet potato, and pomelo peels were collected, and are referred to as fruit wastes in this work. Eggshell wastes were utilized as the calcium precursor. This work or process depicts a remarkably greener route for the production of HAp by using waste materials as precursors.212
Reported phosphate sources are very limited; thus, any effort or mechanism that can recover used phosphates is very attractive for research.213–215 Other sources of vegetable wastes can also be used for phosphate regeneration in the form of calcium hydroxyapatite, as they contain excellent amounts of phosphorus.216–219 This could minimize the excessive dependence on commercial phosphate precursors for hydroxyapatite synthesis.
5 Shortcomings of plant-mediated HAps
5.1 Toxins from plant extracts
There is no clear-cut routine that defines the synthesis of nanoparticles using plant extracts. However, there is a combination of methods and manipulation of reaction conditions in the presence of certain or desired biomolecules for anticipated results.220 Researchers have a fair knowledge of specific plants and their potential effect on synthesized nanoparticles, and therefore incorporate these extracts in preparation procedures. The only problem or issue that sometimes arises is the failure to identify the specific phytochemicals (biomolecules) that molds the formed nanoparticles. This is because some reported procedures used the crude form of plant extracts129,130 without further treatment techniques (e.g., extraction) and separation techniques (e.g., chromatography).156 Plant biomolecules have a chelating effect, thereby helping in PMS. However, not all plant extracts are non-toxic. Although the PMS of HAp is non-expensive, eco-friendly, and devoid of toxic reagents, a careful analysis/extraction of the plant extracts should be done to avoid the plant toxins (cyanogenic glycosides) in the extracts used for PMS. Cyanogenic glycosides are natural toxins that exist in parts of several plants (cashews, apples, apricots, cherries, peaches, plums, quinces, etc.).221–223 The processing or ingestion of these plants could lead to cyanide formation if hydrolysis occurs.224 Most PMS of HAp follows wet process (synthesis) routes. Therefore, there is a high probability of hydrolysis occurring during synthesis. In that regard, extracts used for PMS should be well screened prior to utilization in HAp preparation for drug loading applications and tissue engineering.5.2 Mechanical strength
Wet synthesis of HAp produces powders with poor crystallinity.2 This could be ascribed to the low processing temperatures required for wet procedures. The crystallinity of HAp has a direct effect on the mechanical strength.225 Thus, the higher crystalline nature HAp has good mechanical strength and vice versa.187 In summary of the wet synthesis of HAp, the PMS of HAp does not yield HAps with high crystallinity/mechanical strength.226 There is a major drawback if such HAps are to be used for load-bearing applications (bone/hard tissue regeneration).227 However, substituting Ca2+ in HAp with strontium, terbium, magnesium, and zinc can improve the mechanical properties of such HAps.2286 Conclusion
The write-up summarizes recent plant parts that are mostly employed as reducing or stabilizing agents in nanostructured hydroxyapatite preparation. The sharp contrasts between the benefits of using plant extracts and conventional reducing agents have been discussed. The possibility of producing HAp and other calcium phosphate nanoparticles solely from plants has also been touched on. This review has shed more light on the environmentally friendly nature of synthesizing HAp through plant-mediated methods. It also creates a possible pathway for more insight into the recent techniques that can be combined or manipulated with the aid of plant biomolecules to produce nano-HAp for multifunctional applications, ranging from catalysis to biomedical uses.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to thank the School of Environmental and Chemical Engineering, Postdoctoral Research Foundation of Jiangsu University of Science and Technology, P. R. China.References
- M. Ibrahim, M. Labaki, J. M. Giraudon and J. F. Lamonier, J. Hazard. Mater., 2020, 383, 121139 CrossRef CAS
.
- N. M. Pu'ad, R. A. Haq, H. M. Noh, H. Abdullah, M. Idris and T. Lee, Mater. Today: Proc., 2020, 29, 233–239 Search PubMed
.
- M. I. Kay, R. A. Young and A. S. Posner, Nature, 1964, 204, 1050–1052 CrossRef CAS
.
- H. Le, K. Natesan and S. Pranti-Haran, J. Adv. Ceram., 2015, 4, 237–243 CrossRef CAS
.
- Y. Chen, J. Yu, Q. Ke, Y. Gao, C. Zhang and Y. Guo, Chem. Eng. J., 2018, 341, 112–125 CrossRef CAS
.
- S. Mondal, U. Pal and A. Dey, Ceram. Int., 2016, 42, 18338–18346 CrossRef CAS
.
- C. Heng, X. Zhou, X. Zheng, M. Liu, Y. Wen, H. Huang, D. Fan, J. Hui, X. Zhang and Y. Wei, Mater. Sci. Eng., C, 2018, 91, 556–563 CrossRef CAS
.
- S. Mondal, G. Hoang, P. Manivasagan, H. Kim and J. Oh, Ceram. Int., 2019, 45, 17081–17093 CrossRef CAS
.
- S. Mondal, S. V. Dorozhkin and U. Pal, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10, e1504 Search PubMed
.
- A. Fihri, C. Len, R. S. Varma and A. Solhy, Coord. Chem. Rev., 2017, 347, 48–76 CrossRef CAS
.
- C. Lv, H. Liang, H. Chen and L. Wu, Microchem. J., 2019, 149, 103959 CrossRef CAS
.
- B. Nayak, A. Samant, P. K. Misra and M. Saxena, Mater. Today: Proc., 2019, 9, 689–698 CAS
.
- S. N. Maddila, S. Maddila, S. V. H. S. Bhaskaruni, N. Kerru and S. B. Jonnalagadda, Inorg. Chem. Commun., 2020, 112, 107706 CrossRef CAS
.
- M. Manoj, D. Mangalaraj, P. Meena and A. Yuan, Mater. Res. Express, 2019, 7, 015012 CrossRef
.
