Open Access Article
Open Access ArticleXanthine-based photoaffinity probes allow assessment of ligand engagement by TRPC5 channels†
Claudia C.
Bauer‡
a,
Aisling
Minard‡
ab,
Isabelle B.
Pickles‡
ab,
Katie J.
Simmons
a,
Eulashini
Chuntharpursat-Bon
a,
Matthew P.
Burnham
c,
Nikil
Kapur
d,
David J.
Beech
a,
Stephen P.
Muench
 ef,
Megan H.
Wright
ef,
Megan H.
Wright
 bf,
Stuart L.
Warriner
bf and
Robin S.
Bon
bf,
Stuart L.
Warriner
bf and
Robin S.
Bon
 *af
*af
aLeeds Institute of Cardiovascular and Metabolic Medicine, LIGHT Laboratories, University of Leeds, Leeds LS2 9JT, UK. E-mail: r.bon@leeds.ac.uk
bSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
cDiscovery Sciences, R&B Biopharmaceuticals, AstraZeneca, Alderley Park, UK
dSchool of Mechanical Engineering, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
eSchool of Biomedical Sciences, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
fAstbury Centre for Structural Molecular Biology, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
First published on 18th September 2020
Abstract
TRPC1/4/5 cation channels are emerging drug targets for the treatment of, amongst others, central nervous system (CNS) disorders, kidney disease, and cardiovascular and metabolic disease. Various small-molecule TRPC1/4/5 modulators have been reported, including highly potent xanthine derivatives that distinguish between specific TRPC1/4/5 tetramers. However, tools to profile ligand engagement by TRPC1/4/5 channels in live cells are lacking. Here, we report a set of potent xanthine-based photoaffinity probes that functionally mimic the xanthines Pico145 and AM237. Using these probes, we have developed a photoaffinity labelling protocol for TRPC5 channels, providing the first method for the quantitative assessment of binding interactions of TRPC5 with small molecules in cells. This method could be important for drug discovery efforts targeting the xanthine/lipid binding site of TRPC1/4/5 channels.
Introduction
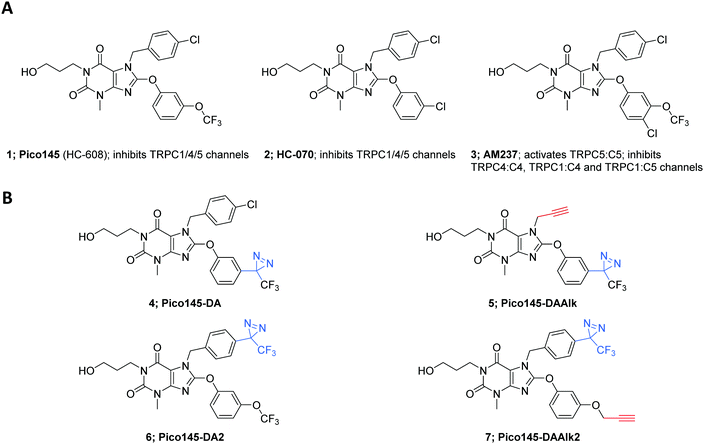
Transient Receptor Potential Canonical (TRPC) proteins are members of the TRP superfamily, which also includes TRPV, TRPA and TRPM.1–8 Like other TRPs, TRPC proteins form tetrameric, non-selective cation channels permeable by Na+ and Ca2+. Based on sequence homology, the human TRPC proteins are divided into two clusters: TRPC1/4/5 and TRPC3/6/7,9 while TRPC2 is encoded by a pseudogene in human.10 TRPC4 and TRPC5 are the most closely related TRPC proteins7 (70% sequence identity; BLAST search8). TRPC1/4/5 proteins can assemble to form homo- or heterotetrameric channels, each with their own biophysical characteristics (e.g., voltage dependence, ion selectivity), cellular function, and pharmacological profile.11 TRPC1 may not form functional homomeric channels, but is an important contributor to heteromeric channels, for example with TRPC4 and/or TRPC5.1,6,9–13 However, there remains a lack of insight into the exact compositions and stoichiometries of native TRPC1/4/5 channels, which may vary depending on, e.g., cell type, tissue and cellular state. The implication of TRPC1/4/5 in a wide range of physiological and pathological mechanisms has driven the search for potent and selective channel modulators for studies in cells, tissues and animal models.11,12 Because disruption of the Trpc4/5 genes13 and global expression of a dominant-negative mutant TRPC514 do not cause catastrophic phenotypes in rodents, TRPC1/4/5 inhibition may be particularly promising as a therapeutic strategy. Indeed, clinical trials with at least two TRPC1/4/5 inhibitors have been announced by Hydra Biosciences/Boehringer Ingelheim (the TRPC4/5 channel inhibitor BI 135889 for treatment of anxiety/CNS disorders) and Goldfinch Bio (the TRPC5 channel inhibitor GFB-887 for genetically-driven kidney disease). The structures of these clinical candidates have not been disclosed to date.Chemically-diverse small-molecule TRPC1/4/5 modulators have been reported, but for most of these the modes-of-action are poorly understood.2,11,12,15,16 Cellular targets may be diverse, and even direct binders may have different TRPC1/4/5 binding sites. The most potent, selective and efficacious TRPC1/4/5 activator is the natural product (−)-englerin A (EA).17–19 Although EA has been used in animals,20 its toxicity19,21 and instability in rodent plasma19 need to be carefully considered in study design. The most promising and well-characterised TRPC1/4/5 inhibitors are two closely-related xanthines, Pico145 1 (also called HC-608) and HC-070 2, which originally appeared in a patent assigned to Hydra Biosciences (Fig. 1).22 Pico145 is the most potent inhibitor of TRPC1/4/5 channels to date,23,24 with the highest potencies against heteromeric channels and high selectivity against 8 related ion channels. Just et al. reported the anxiolytic and antidepressant effects in mice of HC-070.25 As part of this study, activities of HC-070 and Pico145 were confirmed against human, mouse and rat TRPC1/4/5 channels. In addition, both compounds were shown to be >400-fold selective against >100 ion channels, receptors, enzymes, kinases and transporters (>2000-fold for most) and to be orally bioavailable. Therefore, Pico145 and HC-070 are considered valuable chemical probes for functional studies of TRPC1/4/5 channels in cells and animals.21,25–27 In addition, we reported that AM237, a close analogue of Pico145 and HC-070, is an activator of homomeric TRPC5:C5 channels, but an inhibitor of TRPC4:C4, TRPC1:C5 and TRPC1:C4 channels.28
Pico145 is a competitive antagonist of both EA- and AM237-mediated TRPC5:C5 activation.17,28 Pico145 also inhibits TRPC1/4/5 channels activated by carbachol25 and sphingosine-1-phosphate (S1P)23 (which act via G-protein signalling), and by the direct agonist Gd3+, although low concentrations of Pico145 (10 pM) can also potentiate Gd3+-induced currents mediated by TRPC4.23 These data suggest that xanthines such as Pico145 and AM237 occupy a well-defined, high-affinity binding site essential to TRPC1/4/5 channel gating to stabilise open or closed TRPC5 channel states, depending on subunit composition and xanthine substituent pattern. We recently reported cryo-EM structures of human TRPC5:C5 in complex with Pico145, revealing that Pico145 binds to a conserved lipid binding site of TRPC5, where it displaces a bound phospholipid.29 A subsequent report of a cryo-EM structure of TRPC5:C5 in complex with HC-07030 suggested that HC-070 binds to the same site as Pico145, with a near-identical binding pose. These studies highlight targeting of the conserved lipid binding site of TRPC1/4/5 channels as a promising strategy for TRPC1/4/5 drug discovery.
An important step in drug discovery is the assessment of cellular target engagement and occupancy by candidate molecules.31–34 Photoaffinity labelling (PAL)35–37 can be used to identify, characterise and validate interactions of small molecules with membrane proteins and ion channels,38–42 which may readily lose their affinity for non-covalent ligands under various experimental conditions. PAL results in the formation of covalent bonds between a photoaffinity probe and its cellular target(s). The initiation of labelling with a specific wavelength of light enables spatio-temporal control and allows straight-forward control experiments in the absence of light.
The structure and pharmacological profile of a photoaffinity probe must closely resemble those of its parent compound, and the placement of the photoreactive group must favour intermolecular crosslinking over intramolecular rearrangement.40 A high degree of nonspecific labelling is not uncommon in PAL, requiring deconvolution of specific and non-specific labelling events.43,44 PAL on ion channels is particularly challenging: low expression levels, in combination with low photolabelling efficiencies common in PAL, often result in only small quantities of labelled protein. Diazirines are the smallest photoreactive groups suitable for PAL. Upon irradiation with UV light (365 nm), they generate highly reactive carbene species that rapidly insert into proximal C–H or heteroatom–H bonds.37,45 Incorporation of diazirines allows the development of photoaffinity probes through minimal structural variation of small molecules.46–48
We hypothesised that PAL would be a suitable approach to assess cellular target engagement of TRPC1/4/5 modulators. Here, we report the development of a PAL protocol for TRPC5 channels. Modification of Pico145 with diazirines and alkyne handles gave a set of photoaffinity probes that functionally mimic either Pico145 or AM237. Our 2-step labelling protocol based on PAL and copper-catalysed azide–alkyne cycloadditions (CuAAC)49 provided the first proof of a direct interaction between xanthines and TRPC5 in live cells, and allowed the quantitative assessment of TRPC5 binding by Pico145 in human cells. This technology may allow assessment of target engagement of other TRPC1/4/5 modulators and elucidation of their mode-of-action, and thereby underpin TRPC1/4/5 drug discovery targeting the xanthine/lipid binding site.
Results
Development of xanthine-based photoaffinity probes
Analysis of semi-quantitative TRPC5 assay data for 621 compounds described in the patent literature22 revealed the N-7 and C-8 substituents of Pico145 to be the most suitable sites for incorporation of functional handles (Fig. S1, ESI†). Aiming for maximal structural similarity to Pico145 itself, we designed and synthesised a set of four Pico145 photoaffinity probes 4–7 (Fig. 1B), all containing the (trifluoromethyl)(phenyl)diazirine motif commonly used in diazirine photocrosslinkers.37,50 Pico145-diazirine (Pico145-DA; 4) and Pico145-diazirine-2 (Pico145-DA2; 6) are structurally most similar to Pico145 and were made to test the functional effect of incorporation of a diazirine group. The compounds designed for our photocrosslinking studies, Pico145-diazirine-alkyne (Pico145-DAAlk; 5) and Pico145-diazirine-alkyne-2 (Pico145-DAAlk2; 7) incorporate an additional alkyne handle that allows bioorthogonal labelling with various reporter groups, such as biotin or a fluorophore, through CuAAC. The compounds were synthesised, purified to homogeneity, and chemically characterised as described in the ESI,† Methods.Xanthine-based photoaffinity probes are nanomolar TRPC1/4/5 modulators
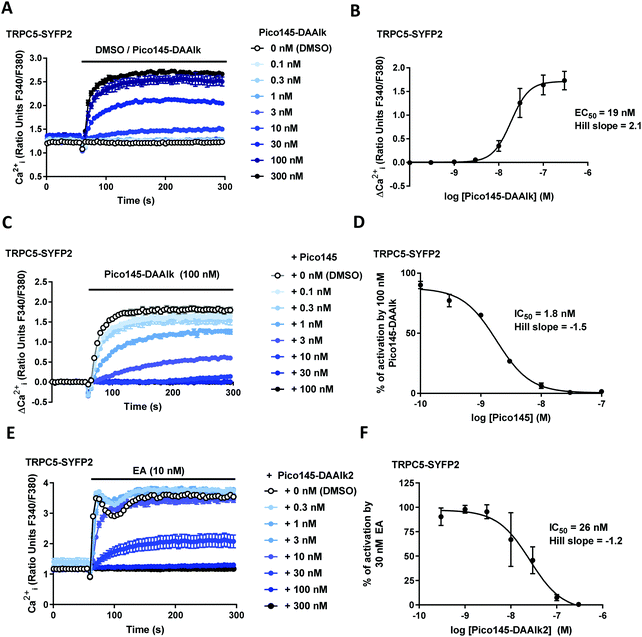
The effects of xanthine photoaffinity probes on TRPC1/4/5 ion channel activity were assessed by intracellular calcium ([Ca2+]i) measurements in HEK T-REx cells expressing tetracycline-inducible TRPC5-SYFP2, TRPC4-SYFP2, TRPC5-C1 or TRPC4-C1, using Fura-2 as the calcium indicator (see Methods). To test TRPC1/4/5 channel inhibition, cells were pre-treated with increasing concentrations of photoaffinity probes 4–7 before starting [Ca2+]i monitoring, followed by application of the TRPC1/4/5 channel activator (−)-englerin A (EA). Because initial experiments with TRPC5-expressing cells revealed concentration-dependent increases in baseline fluorescence upon pre-incubation with 4 and 5 (reminiscent of AM23728) these compounds were further evaluated in activation mode, in which acute effects of application of 4 and 5 on [Ca2+]i were monitored. The results of these experiments are depicted in Fig. 2 and ESI,† Fig. S2–S5. All four photoaffinity probes 4–7 modulated TRPC1/4/5 activities in a concentration-dependent manner, with half-maximal effects at concentrations in the nanomolar range. Unlike Pico145, and similar to AM237, Pico145-DAAlk 5 activated TRPC5:C5 channels (EC50 19 nM; Fig. 2A and B), and inhibited other TRPC1/4/5 channels (IC50 values 46–196 nM; ESI,† Fig. S2). The activation of TRPC5:C5 by Pico145-DAAlk 5 (100 nM) could be inhibited by Pico145 in a concentration-dependent manner (IC50 1.8 nM; Fig. 2C and D). Pico145-DA 4 also activated TRPC5:C5 (EC50 13 nM; ESI,† Fig. S3A and B) and inhibited other TRPC1/4/5 channels (IC50 values 1.4–40 nM; ESI,† Fig. S3C–H), suggesting that the activating effect on TRPC5:C5 is a consequence of incorporation of the diazirine group into the C-8 substituent rather than the alkyne handle on N-7. In contrast, Pico145-DAAlk2 7 inhibited TRPC5:C5 (IC50 26 nM; Fig. 2E and F) as well as other TRPC1/4/5 channels (IC50 values 2.0–17 nM; ESI,† Fig. S4), and similar effects were seen for Pico145-DA2 6 (IC50 values 1.3–18 nM; ESI,† Fig. S5). A comparison of the EC50 and IC50 values, shown in Table 1, suggests that – in the context of TRPC1/4/5 channels – Pico145-DA 4 and Pico145-DAAlk 5 mimic AM237, while Pico145-DA2 6 and Pico145-DAAlk2 7 mimic Pico145. In addition, the four photoaffinity probes retained high potency against TRPC1/4/5 channels compared to AM237 and Pico145. Note that Pico145 potency at TRPC1/4/5 channels can be reduced by higher EA concentrations, suggesting competition of the compounds. Therefore, the different concentrations of EA used as activator in inhibition assays with TRPC4:C4, TRPC4-C1 and TRPC5-C1 may exaggerate differences in IC50 values between compounds.| Compound | TRPC5:C5 EC50 (nM) | TRPC5:C5 IC50 (nM) | TRPC4:C4 IC50 (nM) | TRPC4-C1 IC50 (nM) | TRPC5-C1 IC50 (nM) |
|---|---|---|---|---|---|
| a EA is a negative modulator of Pico145 potency, suggesting that Pico145 is a competitive inhibitor of EA.23 Therefore, lower EA concentrations used to activate TRPC1/4/5 channels may result in lower IC50 values. b 10 nM EA was used as activator. c 30 nM EA was used as activator. d 100 nM EA was used as activator. | |||||
| AM237 328 | 15 | 7c | 3d | 4c | |
| Pico145-DA 4 | 13 | 40c | 1.4c | 4.9c | |
| Pico145-DAAlk 5 | 19 | 45c | 121d | 196c | |
| Pico145 123 | 1.3b | 0.35b | 0.03b | 0.2b | |
| Pico145-DA2 6 | 18b | 16c | 1.3c | 6.1c | |
| Pico145-DAAlk2 7 | 26b | 17c | 2.0c | 6.2c | |
Docking of compounds 4–7 to the Pico145/lipid binding site of TRPC5:C529 revealed potential binding modes. Similar to the best predictions for the binding mode of Pico145,29 these simulations suggest that the xanthine cores of all four compounds 4–7 make π-stacking interactions with F576, while their 3-hydroxypropyl groups make hydrogen bonding interactions with Q573 and W577 (Fig. 3). The top-scoring poses of Pico145-DA2 6 and Pico145-DAAlk2 7 (Fig. 3C and D) were near-identical to that of Pico145 in the cryo-EM structure,29 with their respective OCF3 and O-propargyl groups oriented towards the hydrophobic tunnel made by L528 and V579. In addition, the diazirine groups at the para-position of the N-7 benzyl substituents of 6 and 7 were easily accommodated in the binding pocket. In contrast, the top-scoring poses for Pico145-DA 4 and Pico145-DAAlk 5 suggest that a slight twist of their C-8 substituents is required to avoid potential clashes between their (trifluoromethyl)diazirine groups and nearby residues V610, V579 and L521 (Fig. 3A, B and ESI,† Fig. S6). These simulations are consistent with the hypothesis that AM237 would also need to rearrange its C-8 aryl substituent to avoid a clash with L521,29 and suggest that changes to the C-8 aryl substitution pattern may be a valid strategy for the rational development of xanthine-based TRPC5:C5 activators. However, further structural insight into xanthine-based TRPC5:C5 activation is required, for example through the determination of open channel structures, which have not been reported to date.