- K. Yaemsunthorn and C. Randorn, Int. J. Hydrogen Energy, 2017, 42, 5056–5062 CrossRef CAS
.
- Chitra, R. Laishram, S. Rajput and K. C. Singh, J. Alloys Compd., 2020, 812, 152128 CrossRef CAS
.
- S. Sun, H. Yao, W. Fu, S. Xue and W. Zhang, J. Hazard. Mater., 2020, 386, 121955 CrossRef CAS
.
- T. Bian, B. Xiao, B. Sun, L. Huang, S. Su, Y. Jiang, J. Xiao, A. Yuan, H. Zhang and D. Yang, Appl. Catal., B, 2020, 263, 118255 CrossRef CAS
.
- S. Devasahayam, in Characterization and Biology of Nanomaterials for Drug Delivery, ed. S. S. Mohapatra, S. Ranjan, N. Dasgupta, R. K. Mishra and S. Thomas, Elsevier, 2019, pp. 477–522, DOI:10.1016/b978-0-12-814031-4.00017-9
.
- J. Doran and G. Ryan, Technol. Soc., 2019, 59, 101183 CrossRef
.
- C.-H. Ooi, Y. P. Ling, S.-Y. Pung and F.-Y. Yeoh, Powder Technol., 2019, 342, 725–734 CrossRef CAS
.
- X. Xia, J. Chen, J. Shen, D. Huang, P. Duan and G. Zou, Adv. Powder Technol., 2018, 29, 1562–1570 CrossRef CAS
.
- X. He, K. Tang, X. Li, F. Wang, J. Liu, F. Zou, M. Yang and M. Li, Int. J. Biol. Macromol., 2019, 137, 45–53 CrossRef CAS
.
- S. Utara and J. Klinkaewnarong, Ceram. Int., 2015, 41, 14860–14867 CrossRef CAS
.
- S. Ahmed, M. Ahmad, B. L. Swami and S. Ikram, J. Adv. Res., 2016, 7, 17–28 CrossRef CAS
.
- C. J. Murphy, Science, 2002, 298, 2139–2141 CrossRef CAS
.
- M. V. Arularasu, SN Appl. Sci., 2019, 1, 393 CrossRef
.
- H. A. Abd El-Rehim and A. R. Tartour, Radiat. Phys. Chem., 2018, 153, 208–213 CrossRef CAS
.
- Z. S. Pillai and P. V. Kamat, J. Phys. Chem. B, 2004, 108, 945–951 CrossRef CAS
.
- G. Mpourmpakis, S. Caratzoulas and D. G. Vlachos, Nano Lett., 2010, 10, 3408–3413 CrossRef CAS
.
- A. K. Zak, W. H. A. Majid, M. Darroudi and R. Yousefi, Mater. Lett., 2011, 65, 70–73 CrossRef CAS
.
- A. K. Singh, V. Viswanath and V. C. Janu, J. Lumin., 2009, 129, 874–878 CrossRef CAS
.
- H. R. Rajabi and M. Farsi, Mater. Sci. Semicond. Process., 2016, 48, 14–22 CrossRef CAS
.
- F. Karimi, H. R. Rajabi and L. Kavoshi, Ultrason. Sonochem., 2019, 57, 139–146 CrossRef CAS
.
- P. Banerjee, M. Satapathy, A. Mukhopahayay and P. Das, Bioresour. Bioproces., 2014, 1, 3 CrossRef
.
- L. Fiandra, P. Bonfanti, Y. Piunno, A. P. Nagvenkar, I. Perlesthein, A. Gedanken, M. Saibene, A. Colombo and P. J. N. Mantecca, NanoImpact, 2020, 17, 100195 CrossRef
.
- N. S. Elbialy, S. F. Aboushoushah and W. W. Alshammari, Life Sci., 2019, 230, 76–83 CrossRef CAS
.
- D. Zhang, Y. Xue, J. Chen, X. Guo, D. Yang, J. Wang, J. Zhang, F. Zhang and A. Yuan, J. Nanosci. Nanotechnol., 2020, 20, 2728–2735 CrossRef CAS
.
- P. Song, J. Tao, X. He, Y. Sun, X. Shen, L. Zhai, A. Yuan, D. Zhang, Z. Ji and B. Li, Chem. Eng. J., 2020, 386, 124024 CrossRef CAS
.
- M. Manoj, R. Subbiah, P. Meena, D. Mangalaraj, N. Ponpandian, C. Viswanathan and K. Park, Adv. Sci., Eng. Med., 2016, 8, 216–221 CrossRef CAS
.
- S. Safi, F. Karimzadeh and S. Labbaf, Mater. Sci. Eng., C, 2018, 92, 712–719 CrossRef CAS
.
- Y. Xu, L. An, L. Chen, L. Cao, D. Zeng and G. Wang, Mater. Chem. Phys., 2018, 214, 359–363 CrossRef CAS
.
- R. Sergi, D. Bellucci, R. T. Candidato, L. Lusvarghi, G. Bolelli, L. Pawlowski, G. Candiani, L. Altomare, L. De Nardo and V. Cannillo, Surf. Coat. Technol., 2018, 352, 84–91 CrossRef CAS
.
- N. Monmaturapoj, A. Sri-On, W. Klinsukhon, K. Boonnak and C. Prahsarn, Mater. Sci. Eng., C, 2018, 92, 96–102 CrossRef CAS
.
- L.-j. Chen, T. Chen, J. Cao, B.-l. Liu, C.-s. Shao, K.-c. Zhou and D. Zhang, Trans. Nonferrous Met. Soc. China, 2018, 28, 125–136 CrossRef CAS
.
- M. Manoj, R. Subbiah, D. Mangalaraj, N. Ponpandian, C. Viswanathan and K. Park, Nanobiomedicine, 2015, 2, 2 CrossRef
.