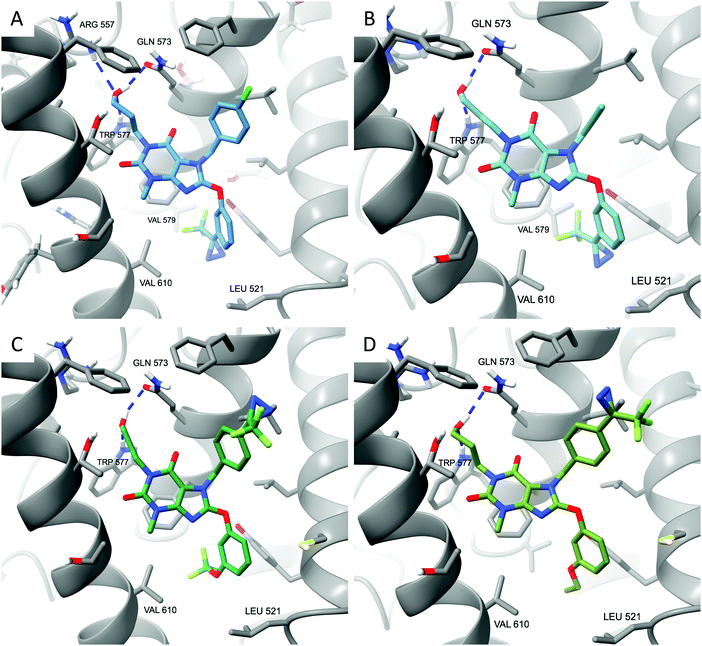 | ||
| Fig. 3 Xanthine-based photoaffinity probes 4–7 have similar predicted binding modes in TRPC5:C5. (A) Top-scoring docked pose of Pico145-DA 4. (B) Top-scoring docked pose of Pico145-DAAlk 5. (C) Top-scoring docked pose of Pico145-DA2 6. (D) Top-scoring docked pose of Pico145-DAAlk2 7. All compounds were docked into a 36 × 36 × 36 Å3 grid around the Pico145 binding site of TRPC5:C5 (PDB: 6YSN). | ||
The high similarity of compounds 4–7 to Pico145 and/or AM237 (in terms of structure, functional effect and potency as TRPC1/4/5 modulators, and predicted binding modes) in combination with their increased predicted aqueous solubility (especially in case of Pico145-DAAlk 5; ESI,† Table S1) are consistent with properties of well-designed xanthine-based photoaffinity probes for cellular ligand engagement studies.
Pico145-DAAlk and Pico145-DAAlk 2 allow photoaffinity labelling of TRPC5 in human cells
To investigate the potential of xanthine-based photoaffinity probes for TRPC ligand engagement studies, we set out to develop a photoaffinity labelling (PAL) protocol, making use of the alkyne handles of the diazirine-alkyne probes Pico145-DAAlk 5 and Pico145-DAAlk2 7 (Fig. 4). In an initial experiment, 300 nM Pico145-DAAlk was used to photolabel and – after CuAAC with TAMRA azide – detect modified proteins by fluorescent gel scanning (ESI,† Fig. S7A). Labelled proteins were only detected in samples that had been subjected to both Pico145-DAAlk 5 and UV irradiation (ESI,† Fig. S7B and C), and included a protein of ∼140 kDa present only in cells that express TRPC5-SYFP2 (ESI,† Fig. S7C and D). This is consistent with direct binding of Pico145-DAAlk 5 to TRPC5-SYFP2. However, the aim of this work was to quantitatively probe interactions between xanthines and TRPC5, and the faint TRPC5-SYFP2 band was difficult to detect against the background of other labelled proteins. Therefore, we modified the photoaffinity labelling protocol to optimise reliable detection of TRPC5–xanthine interactions (see Methods).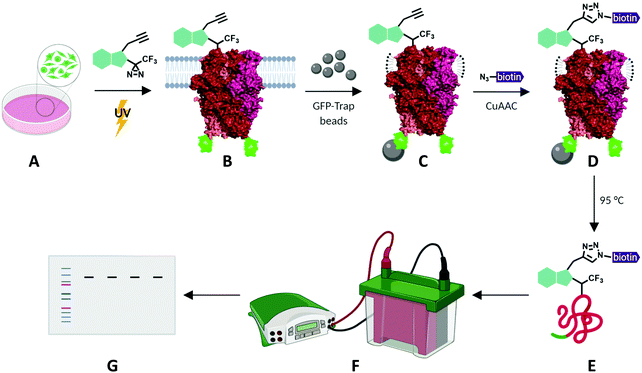 | ||
| Fig. 4 Schematic representation of photoaffinity labelling protocol with Pico145-DAAlk 5 or Pico145-DAAlk2 7. HEK T-REx cells expressing TRPC5-SYFP2 (A) were incubated with photoaffinity probes and irradiated with UV light (B). Following lysis and GFP-Trap immunoprecipitation (C), photolabelled TRPC5-SYFP2 was conjugated to TAMRA biotin azide (D) and eluted from GFP-Trap beads (E). Samples were separated using SDS-PAGE and analysed by western blotting (F and G). TRPC5 representation is based on its cryo-EM structure (PDB: 6AEI).51 | ||
HEK T-REx cells expressing TRPC5-SYFP2 were treated with Pico145-DAAlk 5 (300 nM), and subjected to irradiation with UV light (365 nm; Analytik Jena AG photocrosslinker) for 25 minutes. Cells were then washed and lysed, before immunoprecipitation of TRPC5-SYFP2 using GFP-Trap affinity beads. An on-bead CuAAC reaction with TAMRA biotin azide (ESI,† Fig. S35) was then performed, after which proteins were eluted from the GFP-Trap beads, separated by SDS-PAGE and analysed by western blotting with Streptavidin-HRP to detect biotin-labelled proteins (Fig. 5A-(i)) and – after membrane stripping – anti-GFP to detect TRPC5-SYFP2 (Fig. 5A-(ii)). Input samples were also probed with anti-GFP to confirm successful TRPC5-SYFP2 expression (Fig. 5A-(iii)). This analysis allowed identification (Fig. 5A) and quantification (Fig. 5B) of TRPC5 photolabelling products. The experiments revealed a band at ca. 140 kDa, corresponding to the expected molecular weight of TRPC5-SYFP2 (Fig. 5A, lane 2), which was absent when TRPC5-SYFP2 expression was not induced with tetracycline (Fig. 5A, lane 1). Control experiments demonstrated that Pico145-DAAlk (Fig. 4A, lane 3), UV irradiation (Fig. 5A, lane 7) and the CuAAC reagents (Fig. 5A, lanes 5 and 6) were all essential for detection of photolabelled TRPC5. In addition, in a competition PAL experiment, pre-treatment of cells with Pico145 (10 μM) led to inhibition of TRPC5 labelling (Fig. 5A, lane 4), suggesting that PAL is mediated by a specific molecular interaction between TRPC5 and Pico145-DAAlk 5. Experiments with Pico145-DAAlk2 7 instead of Pico145-DAAlk 5, which were performed using a custom-built LED-based photocrosslinker (see ESI,† Methods for details) that allowed reduction of irradiation time to 1 minute, revealed similar results (ESI,† Fig. S8 and S9). These results suggest that xanthine-based photoaffinity probes 5 and 7 can be used for ligand-directed PAL of TRPC5 protein in human cells, and (through competition assays) for quantitative assessment of target engagement of TRPC5 modulators (see below).
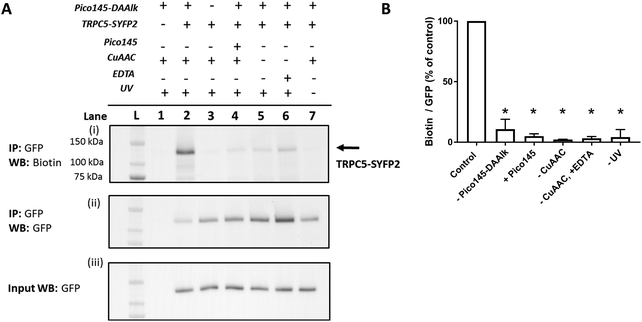 | ||
| Fig. 5 Photoaffinity labelling of TRPC5 with Pico145-DAAlk 5. (A) Representative western blot after experiment depicted in Fig. 3. Samples were blotted with Streptavidin-HRP (i) and anti-GFP (ii; loading control). Western blot with anti-GFP of input (pre-GFP trap) samples was performed to confirm successful GFP-SYFP2 expression in HEK T-REx cells (iii). The data show that photolabelling of TRPC5-SYFP2 in HEK T-REx cells (lane 2) is dependent on expression of TRPC5-SYFP2 (lane 1), treatment with Pico145-DAAlk 5 (300 nM, 30 min, 37 °C; lane 3), UV irradiation (365 nm, Analytik Jena AG photocrosslinker, 25 min; lane 7), and CuAAC (TAMRA biotin azide 100 μM, CuSO4 1 mM, TCEP 1 mM, TBTA 800 μM; 1 h, 37 °C; lanes 5 and 6). In addition, Pico145 (10 μM) inhibits photoaffinity labelling with Pico145-DAAlk 5 (lane 4). (B) Densitometry analysis of data presented in (A). p < 0.05, all compared to control (+TRPC5-SYFP2, +Pico145-DAAlk probe; lane 2; one-way ANOVA with Dunnett's post-test). For ‘–CuAAC’ experiments, CuSO4, TCEP and TBTA were replaced by their corresponding vehicles (water or DMSO) during the CuAAC step of the protocol. Lane L indicates protein molecular weight ladder. Pictures of complete western blots are shown in ESI,† Fig. S10. | ||
Dependence of TRPC5 photolabelling on photoaffinity probe concentration
To investigate the dependence of TRPC5 labelling on photoaffinity probe concentration, the quantitative PAL experiments were performed with either 0.3–300 nM Pico145-DAAlk 5 or 0–1000 nM Pico145-DAAlk2 7. Western blots with Streptavidin-HRP showed concentration-dependent detection of labelled TRPC5 (Fig. 6A, (i)), while reprobing of membranes with anti-GFP, and blotting of input samples, showed consistent pull-down and expression of TRPC5-SYFP2 (Fig. 6A, (ii) and (iii)). Densitometry analysis showed that TRPC5 labelling increased with increasing concentrations of Pico145-DAAlk 5 (Fig. 6A and B) or Pico145-DAAlk2 7 (Fig. 6C and D). Although it would be tempting to fit dose–response curves to these data, it is possible that at higher concentration of photoaffinity probes, non-specific photoaffinity labelling – of other sites than the modulatory binding site(s) – contributes to signal intensity. Because clear signals could be detected at 300 nM of either Pico145-DAAlk 5 or Pico145-DAAlk2 7, this concentration was chosen for further experiments.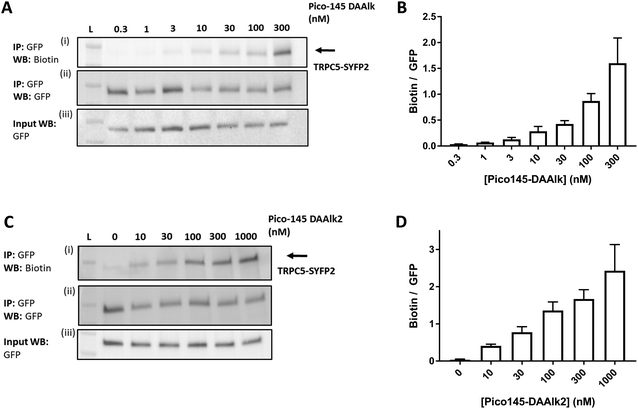 | ||
| Fig. 6 Dependence of TRPC5 photoaffinity labelling on photoaffinity probe concentration. (A and C) Representative western blots showing concentration-dependent TRPC5 photolabelling with Pico145-DAAlk 5 (A) and Pico145-DAAlk2 7 (C). HEK T-REx cells overexpressing TRPC5-SYFP2 were treated with 0.3–300 nM Pico145-DAAlk 5 (A) or 0–1000 nM Pico145-DAAlk2 7 (C) for 30 min at 37 °C and irradiated (365 nm). Following lysis and GFP pull-down, photolabelled TRPC5-SYFP2 was conjugated to TAMRA biotin azide (100 μM) using CuAAC (CuSO4 1 mM, TCEP 1 mM, TBTA 800 μM; 1 h, 37 °C). Samples were separated by SDS-PAGE and blotted with Streptavidin-HRP (i) and anti-GFP (ii; loading control). Western blot with anti-GFP of input (pre-GFP trap) samples was performed to confirm successful GFP-SYFP2 expression in HEK T-REx cells (iii). (B and D) Densitometry analysis of data presented in (A) and (C) respectively, showing mean data ± SEM (n = 3 independent experiments each). Lane L indicates protein molecular weight ladder. Pictures of complete western blots are shown in ESI,† Fig. S11. | ||
Assessment of Pico145 binding to TRPC5 in human cells by photoaffinity labelling
The results described above suggest that our quantitative PAL protocol with Pico145-DAAlk 5 and Pico145-DAAlk2 7, when performed in competition mode, could potentially provide an indirect, quantitative method to assess binding of small molecules to TRPC5 in cells. To test this, HEK T-REx cells expressing TRPC5-SYFP2 were pre-treated with either DMSO or 0–3000 nM Pico145 1, before performing PAL experiments with Pico145-DAAlk 5 (300 nM) or Pico145-DAAlk2 7 (300 nM). TRPC5-SYFP2 pull-down and expression was consistent across samples (Fig. 7A and C), and Pico145 concentration-dependently inhibited TRPC5 labelling by Pico145-DAAlk 5 (IC50 2.5 nM; Fig. 7A and B) and Pico145-DAAlk2 7 (IC50 17 nM; Fig. 7C and D). Interestingly, the IC50 value of Pico145 inhibition of PAL with Pico145-DAAlk 5 (2.5 nM; Fig. 7A and B) is almost identical to the IC50 value of Pico145 inhibition of Pico145-DAAlk 5 evoked calcium entry (1.8 nM; Fig. 2C and D), even when taking into account the 3-fold difference in the concentration of Pico145-DAAlk 5 used in the respective assays (100 nM in calcium recording; 300 nM in PAL). These data suggest that the functional activity of xanthine-based TRPC5 modulators such as Pico145 on TRPC5:C5 channels is directly related to a specific binding event between xanthines and TRPC5 protein.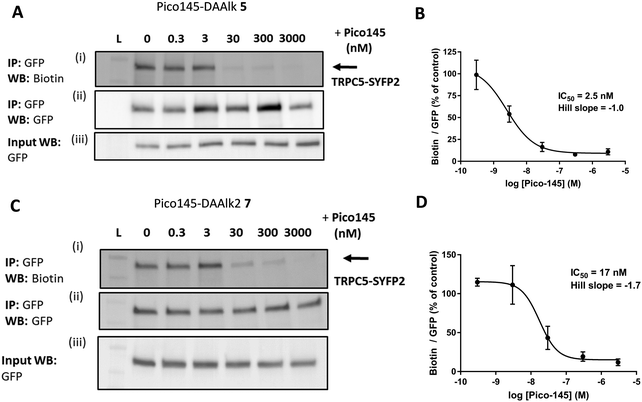 | ||
| Fig. 7 Pico145 concentration-dependently inhibits TRPC5 photoaffinity labelling. (A and C) Representative western blots showing concentration-dependent inhibition by Pico145 of TRPC5 photolabelling with Pico145-DAAlk 5 (A) or Pico145-DAAlk2 7 (C). HEK T-REx cells overexpressing TRPC5-SYFP2 were pre-treated with 0–3000 nM Pico145 (30 min, 37 °C) before being treated with 300 nM Pico145-DAAlk 5 (A) or 300 nM Pico145-DAAlk2 7 (C) for 30 min at 37 °C and irradiated (365 nm). Following lysis and GFP pull-down, TRPC5-SYFP2 was conjugated to TAMRA biotin azide (100 μM) using CuAAC (CuSO4 1 mM, TCEP 1 mM, TBTA 800 μM; 1 h, 37 °C). Samples were separated by SDS-PAGE and blotted with Streptavidin-HRP (i) and anti-GFP (ii; loading control). Western blot with anti-GFP of input (pre-GFP trap) samples was performed to confirm successful GFP-SYFP2 expression in HEK T-REx cells (iii). (B and D) Concentration–response data for experiments (A) and (C) respectively, showing mean data ± SEM (n = 3 independent experiments each). Biotin/GFP was compared to control (0 nM Pico145). Lane L indicates protein molecular weight ladder. Pictures of complete western blots are shown in ESI,† Fig. S12. | ||
Discussion