- M. Manoj, D. Mangalaraj, N. Ponpandian and C. Viswanathan, RSC Adv., 2015, 5, 48705–48711 RSC
.
- M. Yoshimura, P. Sujaridworakun, F. Koh, T. Fujiwara, D. Pongkao and A. Ahniyaz, Mater. Sci. Eng., C, 2004, 24, 521–525 CrossRef
.
- K. Sonoda, Solid State Ionics, 2002, 151, 321–327 CrossRef CAS
.
- S. Pramanik, A. K. Agarwal, K. N. Rai and A. Garg, Ceram. Int., 2007, 33, 419–426 CrossRef CAS
.
- W. Weng, G. Han, P. Du and G. Shen, Mater. Chem. Phys., 2002, 74, 92–97 CrossRef CAS
.
- B. Prélot and T. Zemb, Mater. Sci. Eng., C, 2005, 25, 553–559 CrossRef
.
- J. Chen, Y. Yu, J. Chen, H. Li, J. Ji and D. Liu, Appl. Clay Sci., 2015, 115, 230–237 CrossRef CAS
.
- A. Khan, M. H. Shamsi and T.-S. Choi, Comput. Mater. Sci., 2009, 45, 257–265 CrossRef CAS
.
- M. A. Ashraf, W. Peng, Y. Zare and K. Y. Rhee, Nanoscale Res. Lett., 2018, 13, 214 CrossRef
.
- D. Gopi, N. Bhuvaneshwari, J. Indira and L. Kavitha, Spectrochim. Acta, Part A, 2013, 104, 292–299 CrossRef CAS
.
- F. Mohandes and M. Salavati-Niasari, Mater. Sci. Eng., C, 2014, 40, 288–298 CrossRef CAS
.
- A. Szcześ, Mater. Sci. Eng., C, 2014, 40, 373–381 CrossRef
.
- M. Meskinfam Langroudi, M. Giahi Saravani and A. Nouri, J. Appl. Biomater. Funct. Mater., 2017, 15, e334–e340 Search PubMed
.
- D. Gopi, J. Indira, L. Kavitha, M. Sekar and U. K. Mudali, Spectrochim. Acta, Part A, 2012, 93, 131–134 CrossRef CAS
.
- M. N. Nadagouda, G. Hoag, J. Collins and R. S. Varma, Cryst. Growth Des., 2009, 9, 4979–4983 CrossRef CAS
.
- F. A. Ahrens and A. L. Aronson, Toxicol. Appl. Pharmacol., 1971, 18, 10–25 CrossRef CAS
.
- C. Oviedo and J. Rodríguez, Quim. Nova, 2003, 26, 901–905 CrossRef CAS
.
- M. D. Adak and K. M. Purohit, Trends Biomater. Artif. Organs, 2011, 25, 101–106 Search PubMed
.
- V. H. Ingole, K. Hany Hussein, A. A. Kashale, K. Ghule, T. Vuherer, V. Kokol, J. Y. Chang, Y. C. Ling, A. Vinchurkar, H. N. Dhakal and A. V. Ghule, J. Biomed. Mater. Res., Part A, 2017, 105, 2935–2947 CrossRef CAS
.
- D. S. R. Krishna, A. Siddharthan, S. Seshadri and T. S. S. Kumar, J. Mater. Sci.: Mater. Med., 2007, 18, 1735–1743 CrossRef CAS
.
- Z. Xiao, M. Yuan, B. Yang, Z. Liu, J. Huang and D. Sun, Chemosphere, 2016, 150, 357–364 CrossRef CAS
.
- M. M. Alves, S. M. Andrade, L. Grenho, M. H. Fernandes, C. Santos and M. F. Montemor, Mater. Sci. Eng., C, 2019, 101, 76–87 CrossRef CAS
.
- W. Wei, X. Zhang, J. Cui and Z. Wei, J. Liq. Chromatogr. Relat. Technol., 2010, 33, 1335–1349 CrossRef CAS
.
- G. D'Urso, J. J. Mes, P. Montoro, R. D. Hall and R. C. H. de Vos, Metabolites, 2019, 10, 7 CrossRef
.
- V. Kalaiselvi, R. Mathammal, S. Vijayakumar and B. Vaseeharan, International Journal of Veterinary Science and Medicine, 2018, 6, 286–295 CrossRef CAS
.
- L. Lin, Y. Gu and H. Cui, Food Packag. Shelf Life, 2019, 19, 86–93 CrossRef
.
- G. B. Jegadeesan, K. Srimathi, N. Santosh Srinivas, S. Manishkanna and D. Vignesh, Biocatal. Agric. Biotechnol., 2019, 21, 101354 CrossRef
.
- K. Anand, C. Tiloke, A. Phulukdaree, B. Ranjan, A. Chuturgoon, S. Singh and R. M. Gengan, J. Photochem. Photobiol., B, 2016, 165, 87–95 CrossRef CAS
.
- A. A. Ezhilarasi, J. J. Vijaya, K. Kaviyarasu, M. Maaza, A. Ayeshamariam and L. J. Kennedy, J. Photochem. Photobiol., B, 2016, 164, 352–360 CrossRef CAS
.
- N. Matinise, X. G. Fuku, K. Kaviyarasu, N. Mayedwa and M. Maaza, Appl. Surf. Sci., 2017, 406, 339–347 CrossRef CAS
.
- D. Gopi, N. Bhuvaneshwari, J. Indira, K. Kanimozhi and L. Kavitha, Spectrochim. Acta, Part A, 2013, 107, 196–202 CrossRef CAS
.
- R. S. Jackson, in Wine Science, ed. R. S. Jackson, Academic Press, San Diego, 2014, pp. 347–426, DOI:10.1016/b978-0-12-381468-5.00006-3
.
- M. V. Moreno-Arribas and M. C. Polo, in Encyclopedia of Food Sciences and Nutrition, ed. B. Caballero, Academic Press, Oxford, 2003, pp. 1274–1280, DOI:10.1016/b0-12-227055-x/00232-7
.
- C. M. Loescher, D. W. Morton, S. Razic and S. Agatonovic-Kustrin, J. Pharm. Biomed. Anal., 2014, 98, 52–59 CrossRef CAS
.
- A. Y. Vazquez-Sanchez, P. Aguilar-Zarate, D. B. Muniz-Marquez, J. E. Wong-Paz, R. Rojas, J. A. Ascacio-Valdes and G. C. G. Martinez-Avila, Heliyon, 2019, 5, e03058 CrossRef
.
- L.-T. Wang, Q. Yang, Q. Cui, X.-H. Fan, M.-Z. Dong, M.-Z. Gao, M.-J. Lv, J.-Y. An, D. Meng, X.-H. Zhao and Y.-J. Fu, J. Cleaner Prod., 2020, 244, 118648 CrossRef CAS
.
- N. Nastić, I. Borrás-Linares, J. Lozano-Sánchez, J. Švarc-Gajić and A. Segura-Carretero, J. Ind. Eng. Chem., 2018, 68, 282–292 CrossRef
.
- P. K. Patial and D. S. Cannoo, Nat. Prod. Res., 2019, 1–5, DOI:10.1080/14786419.2019.1696330
.
- D. T. Coxon, Am. Potato J., 1984, 61, 169–183 CrossRef CAS
.
- P. E. Kolattukudy, K. Kronman and A. J. Poulose, Plant Physiol., 1975, 55, 567–573 CrossRef CAS
.
- Seneca, in Alkaloids – Secrets of Life, ed. T. Aniszewski, Elsevier, Amsterdam, 2007, pp. 61–139, DOI:10.1016/b978-044452736-3/50004-0
.
- B. R. Kumar, J. Pharm. Anal., 2017, 7, 349–364 CrossRef
.
- W. Wei, R. Sun, Z. Wei, H. Zhao, H. Li and F. Hu, J. Liq. Chromatogr. Relat. Technol., 2008, 32, 106–124 CrossRef
.
- M. L. Richmond, S. C. Brandao, J. I. Gray, P. Markakis and C. M. Stine, J. Agric. Food Chem., 1981, 29, 4–7 CrossRef CAS
.
- D. Gopi, N. Bhuvaneshwari, L. Kavitha and S. Ramya, Ceram. Int., 2015, 41, 3116–3127 CrossRef CAS
.
- D. B. Gomis, M. J. M. Gutiérrez, M. D. G. Alvarez and J. J. M. Alonso, Chromatographia, 1988, 25, 1054–1058 CrossRef
.
- L. R. Snyder, J. W. Dolan and D. C. Lommen, J. Chromatogr. A, 1989, 485, 65–89 CrossRef CAS
.
- M. Karkacier, M. Erbas, M. K. Uslu and M. Aksu, J. Chromatogr. Sci., 2003, 41, 331–333 CAS
.
- D. Tolkatchev, D. Elnatan, L. Nogara, T. Ly, N. Naber, K. Haak, R. Meech, R. Cooke and A. S. Kostyukova, Arch. Biochem. Biophys., 2018, 659, 75–84 CrossRef CAS
.
- S. Shityakov, E. Bigdelian, A. A. Hussein, M. B. Hussain, Y. C. Tripathi, M. U. Khan and M. A. Shariati, Eur. J. Med. Chem., 2019, 176, 149–161 CrossRef CAS
.
- A. K. Hazra, B. Chakraborty, A. Mitra and T. K. Sur, J. Ayurveda Integr. Med., 2019, 10, 248–254 CrossRef
.
- I. van Die, C. M. W. van Stijn, H. Geyer and R. Geyer, in Glycobiology, ed. M. Fukuda, Academic Press, 2010, vol. 480, pp. 117–140 Search PubMed
.
- Y. Wang, L. Chen, K. Chaisiwamongkhol and R. G. Compton, Food Chem., 2020, 309, 125606 CrossRef CAS
.
- R. Subramanian, S. Sathish, P. Murugan, A. Mohamed Musthafa and M. Elango, J. King Saud Univ., Sci., 2019, 31, 667–673 CrossRef
.
- R. Subramanian, P. Subbramaniyan, J. Noorul Ameen and V. Raj, Arabian J. Chem., 2016, 9, S537–S540 CrossRef CAS
.
- N. Manousi, I. Sarakatsianos and V. Samanidou, in Engineering Tools in the Beverage Industry, ed. A. M. Grumezescu and A. M. Holban, Woodhead Publishing, 2019, pp. 283–314, DOI:10.1016/B978-0-12-815258-4.00010-X
.
- C. F. Poole, in Liquid-Phase Extraction, ed. C. F. Poole, Elsevier, 2020, pp. 1–44, DOI:10.1016/b978-0-12-816911-7.00001-3
.
- S. Suna, C. E. Tamer and G. Özcan-Sinir, in Natural Beverages, ed. A. M. Grumezescu and A. M. Holban, Academic Press, 2019, pp. 361–398, DOI:10.1016/b978-0-12-816689-5.00013-4
.
- M. O. Nafiu, A. A. Hamid, H. F. Muritala and S. B. Adeyemi, in Medicinal Spices and Vegetables from Africa, ed. V. Kuete, Academic Press, 2017, pp. 171–204, DOI:10.1016/b978-0-12-809286-6.00007-8
.
- B. Trusheva, D. Trunkova and V. Bankova, Chem. Cent. J., 2007, 1, 13 CrossRef
.
- O. Jović, I. Habinovec, N. Galić and M. Andrašec, J. Spectrosc., 2018, 2018, 1–9 CrossRef
.
- S. Bajad and V. Shulaev, Trends Anal. Chem., 2007, 26, 625–636 CrossRef CAS
.