Through minimal structural variation of the most potent TRPC1/4/5 inhibitor, Pico145, we have developed a set of potent xanthine-based photoaffinity probes. Pico145-DA 4 and Pico145-DAAlk 5 functionally mimic AM237; they activate TRPC5:C5 channels but inhibit TRPC4:C4, TRPC5-C1 and TRPC4-C1 channels. In contrast, Pico145-DA2 6 and Pico145-DAAlk2 7 functionally mimic Pico145; they inhibit all TRPC1/4/5 channels tested. We used Pico145-DAAlk 5 and Pico145-DAAlk2 7 to develop a new PAL/CuAAC protocol to measure cellular interactions between xanthines and TRPC5 protein. PAL of TRPC5 was successful within the same concentration range as the effects on TRPC5-mediated calcium influx, providing further evidence that xanthines modulate TRPC5 channels through direct interaction with TRPC5 protein, consistent with their behaviour in excised outside-out patch recordings23,28 and with our cryo-EM and site-directed mutagenesis studies.29 To the best of our knowledge, only one previous example of PAL on a TRPC channel had been reported: Kiyonaka et al. used a pyrazole derivative (Pyr-PP) incorporating a diazirine photocrosslinker and a ketone handle (for oxime ligation with a biotin hydroxylamine probe) to suggest that the TRPC3 channel inhibitor Pyr3 directly interacts with TRPC3 protein.39Competition PAL experiments with Pico145 allowed the indirect, quantitative assessment of the relative affinities of Pico145 and photoaffinity probes. The IC50 values determined for Pico145 in calcium recordings (Fig. 2C and D) and in competition PAL experiments (Fig. 7A and B) with Pico145-DAAlk 5 are nearly identical. These results – in combination with the close structural similarities between Pico145 and Pico145-DAAlk 5 – are consistent with the hypothesis that xanthine-based TRPC5:C5 activators (e.g., AM237, Pico145-DA 4, and Pico145-DAAlk 5) and xanthine-based TRPC5:C5 inhibitors (e.g., Pico145) bind to identical site(s) of TRPC1/4/5 channels, leading to stabilisation of open or closed TRPC1/4/5 channel conformations. In addition, our docking studies suggest that, similar to AM237,29 Pico145-DA 4 and Pico145-DAAlk 5 contain structural features associated with TRPC5:C5 activation rather than inhibition (Fig. 3 and ESI,† Fig. S6). Despite extensive experimentation resulting in successful mass spectrometry-based peptide mapping of TRPC5 constructs (up to 99% sequence coverage; not shown), we have not yet been able to determine which peptides/residues are labelled by xanthine-based photoaffinity probes. We hypothesise that this is the result of low efficiency of photolabelling of TRPC5, for which diazirines are notorious.37,52 Our cryo-EM studies revealed that up to four molecules of Pico145 can bind to four lipid binding sites of a TRPC5 tetramer.29 Although it is not yet known how many binding sites need to be occupied for channel modulation, xanthine-based TRPC1/4/5 modulators often display Hill slopes >1 in intracellular calcium measurements (Fig. 2 and ESI,† Fig. S2–S5, and ref. 28), suggesting that a degree of cooperativity may exist.
Our xanthine-based photoaffinity probes were designed to have the closest possible structural similarity to their parent compound, Pico145. The observation that the photoaffinity probes retain potency against all tested homo- and concatemeric TRPC1/4/5 channels implies that they can be used to assess cellular engagement of TRPC1, TRPC4 and TRPC5 in various tetramers. Considering its potency against TRPC4-C1 and TRPC5-C1 (Table 1), Pico145-DAAlk2 7 may be the most suitable photoaffinity probe for heteromeric or concatemeric TRPC1/4/5 channels. In addition, the competition PAL protocol may allow the assessment of target engagement of other TRPC1/4/5 modulators as well, and thereby support studies into their modes-of-action.
Pico145 and HC-070 are the most potent TRPC1/4/5 inhibitors reported to date, and in vitro screening of these compounds against hundreds of other proteins (ion channels, receptors, transporters and enzymes) suggested exquisite selectivity.23,25 Limited aqueous solubility and high plasma protein binding (PPB) have been highlighted as potential draw-backs of xanthine-based TRPC1/4/5 modulators, motivating the development of pyridazone-based TRPC4/5/inhibitors such as GFB-8438.53 Although GFB-8438 has lower potency against TRPC4/5 channels (TRPC4:C4 IC50 = 0.29 μM; TRPC5:C5 IC50 = 0.18–0.28 μM), it has substantially higher solubility and lower PPB than Pico145,53 consistent with our calculations (ESI,† Table S1). However, the xanthines Pico145 and HC-070 have been used successfully for the modulation of TRPC1/4/5 channels in tissues and animals by multiple academic and industrial groups,21,25–27 suggesting that the lower solubility and higher PPB of these xanthines may be off-set by their substantially higher potency. In addition, all four of our xanthine-based photoaffinity probes (and especially Pico145-DAAlk 5 lacking the non-essential 4-chlorobenzyl substituent on N-7) are predicted to have higher aqueous solubility than Pico145 (ESI,† Table S1), suggesting that further modification to the xanthines could address potential solubility issues if needed. Photoaffinity labelling with excess Pico145-DAAlk 5 in HEK T-REx cells suggested modification of multiple proteins (ESI,† Fig. S7), which is consistent with the hydrophobic nature and of Pico145-DAAlk 5 (favouring non-specific interactions with proteins) and the indiscriminate nature of diazirine-based photoaffinity labelling. In future studies, the use of our photoaffinity probes in comparative mass spectrometry-based chemical proteomics workflows (for example using Pico145 as a competitor) may enable the unbiased profiling of true targets and off-targets of xanthine-based TRPC1/4/5 modulators in cells and tissues.
Our PAL protocol provides the first method for the quantitative assessment of ligand engagement by TRPC5 channels, a crucial step in the drug discovery process. Because xanthines bind to a conserved, key modulatory TRPC1/4/5 channel binding site, competition PAL with xanthine-based photoaffinity probes may underpin the development of new TRPC1/4/5 modulators as chemical probes and drug candidates for a wide range of diseases – including those of the CNS, kidney, heart, and cardiovascular system.