- C. Castro-Lopez, J. M. Ventura-Sobrevilla, M. D. Gonzalez-Hernandez, R. Rojas, J. A. Ascacio-Valdes, C. N. Aguilar and G. C. G. Martinez-Avila, Food Chem., 2017, 237, 1139–1148 CrossRef CAS
.
- M. Llompart, C. Garcia-Jares, M. Celeiro and T. Dagnac, in Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, ed. P. Worsfold, C. Poole, A. Townshend and M. Miró, Academic Press, Oxford, 2018, pp. 67–77, DOI:10.1016/b978-0-12-409547-2.14442-7
.
- I. Niyonizigiye, D. Nkurunziza, D. Ngabire, A. T. Gitachew, B. S. Chun and G.-D. Kim, S. Afr. J. Bot., 2020, 128, 231–238 CrossRef CAS
.
- S. C. Mandal, V. Mandal and A. K. Das, in Essentials of Botanical Extraction, ed. S. C. Mandal, V. Mandal and A. K. Das, Academic Press, Boston, 2015, pp. 83–136, DOI:10.1016/b978-0-12-802325-9.00006-9
.
- C. Liu, Y. Huang, W. Shen and J. Cui, Biomaterials, 2001, 22, 301–306 CrossRef CAS
.
- R. Palanivelu, A. Mary Saral and A. Ruban Kumar, Spectrochim. Acta, Part A, 2014, 131, 37–41 CrossRef CAS
.
- S. Ponnusamy, S. Sadhasivam, K. Louis, M. Periasamy and G. Dhanaraj, Int. J. Appl. Ceram. Technol., 2020, 17, 2008–2016 CrossRef CAS
.
- R. Zhou, S. Si and Q. Zhang, Appl. Surf. Sci., 2012, 258, 3578–3583 CrossRef CAS
.
- J. Klinkaewnarong, E. Swatsitang, C. Masingboon, S. Seraphin and S. Maensiri, Curr. Appl. Phys., 2010, 10, 521–525 CrossRef
.
- S. Suzanna, A. Dewi, S. Al and H. Tetiana, 2018.
- S. Mortazavi-Derazkola, M. Salavati-Niasari, H. Khojasteh, O. Amiri and S. M. Ghoreishi, J. Cleaner Prod., 2017, 168, 39–50 CrossRef CAS
.
- D. Hao, Y. Xu, M. Zhao, J. Ma, Y. Wei and X. Wang, J. Photochem. Photobiol., B, 2020, 202, 111674 CrossRef CAS
.
- S. Balu, S. Andra, S. Kannan, M. Vidyavathy S and M. Muthalagu, Mater. Lett., 2020, 259, 126900 CrossRef CAS
.
- B. Sahin, A. Aygun, H. Gunduz, K. Sahin, E. Demir, S. Akocak and F. Sen, Colloids Surf., B, 2018, 163, 119–124 CrossRef CAS
.
- S. Maensiri, P. Laokul, J. Klinkaewnarong, S. Phokha, V. Promarak and S. Seraphin, J. Optoelectron. Adv. Mater., 2008, 10, 161–165 Search PubMed
.
- S. Bose and S. K. Saha, J. Am. Ceram. Soc., 2003, 86, 1055–1057 CrossRef CAS
.
- D. Gopi, K. Kanimozhi, N. Bhuvaneshwari, J. Indira and L. Kavitha, Spectrochim. Acta, Part A, 2014, 118, 589–597 CrossRef CAS
.
- D. Gopi, L. Kavitha, S. Ramya and D. Rajeswari, in Engineering of Nanobiomaterials, ed. A. M. Grumezescu, William Andrew Publishing, 2016, pp. 485–521, DOI:10.1016/b978-0-323-41532-3.00015-4
.
- S. A. Ibraheem, E. A. Audu, M. u. Jaafar, J. A. Adudu, J. T. Barminas, V. Ochigbo, A. Igunnu and S. O. Malomo, Surf. Interfaces, 2019, 17, 100360 CrossRef CAS
.
- M. Sundrarajan, S. Jegatheeswaran, S. Selvam, N. Sanjeevi and M. Balaji, Mater. Des., 2015, 88, 1183–1190 CrossRef CAS
.
- G. S. Kumar, S. Rajendran, S. Karthi, R. Govindan, E. K. Girija, G. Karunakaran and D. Kuznetsov, MRS Commun., 2017, 7, 183–188 CrossRef CAS
.
- S. Nayar and A. Guha, Mater. Sci. Eng., C, 2009, 29, 1326–1329 CrossRef CAS
.
- A. Tampieri, S. Sprio, A. Ruffini, G. Celotti, I. G. Lesci and N. Roveri, J. Mater. Chem., 2009, 19, 4973–4980 RSC
.
- B. Saha, S. K. Yadav and S. Sengupta, Fuel, 2018, 222, 743–752 CrossRef CAS
.
- P. Phatai, C. M. Futalan, S. Kamonwannasit and P. Khemthong, J. Sol-Gel Sci. Technol., 2019, 89, 764–775 CrossRef CAS
.
- V. P. Padmanabhan, S. Balakrishnan, R. Kulandaivelu, S. N. TSN, M. Lakshmipathy, S. Sagadevan, F. Mohammad, H. A. Al-Lohedan, S. Paiman and W. C. Oh, New J. Chem., 2020, 44, 7175–7185 RSC
.
- G. S. Kumar, D. Muthu, G. Karunakaran, S. Karthi, E. K. Girija and D. Kuznetsov, J. Sol-Gel Sci. Technol., 2018, 86, 610–616 CrossRef CAS
.
- C. Jäger, T. Welzel, W. Meyer-Zaika and M. Epple, Magn. Reson. Chem., 2006, 44(6), 573–580 CrossRef
.
- R. N. Panda, M. F. Hsieh, R. J. Chung and T. S. Chin, J. Phys. Chem. Solids, 2003, 64, 193–199 CrossRef CAS
.
- Y.-H. Tseng, C.-Y. Mou and J. C. C. Chan, J. Am. Chem. Soc., 2006, 128, 6909–6918 CrossRef CAS
.
- A. Altemimi, N. Lakhssassi, A. Baharlouei, D. G. Watson and D. A. Lightfoot, Plants, 2017, 6, 42 CrossRef
.
- P. Negi, A. Chauhan, G. Sadia, Y. Rohinishree and R. Ramteke, Food Chem., 2005, 92, 119–124 CrossRef CAS
.
- S. Redondo-Blanco, J. Fernandez, S. Lopez-Ibanez, E. M. Miguelez, C. J. Villar and F. Lombo, J. Food Prot., 2020, 83, 163–171 CrossRef CAS
.
- S. K. Chang, C. Alasalvar and F. Shahidi, Crit. Rev. Food Sci. Nutr., 2019, 59, 1580–1604 CrossRef CAS
.
- D. C. Pham, M. A. Shibu, B. Mahalakshmi and B. K. Velmurugan, Crit. Rev. Food Sci. Nutr., 2019, 1–25, DOI:10.1080/10408398.2019.1699014
.
- D. Singh and P. K. Chaudhuri, Ind. Crops Prod., 2018, 118, 367–382 CrossRef CAS
.
- M. S. Akhtar, J. Panwar and Y.-S. Yun, ACS Sustainable Chem. Eng., 2013, 1, 591–602 CrossRef CAS
.
- S. Phumying, S. Labuayai, C. Thomas, V. Amornkitbamrung, E. Swatsitang and S. Maensiri, Appl. Phys. A, 2012, 111, 1187–1193 CrossRef
.
- R. Jose Varghese, N. Zikalala, E. H. M. Sakho and O. S. Oluwafemi, in Colloidal Metal Oxide Nanoparticles, ed. S. Thomas, A. Tresa Sunny and P. Velayudhan, Elsevier, 2020, pp. 67–82, DOI:10.1016/b978-0-12-813357-6.00006-1
.
- R. Rahmani, M. Gharanfoli, M. Gholamin, M. Darroudi, J. Chamani, K. Sadri and A. Hashemzadeh, Ceram. Int., 2020, 46, 3051–3058 CrossRef CAS
.
- N. Pramanik, A. Tarafdar and P. Pramanik, J. Mater. Process. Technol., 2007, 184, 131–138 CrossRef CAS
.
- C. Li, Powder Technol., 2009, 192, 1–5 CrossRef CAS
.
- M. A. Martins, C. Santos, M. M. Almeida and M. E. Costa, J. Colloid Interface Sci., 2008, 318, 210–216 CrossRef CAS
.
- A. Wang, H. Yin, D. Liu, H. Wu, Y. Wada, M. Ren, Y. Xu, T. Jiang and X. Cheng, Appl. Surf. Sci., 2007, 253, 3311–3316 CrossRef CAS
.
- A. Wang, D. Liu, H. Yin, H. Wu, Y. Wada, M. Ren, T. Jiang, X. Cheng and Y. Xu, Mater. Sci. Eng., C, 2007, 27, 865–869 CrossRef CAS
.
- R. Thaya, B. Malaikozhundan, S. Vijayakumar, J. Sivakamavalli, R. Jeyasekar, S. Shanthi, B. Vaseeharan, P. Ramasamy and A. Sonawane, Microb. Pathog., 2016, 100, 124–132 CrossRef CAS
.
- J. Polte, CrystEngComm, 2015, 17, 6809–6830 RSC
.
- R. Subramanian, S. Sathish, P. Murugan, A. Mohamed Musthafa and M. Elango, J. King Saud Univ., Sci., 2018, 31(4), 667–673 CrossRef
.
- K. Qwele, A. Hugo, S. O. Oyedemi, B. Moyo, P. J. Masika and V. Muchenje, Meat Sci., 2013, 93, 455–462 CrossRef CAS
.
- B. Sreedhar, D. K. Devi, A. S. Neetha, V. P. Kumar and K. V. R. Chary, Mater. Chem. Phys., 2015, 153, 23–31 CrossRef CAS
.
- Y. K. Leong, U. Seah, S. Y. Chu and B. C. Ong, Colloids Surf., A, 2001, 182, 263–268 CrossRef CAS
.
- S. Brown, in Fruit Breeding, ed. M. L. Badenes and D. H. Byrne, Springer, US, Boston, MA, 2012, pp. 329–367, DOI:10.1007/978-1-4419-0763-9_10
.
- D. Arcos and M. Vallet-Regí, J. Mater. Chem. B, 2020, 8, 1781–1800 RSC
.
- M. Bornapour, N. Muja, D. Shum-Tim, M. Cerruti and M. Pekguleryuz, Acta Biomater., 2013, 9, 5319–5330 CrossRef CAS
.
- D. Shepherd and S. M. Best, Biomed. Mater., 2013, 8, 025003 CrossRef
.
- A. Bhattacharjee, A. Gupta, M. Verma, P. A. Murugan, P. Sengupta, S. Matheshwaran, I. Manna and K. Balani, Ceram. Int., 2019, 45, 12225–12233 CrossRef CAS
.
- I. Qayoom, A. K. Teotia, M. Meena, P. Singh, A. Mishra, S. Singh and A. Kumar, Biomed. Mater., 2020, 15, 055015 CrossRef
.
- X. Zhang, Y. Li, G. Lv, Y. Zuo and Y. Mu, Polym. Degrad. Stab., 2006, 91, 1202–1207 CrossRef CAS
.
- Z. Zyman, I. Ivanov, D. Rochmistrov, V. Glushko, N. Tkachenko and S. Kijko, J. Biomed. Mater. Res., 2001, 54(2), 256–263 CrossRef CAS
.
- M. Okazaki, T. Aoba, Y. Doi, J. Takahashi and Y. Moriwaki, J. Dent. Res., 1981, 60, 845–849 CrossRef CAS
.
- D. G. Nelson, J. Dent. Res., 1981, 60(Spec No. C), 1621–1629 CrossRef CAS
.
- R. LeGeros, in Calcium phosphates in oral biology and medicine, Karger Publishers, 1991, vol. 15, pp. 108–129 Search PubMed
.
- M. T. Fulmer, I. C. Ison, C. R. Hankermayer, B. R. Constantz and J. Ross, Biomaterials, 2002, 23, 751–755 CrossRef CAS
.
- J. Wolff, D. Hofmann, W. Amelung, H. Lewandowski, K. Kaiser and R. Bol, Appl. Geochem., 2018, 97, 181–186 CrossRef CAS
.
- X. Chen, X. Jin, J. Tan, W. Li, M. Chen, L. Yao and H. Yang, J. Colloid Interface Sci., 2016, 468, 300–306 CrossRef CAS
.
- Y. Zhang, K. Dong, F. Wang, H. Wang, J. Wang, Z. Jiang and S. Diao, Colloids Surf., B, 2018, 170, 497–504 CrossRef CAS
.
- C. Tian, P. Xu, J. Jiang and C. Han, J. Dispersion Sci. Technol., 2020, 1–9, DOI:10.1080/01932691.2019.1710185
.
- L. Li and L.-J. Zheng, Sci. Adv. Mater., 2020, 12, 110–115 CrossRef CAS
.
- C.-Y. Yu, Y.-M. Wang, N.-M. Li, G.-S. Liu, S. Yang, G.-T. Tang, D.-X. He, X.-W. Tan and H. Wei, Mol. Pharm., 2014, 11, 638–644 CrossRef CAS
.
- W. Zhang, P. Xu and H. Zhang, Trends Food Sci. Technol., 2015, 44, 258–271 CrossRef CAS
.
- Y.-H. Yang, C.-H. Liu, Y.-H. Liang, F.-H. Lin and K. C. W. Wu, J. Mater. Chem. B, 2013, 1, 2447–2450 RSC
.
- D. R. K. Weerasuriya, W. P. S. L. Wijesinghe and R. M. G. Rajapakse, Mater. Sci. Eng., C, 2017, 71, 206–213 CrossRef CAS
.
- K.-W. Wang, Y.-J. Zhu, X.-Y. Chen, W.-Y. Zhai, Q. Wang, F. Chen, J. Chang and Y.-R. Duan, Chem.–Asian J., 2010, 5, 2477–2482 CrossRef CAS
.
- D. Predoi, S. L. Iconaru, N. Buton, M. L. Badea and L. Marutescu, Nanomaterials, 2018, 8, 291 CrossRef
.
- R. K. Das, S. K. Brar and M. Verma, Trends Biotechnol., 2016, 34, 440–449 CrossRef CAS
.
- R. Rajan, K. Chandran, S. L. Harper, S.-I. Yun and P. T. Kalaichelvan, Ind. Crops Prod., 2015, 70, 356–373 CrossRef CAS
.
- A. Kovtun, M. J. Goeckelmann, A. A. Niclas, E. B. Montufar, M.-P. Ginebra, J. A. Planell, M. Santin and A. Ignatius, Acta Biomater., 2015, 12, 242–249 CrossRef CAS
.
- M. L. Badea, S. L. Iconaru, A. Groza, M. C. Chifiriuc, M. Beuran and D. Predoi, Molecules, 2019, 24, 2169 CrossRef CAS
.
- A. Shebi and S. Lisa, Carbohydr. Polym., 2018, 201, 39–47 CrossRef CAS
.
- S. Ponnusamy, S. Sadhasivam, K. Louis, M. Periasamy and G. Dhanaraj, Int. J. Appl. Ceram. Technol., 2020, 17, 2008–2016 CrossRef CAS
.
- C. Li, Q. Yuan, L. He, Z.-J. Qian, C. Zhou and P. Hong, Coatings, 2019, 9, 141 CrossRef
.
- Ö. Elif, Ö. Belma and Ş. İlkay, Mater. Res. Express, 2017, 4, 015601 CrossRef
.
- N. A. M. Barakat, M. S. Khil, A. M. Omran, F. A. Sheikh and H. Y. Kim, J. Mater. Process. Technol., 2009, 209, 3408–3415 CrossRef CAS
.
- R.-X. Sun, Y. Lv, Y.-R. Niu, X.-H. Zhao, D.-S. Cao, J. Tang, X.-C. Sun and K.-Z. Chen, Ceram. Int., 2017, 43, 16792–16798 CrossRef CAS
.
- K. Haberko, M. M. Bućko, J. Brzezińska-Miecznik, M. Haberko, W. Mozgawa, T. Panz, A. Pyda and J. Zarębski, J. Eur. Ceram. Soc., 2006, 26, 537–542 CrossRef CAS
.
- N. A. S. Mohd Pu'ad, P. Koshy, H. Z. Abdullah, M. I. Idris and T. C. Lee, Heliyon, 2019, 5, e01588 CrossRef
.
- H. Susanto, A. Taufiq, S. Sunaryono, S. Soontaranon, Y. A. Hariyanto, A. I. Mawardi, N. G. Adreyanto, D. T. Yunisa, F. Rufiandita, F. Nizarghazi, G. Alifi, L. N. Putri, S. D. M. Kurnia and Sumardi, J. Phys.: Conf. Ser., 2018, 1093, 012007 CrossRef
.
- H. Susanto, A. Taufiq, Sunaryono, A. Imam Mawardi, Y. A. Hariyanto, A. Nicholas Gerry, D. Tri Yunisa, F. Rufiandita, F. Nizarghazi, G. Alifi, P. Lita Neldya, M. Sinta Dewi and Sumardi, IOP Conf. Ser.: Mater. Sci. Eng., 2019, 276, 012005 CrossRef
.
- Q. H. Duong, K. D. Clark, K. G. Lapsley and R. B. Pegg, Food Chem., 2017, 229, 84–92 CrossRef CAS
.
- V. K. Khlestkin, I. V. Rozanova, V. M. Efimov and E. K. Khlestkina, BMC Genet., 2019, 20, 29 CrossRef
.
- D. Domene-López, J. J. Delgado-Marín, I. Martin-Gullon, J. C. García-Quesada and M. G. Montalbán, Int. J. Biol. Macromol., 2019, 135, 845–854 CrossRef
.
- A. A. Shaltout, M. A. Allam and M. A. Moharram, Spectrochim. Acta, Part A, 2011, 83, 56–60 CrossRef CAS
.
- E. Desmidt, K. Ghyselbrecht, A. Monballiu, W. Verstraete and B. D. Meesschaert, Water Sci. Technol., 2012, 65, 1954–1962 CrossRef CAS
.
- L. Seghezzo, G. Zeeman, J. B. van Lier, H. V. M. Hamelers and G. Lettinga, Bioresour. Technol., 1998, 65, 175–190 CrossRef CAS
.
- A. Jiraprasertwong, K. Maitriwong and S. Chavadej, Renewable Energy, 2019, 130, 191–205 CrossRef CAS
.
- L. Zhang, Q. Ban, J. Li and C. Wan, Bioresour. Technol., 2019, 288, 121464 CrossRef CAS
.
- E. Fernandez-Palacios, J. Lafuente, M. Mora and D. Gabriel, Sci. Total Environ., 2019, 688, 1184–1192 CrossRef CAS
.
- L. Zhang, J. De Vrieze, T. L. G. Hendrickx, W. Wei, H. Temmink, H. Rijnaarts and G. Zeeman, Chem. Eng. J., 2018, 334, 2088–2097 CrossRef CAS
.
- G. Lettinga and L. W. Hulshoff Pol, Water Sci. Technol., 1991, 24, 87–107 CrossRef CAS
.
- A. Monballiu, E. Desmidt, K. Ghyselbrecht and B. Meesschaert, J. Environ. Chem. Eng., 2018, 6, 4413–4422 CrossRef CAS
.
- N. A. Mata, P. Ros-Tárraga, P. Velasquez, A. Murciano and P. N. De Aza, Ceram. Int., 2020, 46, 968–977 CrossRef CAS
.
- M. V. B. Santos, A. L. Oliveira, J. A. Osajima and E. C. Silva-Filho, Ceram. Int., 2020, 46, 3811–3817 CrossRef CAS
.
- P. Pankaew and P. White, J. Nanosci. Nanotechnol., 2020, 20, 81–86 CrossRef CAS
.
- S.-C. Wu, H.-K. Tsou, H.-C. Hsu, S.-K. Hsu, S.-P. Liou and W.-F. Ho, Ceram. Int., 2013, 39, 8183–8188 CrossRef CAS
.
- E. Desmidt, K. Ghyselbrecht, Y. Zhang, L. Pinoy, B. Van der Bruggen, W. Verstraete, K. Rabaey and B. Meesschaert, Crit. Rev. Environ. Sci. Technol., 2014, 45, 336–384 CrossRef
.
- K. C. van Dijk, J. P. Lesschen and O. Oenema, Sci. Total Environ., 2016, 542, 1078–1093 CrossRef CAS
.
- J. Driver, D. Lijmbach and I. Steen, Environ. Technol., 2010, 20, 651–662 CrossRef
.
- W. Somsub, R. Kongkachuichai, P. Sungpuag and R. Charoensiri, J. Food Compos. Anal., 2008, 21, 187–197 CrossRef CAS
.
- E. Ruiz, M. I. Santillana, M. T. Nieto and I. Sastre, J. Liq. Chromatogr., 1995, 18, 989–1000 CrossRef CAS
.
- R. A. Molins, Phosphates in food, CRC Press, 1990 Search PubMed
.
- C. Lintas, Options Méditerranéennes, 1992, 19, 79–87 Search PubMed
.
- R. K. Das and S. K. Brar, Nanoscale, 2013, 5, 10155–10162 RSC
.
- M. R. Haque and J. H. Bradbury, Food Chem., 2002, 77, 107–114 CrossRef
.
- J. E. Poulton, Plant Physiol., 1990, 94, 401 CrossRef CAS
.
- I. F. Bolarinwa, C. Orfila and M. R. A. Morgan, Food Chem., 2014, 152, 133–139 CrossRef CAS
.
- I. F. Bolarinwa, M. O. Oke, S. A. Olaniyan and A. S. Ajala, Toxicology – new aspects to this conundrum, A Review of Cyanide Glycolysis in Edible Plants, Intech Open, 2016, ch. 8, pp. 171–191, DOI:10.5772/64886
.
- X. Zhao, S. de Juan, F. R. Guerrero, Z. Li, J. Llorca and D.-Y. Wang, Polym. Degrad. Stab., 2016, 127, 2–10 CrossRef CAS
.
- M. Sadat-Shojai, M.-T. Khorasani, E. Dinpanah-Khoshdargi and A. Jamshidi, Acta Biomater., 2013, 9, 7591–7621 CrossRef CAS
.
- S. Kumar, C. Gautam, B. S. Chauhan, S. Srikrishna, R. S. Yadav and S. B. Rai, Ceram. Int., 2020, 46, 16235–16248 CrossRef CAS
.
- D. Gopi, S. Ramya, D. Rajeswari, P. Karthikeyan and L. Kavitha, Colloids Surf., A, 2014, 451, 172–180 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2020 |