Methods
Abbreviations and definitions
TRPC, Transient Receptor Potential Canonical; EA, (−)-englerin A; S1P, sphingosine-1-phosphate; HEK T-REx cells; human embryonic kidney 293 cells stably expressing the tetracycline repressor protein, allowing tetracycline-inducible recombinant protein over-expression. TRPC1, TRPC4 and TRPC5 denote the different proteins or channels incorporating them; TRPC1/4/5 denotes channels composed of TRPC1, TRPC4 and/or TRPC5 (homo- or heteromeric; any ratio); TRPC4:C4 and TRPC5:C5 denote specific homomeric channels; TRPC1:C4 and TRPC1:C5 denote heteromeric channels formed by TRPC1 and either TRPC4 or TRPC5 (any ratio). TRPC4-C1 and TRPC5-C1 denote (channels composed of) recombinant, concatemeric proteins (fusions of TRPC1 at the C-terminus of either TRPC4 or TRPC5 through a short linker);18,23,28,54 [Ca2+]i, intracellular concentration of Ca2+.Synthesis and characterization
Full synthetic procedures and characterisation data are available in the ESI,† Methods.Cell culture and expression systems
HEK T-REx cells expressing inducible TRPC5-SYFP2, TRPC4-SYFP2, TRPC5-C1 and TRPC4-C1 have been described previously.18,23,28,54 Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM), supplemented with foetal bovine serum (FBS; 10%), penicillin–streptomycin (100 units per ml; 100 μg ml−1) and blasticidin (10 μg ml−1) to maintain expression of the tetracycline repressor, and zeocin (400 μg ml−1) to maintain incorporation of the relevant TRPC plasmid. Cells were kept in a humidified incubator at 37 °C at 5% CO2. Expression of TRPC protein was induced by the addition of 1 μg ml−1 tetracycline (Merck, Gillingham, Dorset, UK) to the culture medium 24 h before experiments.Intracellular Ca2+ measurements
[Ca2+]i recordings were carried out using the ratiometric Ca2+ dye Fura-2. 24 h before experiments, HEK T-REx cells stably transfected with relevant TRPC plasmids were plated onto black, clear-bottom, poly-D-lysine coated 96-well plates at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. Cells were induced with 1 μg ml−1 tetracycline at this point. To load cells with the Fura-2 dye, media was removed and cells were incubated with standard bath solution (SBS) containing 2 μM Fura-2 acetoxymethyl ester (Fura-2 AM; ThermoFisher Scientific, Waltham, MA, USA) and 0.01% pluronic acid (Merck) for 1 h at 37 °C. SBS contained (in mM): NaCl 135, KCl 5, glucose 8, HEPES 10, CaCl2 1.5 and MgCl2 1.2. After this incubation, cells were washed with fresh SBS and incubated at room temperature for a further 30 min. Immediately prior to the experiment, SBS was removed, and recording buffer (SBS + 0.01% pluronic acid + 0.01% DMSO) was added to cells. When assaying cells in inhibition mode, cells were washed briefly, twice, with SBS after Fura-2 AM incubation, and then incubated in recording buffer with inhibitors or DMSO for 30 min at rt. [Ca2+]i was measured in the FlexStation3 (Molecular Devices, Wokingham, UK), by alternating excitation at 340 nm and 380 nm, with an emission at 510 nm. Measurements were performed at rt for 5 min at 5 s intervals. Compounds used or tested as TRPC channel activators were dissolved at 2× final concentration in compound buffer (SBS + 0.01% pluronic acid) and added to cells after recording for 60 s. When testing compounds as inhibitors, inhibitors or DMSO were added to the compound buffer at the same concentration as in the recording buffer.
000 cells per well. Cells were induced with 1 μg ml−1 tetracycline at this point. To load cells with the Fura-2 dye, media was removed and cells were incubated with standard bath solution (SBS) containing 2 μM Fura-2 acetoxymethyl ester (Fura-2 AM; ThermoFisher Scientific, Waltham, MA, USA) and 0.01% pluronic acid (Merck) for 1 h at 37 °C. SBS contained (in mM): NaCl 135, KCl 5, glucose 8, HEPES 10, CaCl2 1.5 and MgCl2 1.2. After this incubation, cells were washed with fresh SBS and incubated at room temperature for a further 30 min. Immediately prior to the experiment, SBS was removed, and recording buffer (SBS + 0.01% pluronic acid + 0.01% DMSO) was added to cells. When assaying cells in inhibition mode, cells were washed briefly, twice, with SBS after Fura-2 AM incubation, and then incubated in recording buffer with inhibitors or DMSO for 30 min at rt. [Ca2+]i was measured in the FlexStation3 (Molecular Devices, Wokingham, UK), by alternating excitation at 340 nm and 380 nm, with an emission at 510 nm. Measurements were performed at rt for 5 min at 5 s intervals. Compounds used or tested as TRPC channel activators were dissolved at 2× final concentration in compound buffer (SBS + 0.01% pluronic acid) and added to cells after recording for 60 s. When testing compounds as inhibitors, inhibitors or DMSO were added to the compound buffer at the same concentration as in the recording buffer.
Docking studies
The region of the TRPC5:C5 tetrameric structure (PDB: 6YSN) assigned for docking studies was chosen based upon residues known to be crucial for Pico145 binding (Y524, L572, Q573, F576, W577, V579, F599, Y603 and V614). A 36 Å clip of the TRPC5 structure around these residues was termed as the receptor for docking studies using Glide (Schrödinger Release 2020-1, Glide, Schrödinger, LLC, New York, NY, 2020).55 The TRPC5 pdb file was prepared using the Protein Preparation Wizard in the Maestro Graphical User Interface (GUI). This aimed to remove any steric clashes of amino acid side chains and optimise the position of hydrogen atoms to facilitate docking studies. The receptor grid was generated using Schrödinger software, allowing docking of ligands in a 36 × 36 × 36 Å3 ‘grid’. Photoaffinity probes 4–7 were imported into the Maestro GUI and prepared using the LigPrep module to produce an energy minimised 3D structure. Docking of photoaffinity probes was carried out using the Glide module of Schrödinger software using the XP mode with flexible ligand sampling and biased sampling of torsions for all predefined functional groups. Epik state penalties were added to the docking score. A maximum of 10 poses were requested in the output file for each ligand and post-docking minimisation was carried out. Docking of probes into the defined Pico145 binding site of TRPC5:C5 produced three poses for each compound with similar predicted binding energies. These poses were almost identical with the lowest energy pose making key hydrogen bonding interactions between the flexible hydroxypropyl groups and the side chain of Q573. Critical π-stacking interactions were seen between the xanthine cores and F576, and between the C-8 aryl substituents and Y524. To modify the prepared Pico145 structure to give Pico145-DA (in ESI,† Fig. S6), the build tool in the Maestro GUI was used to swap the OCF3 for a diazirine-CF3 moiety and this section of the ligand was minimised, leaving stationary the remaining core of the ligand and amino acids in proximity to the group. After minimisation, the proximity of the diazirine moiety to nearby residues was measured.Calculation of predicted solubility
The predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S and log
S and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values for energy-minimised versions of Pico145, AM237, photoaffinity probes 4–7 and GFB-8438 (ESI,† Table S1) were calculated using QikProp (Schrödinger Release 2020-1, Glide, Schrödinger, LLC, New York, NY, 2020).
P values for energy-minimised versions of Pico145, AM237, photoaffinity probes 4–7 and GFB-8438 (ESI,† Table S1) were calculated using QikProp (Schrödinger Release 2020-1, Glide, Schrödinger, LLC, New York, NY, 2020).
Photoaffinity labelling protocol
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C and transferred to fresh tubes. At this stage, input samples were taken by removal of 20 μl of lysate. To these input samples, 4× loading buffer (BioRad, Watford, UK) + 10% beta-mercaptoethanol was added, and samples were boiled at 95 °C for 10 min, then stored at −20 °C. Protein concentrations for the remaining lysates were quantified using bicinchoninic acid (ThermoFisher Scientific), followed by immunoprecipitation of TRPC5-SYFP2. TRPC5-SYFP2 was immunoprecipitated using GFP-Trap agarose (Chromotek, Planegg, Germany) according to manufacturer's instructions. Samples were adjusted to 2.5 mg protein and equal volume in dilution buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), before addition of 20 μl washed bead slurry per sample. Immunoprecipitations were incubated with end-over-end mixing at 4 °C for 3 h, followed by 3 washes in wash buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), to remove non-specific binders. Following removal of wash buffer, immunoprecipitated proteins were resuspended in 74 μl PBS + 1% SDS. Proteins were then tagged with biotin by on-bead copper-catalysed azide–alkyne cycloaddition (CuAAC). Reagents (TAMRA biotin azide 100 μM, CuSO4 1 mM, TCEP 1 mM, TBTA 800 μM) were added to immunoprecipitated proteins and reactions incubated at 37 °C for 1 h with shaking at 250 rpm. Reactions were quenched by addition of 10 mM EDTA, and precipitations washed 3× in wash buffer (3000 g, 5 min). After removal of the last wash, GFP-Trap beads were resuspended in 45 μl 2% SDS in PBS and 25 μl 4× loading buffer containing 10% (v/v) betamercaptoethanol, and boiled for 95 °C for 10 min to dissociate protein from GFP-Trap beads, and stored at −20 °C prior to SDS-PAGE.
000g for 10 min at 4 °C and transferred to fresh tubes. At this stage, input samples were taken by removal of 20 μl of lysate. To these input samples, 4× loading buffer (BioRad, Watford, UK) + 10% beta-mercaptoethanol was added, and samples were boiled at 95 °C for 10 min, then stored at −20 °C. Protein concentrations for the remaining lysates were quantified using bicinchoninic acid (ThermoFisher Scientific), followed by immunoprecipitation of TRPC5-SYFP2. TRPC5-SYFP2 was immunoprecipitated using GFP-Trap agarose (Chromotek, Planegg, Germany) according to manufacturer's instructions. Samples were adjusted to 2.5 mg protein and equal volume in dilution buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), before addition of 20 μl washed bead slurry per sample. Immunoprecipitations were incubated with end-over-end mixing at 4 °C for 3 h, followed by 3 washes in wash buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), to remove non-specific binders. Following removal of wash buffer, immunoprecipitated proteins were resuspended in 74 μl PBS + 1% SDS. Proteins were then tagged with biotin by on-bead copper-catalysed azide–alkyne cycloaddition (CuAAC). Reagents (TAMRA biotin azide 100 μM, CuSO4 1 mM, TCEP 1 mM, TBTA 800 μM) were added to immunoprecipitated proteins and reactions incubated at 37 °C for 1 h with shaking at 250 rpm. Reactions were quenched by addition of 10 mM EDTA, and precipitations washed 3× in wash buffer (3000 g, 5 min). After removal of the last wash, GFP-Trap beads were resuspended in 45 μl 2% SDS in PBS and 25 μl 4× loading buffer containing 10% (v/v) betamercaptoethanol, and boiled for 95 °C for 10 min to dissociate protein from GFP-Trap beads, and stored at −20 °C prior to SDS-PAGE.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000; ThermoFisher Scientific) or anti-GFP (input, 1
5000; ThermoFisher Scientific) or anti-GFP (input, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8000; Abcam, Cambridge, UK in blocking buffer, overnight at 4 °C). For input samples membranes were washed in PBS-T (6 × 5 min, rt) before incubation in secondary antibody (anti-mouse HRP, 1
8000; Abcam, Cambridge, UK in blocking buffer, overnight at 4 °C). For input samples membranes were washed in PBS-T (6 × 5 min, rt) before incubation in secondary antibody (anti-mouse HRP, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000; ThermoFisher Scientific) for 1 h at rt, and further washing in PBS-T. For PAL samples, membranes were washed in PBS-T (6 × 5 min, rt). Following washing, blots were imaged using ECL (ThermoFisher Scientific) and G-box (Syngene, Bangalore, India). To reprobe PAL membranes, membranes were stripped in Restore Stripping buffer (20 min, rt; ThermoFisher Scientific), washed in PBS (3 × 5 min, rt), and blocked in 5% milk in PBS-T. Membranes were then incubated with anti-GFP (1
5000; ThermoFisher Scientific) for 1 h at rt, and further washing in PBS-T. For PAL samples, membranes were washed in PBS-T (6 × 5 min, rt). Following washing, blots were imaged using ECL (ThermoFisher Scientific) and G-box (Syngene, Bangalore, India). To reprobe PAL membranes, membranes were stripped in Restore Stripping buffer (20 min, rt; ThermoFisher Scientific), washed in PBS (3 × 5 min, rt), and blocked in 5% milk in PBS-T. Membranes were then incubated with anti-GFP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8000, 2 h, rt), washed with PBS-T, incubated with anti-mouse HRP (1
8000, 2 h, rt), washed with PBS-T, incubated with anti-mouse HRP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, 1 h, rt; ThermoFisher Scientific), washed with PBS-T and imaged using ECL and G-box. Blots were quantified using ImageJ.
5000, 1 h, rt; ThermoFisher Scientific), washed with PBS-T and imaged using ECL and G-box. Blots were quantified using ImageJ.
Author contributions
AM, IBP and RSB designed and synthesised photoaffinity probes. CCB, AM and IBP carried out calcium measurements and photoaffinity labelling experiments. IBP and KJS performed docking studies. KJS performed calculations of compound solubilities. CCB, AM, IBP, KJS, ECB and RSB analysed data. NK developed the LED-based photocrosslinker. MPB, DJB, SPM, MHW and SLW advised on experimental design and data interpretation, and generated research funds. RSB conceived and led the project, and generated research funds. CCB, IBP, ECB, NK and RSB made figures. CCB, IBP, KJS and RSB wrote the manuscript. All authors commented on the manuscript.Conflicts of interest
DJB is an inventor on the following patent applications: 1) PCT/GB2018/050369. TRPC ion channel inhibitors for use in therapy. Filing date: 9th February 2018. Inventors: David J Beech, Richard J Foster, Sin Ying Cheung and Baptiste M Rode; 2) 62/529,063. Englerin derivatives for treatment of cancer. Filing date: 6th July 2017. Inventors: John A Beutler, Antonio Echavarren, William Chain, David Beech, Zhenhua Wu, Jean-Simon Suppo, Fernando Bravo and Hussein Rubaiy.Acknowledgements
This work was supported by the BBSRC (BB/P020208/1), the BHF (PG/19/2/34084), a BBSRC/AstraZeneca iCASE PhD studentship (BB/L015676/1) to AM, and a Wellcome Trust PhD studentship (102174/B/13/Z) to IBP and a Wellcome Trust Award to DJB (10044/Z/15/Z). We thank Lucia Pedicini, Katarina Miteva and Lynn McKeown for discussions, Martin Walko for donation of a sample of diazirine building block 22, and Devon Legge for donation of a sample of TAMRA biotin azide used in pilot photoaffinity labelling experiments. Docking studies in this work were undertaken on ARC3, part of the High Performance Computing facilities at the University of Leeds, UK. Fig. 4 and the TOC figure were produced using biorender.com.References
- J. Abramowitz and L. Birnbaumer, Physiology and pathophysiology of canonical transient receptor potential channels, FASEB J., 2009, 23, 297–328 Search PubMed.
- R. S. Bon and D. J. Beech, In pursuit of small molecule chemistry for calcium-permeable non-selective TRPC channels – mirage or pot of gold?, Br. J. Pharmacol., 2013, 170, 459–474 Search PubMed.
- K. Venkatachalam and C. Montell, TRP Channels, Annu. Rev. Biochem., 2007, 76, 387–417 Search PubMed.
- C. Montell, The TRP superfamily of cation channels, Sci. STKE, 2005, re3 Search PubMed.
- T. Voets, K. Talavera, G. Owsianik and B. Nilius, Sensing with TRP channels, Nat. Chem. Biol., 2005, 1, 85–92 Search PubMed.
- C. Montell, et al., A Unified nomenclature for the superfamily of TRP cation channels, Mol. Cell, 2002, 9, 229–231 Search PubMed.
- X. Zhu, et al., trp, a novel mammalian gene family essential for agonist-activated capacitative Ca2+ entry, Cell, 1996, 85, 661–671 Search PubMed.
- P. D. Wes, et al., TRPC1, a human homolog of a Drosophila store-operated channel, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 9652–9656 Search PubMed.
- D. E. Clapham, L. W. Runnels and C. Strübing, The trp ion channel family, Nat. Rev. Neurosci., 2001, 2, 387–396 Search PubMed.
- B. Vannier, et al., Mouse trp2, the homologue of the human trpc2 pseudogene, encodes mTrp2, a store depletion-activated capacitative Ca2+ entry channel, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 2060–2064 Search PubMed.
- A. Minard, et al., Remarkable progress with small-molecule modulation of TRPC1/4/5 channels: implications for understanding the channels in health and disease, Cells, 2018, 7, 52 Search PubMed.
- H. Wang, et al., TRPC channels: structure, function, regulation and recent advances in small molecular probes, Pharmacol. Ther., 2020, 107497 Search PubMed.
- S. Suresh Babu, et al., Mechanism of Stretch-Induced Activation of the Mechanotransducer Zyxin in Vascular Cells, Sci. Signaling, 2012, 5, ra91 Search PubMed.
- P. Sukumar, et al., Constitutively active TRPC channels of adipocytes confer a mechanism for sensing dietary fatty acids and regulating adiponectin, Circ. Res., 2012, 111, 191–200 Search PubMed.
- M. Gautier, et al., New insights into pharmacological tools to TR(i)P cancer up, Br. J. Pharmacol., 2014, 171, 2582–2592 Search PubMed.
- S. Sharma and C. R. Hopkins, A review of transient receptor potential channel (TRPC) modulators and diseases, J. Med. Chem., 2019, 62, 7589–7602 Search PubMed.
- Y. Akbulut, et al., (−)-Englerin A is a potent and selective activator of TRPC4 and TRPC5 calcium channels, Angew. Chem., Int. Ed., 2015, 54, 3787–3791 Search PubMed.
- M. J. Ludlow, et al., (−)-Englerin A-evoked cytotoxicity is mediated by Na+ influx and counteracted by Na+/K+ – ATPase, J. Biol. Chem., 2017, 292, 723–731 Search PubMed.
- C. Carson, et al., Englerin A agonizes the TRPC4/C5 cation channels to inhibit tumor cell line proliferation, PLoS One, 2015, 1, 1–21 Search PubMed.
- X. Wang, et al., TRPC5 does not cause or aggravate glomerular disease, J. Am. Soc. Nephrol., 2018, 29, 409–415 Search PubMed.
- S. Y. Cheung, et al., TRPC4/TRPC5 channels mediate adverse reaction to the cancer cell cytotoxic agent (−)-Englerin A, Oncotarget, 2018, 9, 29634–29643 Search PubMed.
- B. L. Chenard and R. J. Gallaschun, Substituted xanthines and methods of use thereof, International patent WO/2014/143799, 2014.
- H. N. Rubaiy, et al., Picomolar, selective, and subtype-specific small-molecule inhibition of TRPC1/4/5 channels, J. Biol. Chem., 2017, 292, 8158–8173 Search PubMed.
- H. N. Rubaiy, M. J. Ludlow, R. S. Bon and D. J. Beech, Pico145 – powerful new tool for TRPC1/4/5 channels, Channels, 2017, 11, 362–364 Search PubMed.
- S. Just, et al., Treatment with HC-070, a potent inhibitor of TRPC4 and TRPC5, leads to anxiolytic and antidepressant effects in mice, PLoS One, 2018, 1, 1–32 Search PubMed.
- S. Lepannetier, et al., Activation of TRPC1 channel by metabotropic glutamate receptor mGluR5 modulates synaptic plasticity and spatial working memory, Front. Cell. Neurosci., 2018, 12, 318 Search PubMed.
- Y. Yu, et al., Development of a carbon-11 PET radiotracer for imaging TRPC5 in the brain, Org. Biomol. Chem., 2019, 17, 5586–5594 Search PubMed.
- A. Minard, et al., Potent, selective, and subunit-dependent activation of TRPC5 channels by a xanthine derivative, Br. J. Pharmacol., 2019, 176, 3924–3938 Search PubMed.
- D. J. Wright, et al., Cryo-EM structures of human TRPC5 reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site, bioRxiv, 2020 DOI:10.1101/2020.04.17.047456.
- K. Song, et al., Structural basis for human TRPC5 channel inhibition by two distinct inhibitors, bioRxiv, 2020 DOI:10.1101/2020.04.21.052910.
- G. M. Simon, M. J. Niphakis and B. F. Cravatt, Determining target engagement in living systems, Nat. Chem. Biol., 2013, 9, 200–205 Search PubMed.
- M. Schürmann, P. Janning, S. Ziegler and H. Waldmann, Small-molecule target engagement in cells, Cell Chem. Biol., 2016, 23, 435–441 Search PubMed.
- M. Schenone, V. Dančík, B. K. Wagner and P. A. Clemons, Target identification and mechanism of action in chemical biology and drug discovery, Nat. Chem. Biol., 2013, 9, 232–240 Search PubMed.
- M. E. Bunnage, A. M. Gilbert, L. H. Jones and E. C. Hett, Know your target, know your molecule, Nat. Chem. Biol., 2015, 11, 368–372 Search PubMed.
- V. Chowdhry and F. H. Westheimer, Photoaffinity labeling of biological systems, Annu. Rev. Biochem., 1979, 48, 293–325 Search PubMed.
- E. Smith and I. Collins, Photoaffinity labeling in target- and binding-site identification, Future Med. Chem., 2015, 7, 159–183 Search PubMed.
- G. W. Preston and A. J. Wilson, Photo-induced covalent cross-linking for the analysis of biomolecular interactions, Chem. Soc. Rev., 2013, 42, 3289–3301 Search PubMed.
- K. J. Gregory, et al., Clickable Photoaffinity ligands for metabotropic glutamate receptor 5 based on select acetylenic negative allosteric modulators, ACS Chem. Biol., 2016, 11, 1870–1879 Search PubMed.
- S. Kiyonaka, et al., Selective and direct inhibition of TRPC3 channels underlies biological activities of a pyrazole compound, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 5400–5405 Search PubMed.
- G. M. S. Yip, et al., A propofol binding site on mammalian GABAA receptors identified by photolabeling, Nat. Chem. Biol., 2013, 9, 715–720 Search PubMed.
- K. A. Woll, et al., Photoaffinity ligand for the inhalational anesthetic sevoflurane allows mechanistic insight into potassium channel modulation, ACS Chem. Biol., 2017, 12, 1353–1362 Search PubMed.
- M. Soethoudt, et al., Selective photoaffinity probe that enables assessment of cannabinoid CB2 receptor expression and ligand engagement in human cells, J. Am. Chem. Soc., 2018, 140, 6067–6075 Search PubMed.
- J. Park, M. Koh, J. Y. Koo, S. Lee and S. B. Park, Investigation of specific binding proteins to photoaffinity linkers for efficient deconvolution of target protein, ACS Chem. Biol., 2016, 11, 44–52 Search PubMed.
- T. Kambe, B. E. Correia, M. J. Niphakis and B. F. Cravatt, Mapping the protein interaction landscape for fully functionalized small-molecule probes in human cells, J. Am. Chem. Soc., 2014, 136, 10777–10782 Search PubMed.
- R. A. G. Smith and J. R. Knowles, Aryldiazirines. Potential reagents for photolabeling of biological receptor sites, J. Am. Chem. Soc., 1973, 95, 5072–5073 Search PubMed.
- L. Dubinsky, B. P. Krom and M. M. Meijler, Diazirine based photoaffinity labeling, Bioorg. Med. Chem., 2012, 20, 554–570 Search PubMed.
- J. R. Hill and A. A. B. Robertson, Fishing for drug targets: a focus on diazirine photoaffinity probe synthesis, J. Med. Chem., 2018, 61, 6945–6963 Search PubMed.
- J. Das, Aliphatic diazirines as photoaffinity probes for proteins: recent developments, Chem. Rev., 2011, 111, 4405–4417 Search PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 Search PubMed.
- J. Brunner, H. Senn and F. M. Richards, 3-Trifluoromethyl-3-phenyldiazirine. A new carbene generating group for photolabeling reagents, J. Biol. Chem., 1980, 255, 3313–3318 Search PubMed.
- J. Duan, et al., Cryo-EM structure of TRPC5 at 2.8-Å resolution reveals unique and conserved structural elements essential for channel function, Sci. Adv., 2019, 5, eaaw7935 Search PubMed.
- G. W. Preston, S. E. Radford, A. E. Ashcroft and A. J. Wilson, Covalent cross-linking within supramolecular peptide structures, Anal. Chem., 2012, 84, 6790–6797 Search PubMed.
- M. Yu, et al., Discovery of a potent and selective TRPC5 inhibitor, efficacious in a focal segmental glomerulosclerosis model, ACS Med. Chem. Lett., 2019, 10, 1579–1585 Search PubMed.
- H. N. Rubaiy, et al., Tonantzitlolone is a nanomolar potency activator of transient receptor potential canonical 1/4/5 channels, Br. J. Pharmacol., 2018, 175, 3361–3368 Search PubMed.
- R. A. Friesner, et al., Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes, J. Med. Chem., 2006, 49, 6177–6196 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis and characterisation of compounds, design of LED photocrosslinker setup, and additional data table and figures. See DOI: 10.1039/d0cb00126k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |