Pushing the limits of electrochemistry toward challenging applications in clinical diagnosis, prognosis, and therapeutic action
P.
Yáñez-Sedeño
,
S.
Campuzano
and
J. M.
Pingarrón
 *
*
Departamento de Química Analítica, Facultad de CC. Químicas, Universidad Complutense de Madrid, E-28040 Madrid, Spain. E-mail: pingarro@quim.ucm.es
First published on 14th January 2019
Abstract
Constant progress in the identification of biomarkers at different molecular levels in samples of different natures, and the need to conduct routine analyses, even in limited-resource settings involving simple and short protocols, are examples of the growing current clinical demands not satisfied by conventional available techniques. In this context, the unique features offered by electrochemical biosensors, including affordability, real-time and reagentless monitoring, simple handling and portability, and versatility, make them especially interesting for adaptation to the increasingly challenging requirements of current clinical and point-of-care (POC) diagnostics. This has allowed the continuous development of strategies with improved performance in the clinical field that were unthinkable just a few years ago. After a brief introduction to the types and characteristics of clinically relevant biomarkers/samples, requirements for their analysis, and currently available methodologies, this review article provides a critical discussion of the most important developments and relevant applications involving electrochemical biosensors reported in the last five years in response to the demands of current diagnostic, prognostic, and therapeutic actions related to high prevalence and high mortality diseases and disorders. Special attention is paid to the rational design of surface chemistry and the use/modification of state-of-the-art nanomaterials to construct electrochemical bioscaffolds with antifouling properties that can be applied to the single or multiplex determination of biomarkers of accepted or emerging clinical relevance in particularly complex clinical samples, such as undiluted liquid biopsies, whole cells, and paraffin-embedded tissues, which have scarcely been explored using conventional techniques or electrochemical biosensing. Key points guiding future development, challenges to be addressed to further push the limits of electrochemical biosensors towards new challenging applications, and their introduction to the market are also discussed.
1. Introduction
Tremendous progress is taking place in the search for new biomarkers for diagnostic, prognostic, and therapeutic actions. The limitations of currently available methodologies, both in clinical validation and device implementation to meet the requirements of hospital routine and point-of-care testing (POCT), have resulted in continuing demand for the development of new methodologies able to solve these important problems.Specific challenges in this area include the different molecular levels of some biomarkers, the need for simultaneous determination of several biomarkers (with the same or different natures, and sometimes very different clinical ranges of interest) to improve diagnosis and prognosis reliability and therapeutic efficiency, the demand for real-time determination outside the hospital environment, and sample complexity.
In this context, it is important to recognize that current methodologies routinely used in hospitals have improved considerably and can offer, in centralized environments and using perfectly established protocols, attractive characteristics (high sensitivity and sampling frequency) for the determination of certain biomarkers in certain sample types. However, some methodologies provide only semiquantitative results and most involve complex, long, and expensive protocols that are limited to skilled personnel in central laboratories. Furthermore, these techniques are not applicable to biomarker multidetermination and their application to other types of samples is not straightforward, requiring exhaustive studies to fine-tune sample preparation protocols and ensure reliable determination.
Considerable recent progress in electrochemical biosensors has resulted in tremendous possibilities for the single or multiplexed determination of biomolecules of different molecular types in clinical samples of very different natures using similar protocols, even using reagentless real-time strategies, making them complementary tools to conventional methods. Furthermore, the simple handling, portability, and low cost of the instrumentation required make electrochemical biosensors particularly attractive compared with available methodologies currently used in clinical and point-of-care diagnostics.
The following sections briefly introduce the types and main characteristics of biomarkers considered of current relevance in clinical diagnosis, prognosis, and therapeutic actions. The requirements for their determination (concentration level and sample type), currently available determination methods and their limitations are discussed. Furthermore, a critical review of selected examples reported in the last five years shows how limits have been pushed to particularly complex applications, and the unique opportunities offered by electrochemical biosensor platforms for diagnostic, prognostic, and therapeutic actions of great relevance in clinics. General conclusions to guide non-specialists or researchers, and remaining challenges, opportunities, and prospects are discussed in the final section.
2. Biomarkers of relevance in clinical diagnosis, prognosis, and therapeutic action
To attack and stop certain diseases, they must be reliably detected well in advance to allow the implementation of efficient therapeutic actions to improve patient life quality and facilitate long-term survival. Accordingly, in recent years, much effort has been directed toward the identification and validation of biomarkers related to the diagnosis, prognosis, and therapy of relevant diseases, and the development of methodologies that allow reliable determination in samples of different natures using simple protocols and reduced analysis times.The US Food and Drug Administration (FDA) defines biomarkers as “any measurable diagnostic indicator used to assess the risk or presence of disease” or, more specifically, “any characteristic indicative of a normal biological process, pathogen, or pharmacological response to a therapeutic intervention, which can be measured and evaluated in an objective manner”.1 Biomarkers include a wide variety of molecules of different molecular levels: DNAs at the genetic level, RNAs (mRNAs and miRNAs) at the regulatory level, proteins (such as enzymes, autoantibodies, and extracellular receptors) at the functional level, and other small molecules at the metabolic level.2 Biomarkers have been detected in minimally invasive samples of liquid biopsies (blood, serum, plasma), secretions (saliva, sputum, faeces, urine), and other bodily fluids,3 in addition to fresh or paraffin-fixed solid tissue samples accessible through solid biopsies or surgical practices.4
Among biomarkers considered relevant for the diagnosis, prognosis and therapy of cancer, cardiovascular, and autoimmune diseases, inflammation processes, and metabolic disorders of great prevalence in society, outstanding examples include epigenetic biomarkers (miRNAs and DNA methylations) in both liquid and solid biopsies, mutations in circulating tumoral DNA, circulating protein biomarkers (such as antigens associated with tumors, autoantibodies, and soluble fraction of extracellular receptors), and protein receptors (nuclear or extracellular) in exosomes, cells, and solid biopsies.
Owing to the complexity of biological samples and large heterogeneity in diseases/disorders, diagnoses and prognoses based on individual levels of these biomarkers are often associated with a high probability of false positives or, even worse, false negatives. Therefore, results are often unreliable, leading to inefficient therapies. In contrast, measuring panels of biomarkers (at single or multiple molecular levels) has provided more accurate information than individual determinations, showing enormous potential for the early detection of associated diseases and allowing personalized therapy, while simplifying diagnosis by providing more information in less time.2,5
3. Conventional methods for clinical biomarker determination
Genomic methods (DNA sequencing, polymerase chain reaction (PCR) amplification detection, DNA microarray assay) and proteomic methods (conventional enzyme linked immunosorbent assays, ELISAs) are routinely used in hospitals to determine clinical biomarkers in clinical samples. However, these methods involve multi-step and time-consuming processes that require special operating staff, high costs, and instrumentation that is difficult to miniaturize. This significantly limits their applicability for POC diagnosis.6,7 Notably, these conventional methodologies cannot be used for the simultaneous determination of biomarkers at different molecular levels or of biomarkers at the same molecular level, but in very different clinical ranges.In this context, electrochemical biosensing of multilevel analytes, discussed in more detail in the following section, is emerging as a highly interesting alternative to overcome most of the aforementioned limitations. This technique allows rapid, affordable, and selective single or multiplexed determination (even in different clinical ranges or at different molecular levels) of target biomolecules using simple protocols and portable instrumentation of ready implementation in POC and in-field testing devices.
4. Electrochemical biosensing in clinical diagnosis, prognosis, and therapeutic action
In recent years, electrochemical biosensors have shown great potential for reliable determination in analyte samples of varying complexity related to diagnosis, prognosis, and therapeutic actions of diseases and disorders of high incidence and poor prognosis.8–35The high sensitivity and selectivity required for these challenging applications is achieved mainly in combination with the use of nanomaterials19–21,23,26,30,36–38 and/or different target nucleic acid amplification strategies.39,40 These strategies usually involve time-consuming and tedious protocols, and are difficult to implement in portable devices. Therefore, a constant and particularly relevant objective is to design affordable, quick, and simple methods to accurately determine target biomolecules while meeting the required demands of sensitivity and selectivity. Recently, an exhaustive comparison conducted using 11 assay test configurations for determination of the same synthetic target DNA demonstrated the possibility of developing very sensitive strategies without using nanomaterials or target nucleic acid amplification. The assay sensitivity could be tailored to more than three orders of magnitude and accomplished selective detection in scarcely treated complex samples, such as raw mitochondrial lysates.41 The tested configurations, implemented on the surface of magnetic beads (MBs), involved the formation of hybrids of different lengths coupled to different assay formats, and enzymatic labeling strategies followed by amperometric transduction upon magnetic capture of the resulting magnetic bioconjugates at screen-printed electrodes (SPCEs) using the system HRP/H2O2/HQ.42
Recent studies have also coupled screen-printed electrodes with magnetic micro- and nanoparticles (MBs and MNPs, respectively) as solid supports43,44 or used rational design of surface chemistry using diazonium salt chemistry,45,46 in connection with conventional bioreceptors or other little-explored high affinity bioreceptors (single-domain antibodies,47 and antibodies or viral proteins for specific RNA duplexes48). These approaches provided interesting routes for the development of affinity biosensing strategies with excellent performance, even in complex samples, and involving simple and rapid protocols. Using MBs as a solid support avoids applying and optimizing laborious protocols for modifying electrode surfaces, and improves the analytical performance of the resulting biosensors in terms of sensitivity, test time, and the minimization of sample matrix effects.49–55 Other applications also take advantage of the easy handling, modification, and manipulation of magnetic particles for the preparation of magnetic bioconjugates used as advanced labels for signal amplification.43
Additionally, screen-printed electrodes (SPEs) are recommended for many applications because they can be inexpensively mass-produced56 from various materials in different geometries and miniaturized and multiplexed formats. Their planar shape facilitates the incorporation of magnetic bioconjugates on their surface in a stable and reproducible manner through simple magnetic attraction. Furthermore, their reduced dimensions allow the use of small sample volumes and increase their potential for POC tests by allowing electrochemical instrumentation to be scaled down to small pocket-size devices. Indeed, these disposable electrodes can be used with most commercially available instruments, but also benchtop and portable instruments equipped with USB ports, laptops, or palm devices provided by various companies (PalmSens, DropSens, and others),57 or with wireless portable low-cost potentiostats recently developed at laboratory level.29
Other attractive biosensing approaches involve using integrated formats by exploiting rational surface chemistry through diazonium salt grafting.45,46 This simple, rapid, and versatile chemistry is a powerful tool for covalent immobilization of a wide range of biomolecules or nanomaterials onto different electrode surfaces in a stable and reproducible manner. The ability of this method to modify closely-spaced electrodes with different biological entities, which is ideal for multiplexing purposes, has also been demonstrated.58,59
Much effort has been made toward the coupling of electrochemical biosensors with isothermal nucleic acids amplification strategies faster than conventional PCR. These strategies include amplification assisted by different enzymes, including endonucleases, recombinases, helicases, or strand displacement polymerases, such as helicase-dependent amplification (HDA), loop-mediated isothermal amplification (LAMP), isothermal exponential amplification reaction (EXPAR), rolling-circle amplification (RCA), and strand displacement amplification (SDA). Enzyme-free strategies include hybridization chain reaction (HCR). These approaches are attractive methodologies for the determination of analytes of different molecular levels in highly complex samples, and are suitable for implementation in POC devices.24,60–63
Despite significant progress toward electrochemical biosensors for single and/or multiplexed determination of biomarkers, direct determination in complex media and continuous operation in biological matrices remain important challenges due to the occurrence of (bio)fouling through non-specific adsorption of proteins and other biological materials such as cells, cell fragments, and DNA/RNA. Therefore, the development of (bio)sensing interfaces that can combine high sensitivity and an antifouling ability is essential for expanding the practical applicability of biosensors and achieving more reliable measurements.64,65 Minimization of electrode surface (bio)fouling has been achieved using nanomaterials (carbon materials, metallic nanoparticles, and nanoporous electrodes66) and biomaterials such as polymers (PEG, conducting, zwitterionic and pH-responsive), hydrogels, peptides, and thiolated DNA self-assembled monolayers (thioaromatic, ternary and tetrahedral DNA nanostructures).65,67 Recently, Wang and coworkers reported the smart use of commercial methacrylate polymeric coatings with pH-responsive behavior to overcome fouling issues and ensure biocatalytic properties in protein-rich and low-pH environments. This group developed electrochemical biosensors with excellent performance for direct glucose monitoring in untreated blood and saliva,68 and gastrointestinal (GI) fluids.69
Furthermore, there is increasing demand for the development of electrochemical biosensing methodologies suitable for real-time, continuous, and direct monitoring of specific molecules in samples. In this context, biosensors that employ electrochemistry to monitor analyte-binding-induced changes in the rigidity of redox-tagged nucleic probes attached to an interrogating electrode have emerged as a new clinical trend owing to their appealing attributes. Folding-based nucleic acid sensors have been applied to the determination of nucleic acids, proteins, small molecules, and ions. These biosensors exhibit a rapid response (seconds to minutes) and are sensitive, reagentless, easily reusable, and less prone to fouling issues because the signaling mechanism relies on a specific, binding-induced conformational change and not target adsorption on the sensor surface.70 Reagentless electrochemical affinity sensors, classified according to the type of bioreceptor in E-DNA (using specific DNA sequences), E-AB (aptamers), and E-PB (peptides), have been reported for real-time and/or continuous biosensing of clinically relevant analytes in challenging samples such as serum, urine, and saliva, or even in vivo.71–73 The LODs achieved for target DNAs are of fM order (in whole blood)74 or even aM order (10% diluted human serum),75 and in the pM range for autoantibodies76 and proteins.77
Nucleic acid biosensing scaffolds prepared using tetrahedral DNA nanostructures allow significantly higher sensitivity (owing to precise control of the immobilized probes nanoscale spacing78) and stability (tetrahedron moves slower than monothiolated probes79), and are less prone to non-specific adsorptions than biosensors constructed with single point-tethered oligonucleotides.80 Tetrahedral bioscaffolds have been used to immobilize DNA probes,78,79 antibodies,81 and aptamers,79,82 and were successfully applied to the determination of DNAs,78,79 miRNAs,83 proteins (prostate specific antigen (PSA)81), and small molecules (cocaine82) in particularly fouling samples, such as serum.
Some recent articles have critically reviewed the latest advances, current trends, and existing challenges in electrochemical affinity biosensors mainly for circulating biomarkers of accepted clinical relevance at different molecular levels.6,7,84–87 In the following sections, selected examples of electrochemical biosensing platforms for single or multiplexed determination of biomarkers related to diagnosis, prognosis, and therapy efficiency, which have shown suitability for use in particularly challenging applications (some of which pioneer employing electrochemical biosensors), such as the determination of extracellular receptors in whole cells or the determination of biomarkers of protein and genetic nature directly in protein extracts and genetic material extracted from paraffin-embedded tumors, are critically discussed. The selected representative examples are classified depending on the type of sample analyzed and, owing to the large amount of literature on this topic, are limited to the last five years. These examples are focused on the determination of biomarkers for prevalent diseases (cancer, cardiovascular, and autoimmune diseases, inflammation processes, and metabolic disorders). Finally, current trends and future perspectives are discussed.
4.1. Electrochemical biosensing in liquid biopsies
A large number of electrochemical biosensors have been reported for the detection of biomarkers in biological fluids. In recent years, biosensors prepared from electrode platforms modified with various materials (including polymers, metal nanoparticles, and carbon nanomaterials), and using different strategies for the immobilization of bioreagents and signal amplification, have been applied to biomarkers related to many diseases, especially cancer, and cardiovascular and inflammatory processes. Considering the many highly relevant and original examples that should be considered, and with the aim to highlight challenging applications in this review, only electrochemical biosensors with tested abilities to determine a target biomarker (preferably endogenous and not spiked content) in real fluids such as serum, saliva, urine, or whole blood are discussed. To provide as much information as possible, Table 1 summarizes the characteristics of different methods involving electrochemical biosensors reported in the reviewed period classified by disease-type and sample analyzed. Selected methods are discussed in detail in this section.| Biomarker/disease | Biosensor | Method | Analytical characteristics/sample | Ref. |
|---|---|---|---|---|
| Abbreviations: AA, ascorbic acid; ACV, alternating current voltammetry; AF, atrial fibrillation; AFP, alpha fetoprotein; AMI, acute myocardial infarction; APO-A1, apolipoprotein A1; AXL, receptor tyrosine kinase; BNP, brain natriuretic peptide; CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; CMA, 4-carboxymethylaniline; CNFE, carbon nanofiber electrode; CNTs, carboxylated carbon nanotubes; CP, ceruloplasmin; cTnI, cardiac troponin I; CVD, cardiovascular disease; dBSA, denatured bovine serum albumin; Den, G3 PAMAM dendrimer; DMD, Duchenne muscular dystrophy; DTSP, 3,3′-dithiodipropionic acid di-(N-hydroxysuccinimide ester); EFGR, epithelial growth factor receptor; EIS, electrochemical impedance spectroscopy; EpCAM, epithelial cell adhesion molecule; ERα, estrogen receptor alpha; Fc, ferrocene; Fc-P1, ferrocene labeled hairpin probe; Fib, fibrinogen; Gr, graphene; HF, heart failure; HQ, hydroquinone; HRP, horseradish peroxidase; LOx, lactate oxidase; MB, methylene blue; Mb, myoglobin; MbBA, mioglobin binding aptamer; MCH, 6-mercapto-1-hexanol; MHA, 6-mercaptohexanoic acid; mMWCNTs, magnetic multiwalled carbon nanotubes; MGCE, magnetic glassy carbon electrode; MNPs, magnetic nanoparticles; MS, metabolic syndrome; μFED, microfluidic electrochemical array device; N-prGO, nitrogen-doped reduced graphene oxide; NSCL, non-small cell lung cancer; PAMAM, poly(amidoamine); PB, Prussian blue; PHA, 6-phosphonohexanoic acid; PL, polylysine; pPPA, poly(pyrrolepropionic acid); pPy, polypyrrole; PS67-b-PAA27, poly(styrene)-block-poly(acrylic acid); PVDF, polyvinylidene fluoride; PWE, paper working electrode; py-COOH, pyrene carboxylic acid; py-PEG, poly(ethylene glycol)-modified pyrene; rGO, reduced graphene oxide; SMA, spinal muscular atrophy; SMN1, survival motor neuron 1; SPCE, screen-printed carbon electrode; SWV, square-wave voltammetry; SWCNHs, single-walled carbon nanohorns; TCEP, tris(2-carboxyethyl)phosphine; TCPP, meso-tetra(4-carboxyphenyl) porphyrin; TGF-β1, transforming growth factor beta 1; TMB, 3,3′,5,5′-tetramethyl benzidine; TNF-α, tumor necrosis factor alpha; VCAM-1, vascular cell adhesion molecule-1. | ||||
| Determinations in serum | ||||
| cTnI/AMI | cTnI–anti-cTnI–Den/TMB/MHA/AuE label-free immunosensor | EIS; TMB | LOD: 0.28 pg mL−1; LR: 1.0 pg mL−1 to 1.0 μg mL−1/spiked serum | 91 |
| cTnI/AMI | Aptasensor; Fc–SiNPs as label | SWV | LOD: 1.0 pM; LR: 1–10 nM/serum | 90 |
| cTnI, cTnT | Dual flexible nano-ZnO–Abs multiplex immunosensor | EIS | LOD: 1 pg mL−1; LR: 0.1–105 pg mL−1; cTnI; 0.1 pg mL−1 cTnT/serum | 92 |
| CRP/AF | Anti-CRPHS-C11-(EG)3-OCH2-COOH/GDD/GCE | EIS, Fe(CN)63−/4− | LOD: 176 pM; LR: 0.5–70 nM/serum | 94 |
| CRP/CVD | Sandwich-type aptasensor; Zn2+/anti-CRP-AuNPs/SiO2 microspheres as immunoprobes; CRP apt. immob. onto AuNPs modified electrode | SWV (Zn2+ reduction) | LOD: 1.7 pg mL−1; LR: 5 pg mL−1–125 ng mL−1/serum | 95 |
| NT-proBNP, CRP | Dual SPCE. competitive and sandwich magnetoimmunosensors | Amperometry, H2O2/TMB −100 mV vs. Ag | LOD: 0.47 ng mL−1; LR: 2.0–100 ng mL−1 (CRP); DR: 2.5–504 ng mL−1 (NT-pro BNP)/certified CRP serum spiked with NT-proBNP | 10 |
| Mb/AMI | Aptasensor; Fc–MbBA–AuNPs/TCPP/Gr/GCE | DPV; Fc | LOD: 6.7 × 10−12 M; LR: 2.0 × 10−11–7.7 × 10−7 M/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted serum 100 diluted serum |
96 |
| Mb/AMI | Anti-Mb–AuNPs@rGO/SPE label-free immunosensor | DPV; reduction of Fe(III) in the heme group of Mb | LOD: 0.67 ng mL−1; LR: 1–1400 ng mL−1/spiked serum | 157 |
| Mb/AMI | Anti-Mb-3D HOOC-Pt(MPA) NPs/ITO label-free immunosensor | EIS; Fe(CN)63−/4− | LOD: 1.70 ng mL−1; LR: 0.01–1 μg mL−1 | 122 |
| AXL/HF | pPPA/SPCE sandwich-type-immunosensor | Amperometry, H2O2/HQ −200 mV vs. Ag | LOD: 337 pg mL−1; LR: 7–700 pg mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 diluted serum 2 diluted serum |
103 |
| BNP/HF | AuNPs; Ps-S-Phe-SPCE sandwich-type immunosensor | Amperometry, H2O2/HQ −200 mV vs. Ag | LOD: 4 pg mL−1; LR: 0.014–15 ng mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted serum 10 diluted serum |
30 |
| Lp(a)/CVD | Anti-Lp(a)–N-[Nα,Nα-bis(carboxymethyl)-lysine]-12-mercaptododecanamide (HS-NTA)-SPCE sandwich-type immunosensor | Amperometry, H2O2/TMB −100 mV vs. Ag | LOD: 8 ng mL−1; LR: 0.02–10 μg mL−1/spiked serum, certified Lp(a) serum | 9 |
| CD105/cancer, EB | pPPA/SPCE sandwich-type-immunosensor | Amperometry, H2O2/HQ −200 mV vs. Ag | LOD: 140 pg mL−1; LR: 0.18–20 ng mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 diluted serum 50 diluted serum |
25 |
| miRNA-21 | Fe3O4/CeO2 @Au MNPs-S1 enzyme-free biosensor nanocatalyst and catalytic hairpin assembly | DPV, MB | LOD: 0.33 fM; LR: 1 fM–1 nM | 108 |
| miRNA125a | Carbon black-modified pGE | EIS; Fe(CN)63−/4− | LOD: 10 pM; LR: 1 nM–2 μm/serum | 158 |
| miRNA-155/breast cancer | Anti-miRNA-155-AuSPE label-free aptasensor | SWV, EIS; Fe(CN)63−/4− | LOD: 5.7 aM; LR: 10 aM–1.0 nM/serum | 159 |
| EGFR, VEGF/cancer | Antigen-MIPs/SPAuE and Abs-conjugated Cu(II) or Cd(II)-nano-liposomes as signal tags | PSA | LOD: 0.01 pg mL−1 (EGFR), 0.005 pg mL−1 VEGF); LR: 0.05 pg mL−1–50 ng mL−1 (EGFR), 0.01–7000 pg mL−1 (VEGF) | 111 |
| HER2/breast cancer | HER2 apt-AuIDE arrays on SiO2 wafers; aptasensor | Capacitance | LR: 200 pg mL−1 to 2 ng mL−1/spiked serum | 160 |
| HER2/breast cancer | Glut/Chit/rGO/GCE label-free aptasensor | DPV, MB | LOD: 0.21 ng mL−1; LR: 0.2–2 ng mL−1; 2–75 ng mL−1/serum | 112 |
| HER2/breast cancer | Sandwich-type label-free immunosensor with Ag enhancement; anti-HER2/APTMS–Fe3O4/GCE; anti-HER2/hydrazine@AuNPs/APTMS–Fe3O4 as label | DPV stripping Ag | LOD: 0.02 pg mL−1; LR: 0.5 pg mL−1–50.0 ng mL−1/patient serum | 113 |
| ERα | μFED; DNA-ERE-AuNPs-GSH-PDDA/SPCE anti-ERα-MP-HRP as the label | Amperometry, C −200 mV vs. Ag/AgCl | LOD: 10.0 fg mL−1/calf serum | 114 |
| PDGF-BB | Sandwich Apt2–AuNPs–Fc MoS2 label-free aptasensor | DPV; Fe(CN)63−/4− | LOD: 0.3 pM; LR: 0.001–10 nM/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted serum 100 diluted serum |
116 |
| PDGF-BB | Aptasensor, HRP–avidin–biotin–cDNA/AuNPs/MoSe2-Gr/GCE; aptamer–PDGF–BB-sdNA + cDNA+ avidin–HRP; ExoIII aided signal amplification | DPV; H2O2/HQ | LOD: 20 fM; LR: 0.0001–1 nM | 117 |
| PDGF-BB/serum | AuNPs α-CD | SWV | LOD: 0.52 nM; LR: 0.52–1.52 nM | 161 |
| VEGF/cancer | Sandwich apta-immunosensor: MBs–VEGF aptamer–VEGF–anti-VEGF–microAuE | Capacitance | LR: 5 pg mL−1–1 ng mL−1/serum | 110 |
| p53/colorectal cancer | p53-anti–p53/Star-PGMA/ITO label-free immunosensor | EIS; Fe(CN)63−/4− | LOD: 7 fg mL−1; LR: 0.02–4 pg mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 diluted serum 1000 diluted serum |
127 |
| TGF-β1/immune, inflammatory | IgG-/MWCNTs/SPCE sandwich-type-immunosensor | Amperometry −200 mV vs. Ag | LOD: 1.3 pg mL−1; LR: 5–200 pg mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted serum 100 diluted serum |
119 |
| TNF-α | Aptasensor; aptamer–Au–Gr/Chit/SPCE; Ag@Pt–Gr as label | DPV; catechol | LOD: 1.64 pg mL−1; DR: 5.0–70 pg mL−1/spiked serum | 120 |
| TNF-α | Label-free immunosensor anti-TNF–C60–CNTs–IL | DPV; catechol | LOD: 2.0 pg mL−1; DR: 5.0–75 pg mL−1/serum | 162 |
| TNF-α | Aptasensor; H1H2/MB/HT/S1S2-AuNPs/SWCNHs/GCE; CB/AuNPs@Chit/GCE; RecJf exonuclease for recycling and DSN; intercalated MB as electrochemical probe | DPV; MB | LOD: 0.5 pg mL−1; LR: 0.001–100 ng mL−1/serum | 121 |
| Determinations in saliva | ||||
| α-Amylase/mental diseases | Starch/Fe(CN)63−/chip (smartphone) | Potentiometry; ΔE Fe(CN)63− | LOD: 0.12 U mL−1; LR: 30–1000 U mL−1/saliva | 124 |
| Cortisol/stress | PS67-b-PAA27 polymer/Gr/DTSP/AuE | EIS; Fe(CN)63−/4− | LOD: 3 pg mL−1; LR: 3 pg mL−1–10 μg mL−1/saliva | 125 |
| IL-8mRNA and IL-8/oral cancer | SPCE dual magnetoimmunosensor | Amperometry; −200 mV vs. Ag | LOD: 0.21 nM (IL-8mRNA); 72.4 pg mL−1 (IL-8); LR: 0.32–7.5 nM (IL-8 mRNA); 87.9–5000 pg mL−1 (IL-8)/saliva | 16 |
| IL-8/cancer | PHA/ITO label free immunosensor | EIS; Fe(CN)63−/4− | LOD: 6 fg mL−1; LR: 0.02–3 pg mL−1/1/200 diluted saliva | 118 |
| IL-8/cancer | SPGMA/PVDF/ITO label-free immunosensor | EIS; Fe(CN)63−/4− | LOD: 3.3 fg mL−1; LR: 0.01–3 pg mL−1/1/50 diluted saliva | 128 |
| ORAOVI/oral cancer | MCH/MB-PPI/AuE hybrid; target DNA with FcP1 probe; exonuclease III amplification | ACV, MB, Fc | LOD: 12.8 fM; LR: 0.02–2 pg mL−1/spiked 1/10 diluted saliva | 129 |
| EpCAM/cancer | EpCAM-driven toehold-mediated DNA recycling amplification. | SWV, MB | LOD: 20 pg mL−1; LR: 0.1–20 ng mL−1/saliva, serum, urine | 163 |
| NSCLC/EGFR mutations | pPy/AuNPs/PWE μ-fluidic DNA biosensor/hybridization | DPV, HRP/H2O2/MB | LOD: 0.167 nM; LR: 5–500 nM | 132 |
| CYFRA-21/oral cancer | APTES/nHfO2/ITO; label-free immunosensor | DPV; Fe(CN)63−/4− | LOD: 0.21 ng mL−1; LR: 2–18 ng mL−1/saliva | 134 |
| CYFRA21-1/NSCLC | 3D graphene–AgNPs | DPV; Fe(CN)63−/4− | LOD: 10−14 M; LR: 10−14–10−7 M/serum | 131 |
| Human fetuin (HFA)/cancer, type 2 diabetes | Strept–Phe-SPCE; sandwich-type immunosensor; anti-HFA (HRP)–mMWCNTs as carrier | Amperometry −200 mV vs. Ag | LOD: 16 pg mL−1; LR: 20–2000 pg mL−1/1/100 diluted saliva | 23 |
| IL-6/oral cancer | SPCE sandwich-type magnetoimmunosensor | Amperometry −200 mV vs. Ag | LOD: 0.39 pg mL−1; LR: 1.75–500 pg mL−1/saliva | 137 |
| TNF-α/HF | CMA-grafted-microAuEs label-free immunosensor | EIS; Fe(CN)63−/4− | LOD: 1 pg mL−1; LR: 1–100 pg mL−1/saliva, artificial saliva | 140 |
| TNF-α/HF | CMA/AuE sandwich-type immunosensor | Amperometry 100 mV vs. SCE | LOD: 1 pg mL−1; LR: 1–30 pg mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted saliva 100 diluted saliva |
141 |
| cTnI/CVD | (py-COOH)(py-PEG)N-prGO; label-free aptasensor | DPV; Fe(CN)64− | LOD: 1 pg mL−1; LR: 1 pg mL−1–100 ng mL−1/saliva | 144 |
| IL-1β, TNF-α/inflammation | HOOC-Phe-DWCNTs/d-SPCE sandwich-type immunosensor | Amperometry −200 mV vs. Ag | LOD: 0.38 pg mL−1 (IL-1β); 0.85 pg mL−1 (TNF-α) LR: 0.5–100 pg mL−1 (IL-β1); 1–200 pg mL−1 (TNF-α)/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 diluted saliva 50 diluted saliva |
19 |
| TGF-β1/immune, inflammatory | Strept-4-carboxyphenyl-grafted-SPCE sandwich-type immunosensor/V-Phe-SWCNT(-HRP)-anti-TGF as carrier tag | Amperometry −300 mV vs. Ag | LOD: 0.95 pg mL−1; LR: 2.5–1000 pg mL−1/saliva | 20 |
| Lactate/stress level | PB/PPD/PET flexible SPEs; LOx enzyme biosensor | Amperometry 0.042 V (vs. Ag/AgCl) | LOD: 0.050 mM; LR: 0.1–1.0 mM/saliva | 145 |
| Lactate/resp. insuf., HF, metab. disorders | Nafion/PB/SPEs; LOx enzyme biosensor | Amperometry −50 mV vs. Ag | LOD: 0.01 mM; LR: 0.025–0.25 mM/saliva | 146 |
| Determinations in urine | ||||
| Fib/CVD | Fib-SWCNHs/SPCEM indirect competitive immunosensor | Amperometry, H2O2/HQ −200 mV vs. Ag | LOD: 58 ng mL−1; LR: 0.1–100 μg mL−1/Liquichek urine | 147 |
| TGF-β1/kidney diseases | SPCE sandwich-type magnetoimmunosensor | Amperometry, H2O2/HQ −200 mV vs. Ag | LOD: 10 pg mL−1; LR: 15–3000 pg mL−1/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 diluted Liquichek urine 3 diluted Liquichek urine |
148 |
| miR-21/bladder and prostate cancer | Hybridization of specific DNA immob. onto chlorinated naphtalene sulfonic acid/GCE | Coulometry; Fe(CN)63−/4− | LR: 10 fM–10 nM/urine | 149 |
| APO-A1/bladder cancer | Anti-APO-A1-ITO; sandwich-type immunosensor with AP-anti-APO-A1 | Chronocoulometry; AA as enzyme product | LOD: 1 pM; LR: 1 pM–100 nM/urine | 151 |
| VCAM1/systemic lupus erythematosus | Microfluidic sandwich-type immunosensor; anti-VCAM1-DSP-gold microelectrode | EIS | LR: 8 fg mL−1 to 800 pg mL−1/urine | 153 |
| Determinations in whole blood | ||||
| AFP/liver cancer | Anti-AFP–Fe3O4–PL-Hep anticoag. MNPs/MGCE; label-free immunosensor | DPV; Fe(CN)63−/4− | LOD: 0.072 ng mL−1; LR: 0.1–100 ng mL−1/whole blood | 155 |
| SMN1/SMA; CTFR/CF; DMD proteins/DMD | Multiplexed immunosensor; covalent immob. of three Abs onto carboxyphenyldiazonium–CNFE | SWV, Fe(CN)63−/4− | LODs: 0.9 (SMN1), 0.7 (CTFR), 0.74 (DMD) pg mL−1/whole blood spiked samples | 156 |
Along with cancer biomarkers, biomarkers for cardiovascular diseases (CVDs) are the most frequently addressed because they are crucial for diagnosis and management processes. Recent progress in the detection of CVD markers using electrochemical biosensors has been well-reviewed.88 Advances in the design of biointerfaces involving nanomaterials for the electrochemical biosensing of cardiovascular biomarkers have also been reviewed by Farzin et al.89 The various serum biomarkers for CVDs analyzed by electrochemical biosensors include cardiac troponins cTnI and cTnT90–92 (reviewed by Abdorahim et al.93), C-reactive protein (CRP),94,95 and myoglobin (Mb).96 Furthermore, emerging biomarkers, such as receptor tyrosine kinase (AXL),97 both B-type natriuretic peptide (BNP) and N-terminal pro B-type natriuretic peptide (NT-proBNP),98 and lipoprotein a (Lp(a)),99 have been targeted with electrochemical biosensors.
Cardiac troponins (I and T) are well established protein biomarkers associated with heart muscle damage and with a high specificity for diagnosing myocardial infarction. As an example of a cTnI biosensor, Akter et al.91 developed a highly sensitive dendrimer-coupled impedimetric immunosensor for label-free and reagent-free cTnI detection in human serum. The design used a carboxylic acid-functionalized third generation (G3) poly(amidoamine) (PAMAM) dendrimer on a gold electrode modified with a 6-mercaptohexanoic acid (MHA) self-assembled monolayer (SAM) and 3,3′,5,5′-tetramethylbenzidine (TMB), and anti-cTnI antibody was covalently immobilized on the electrode surface. After capturing the antigen, impedance monitoring was conducted using TMB as an internal surface redox couple to generate the signal and avoid using external electroactive reagents. Owing to the high loading of antibodies immobilized on the large number of carboxylic groups at the dendrimer, high sensitivity was obtained, with a LOD of 11.7 fM cTnI.
Using ferrocene-modified silica nanoparticles (Fc–SiNPs) characterized by their high stability, easy modification, electroactivity, and great signal amplification via electron transfer, Jo et al.90 proposed an aptamer-based label-free detection platform for cTnI. Single-stranded DNA (ssDNA) aptamers against cTnI were selected using the SELEX method and the corresponding binding affinities were measured by surface plasmon resonance (SPR). The fundamentals of this electrochemical detection are summarized in Fig. 1. In the absence of cTnI, the aptamer-SAM allowed easy access of Fc–SiNPs to the electrode surface, resulting in a large voltammetric response. However, in the presence of cTnI, the specific interaction between cTnI and aptamers prevented Fc–SiNP approach. Therefore, the oxidation peak current significantly decreased depending on the analyte concentration.
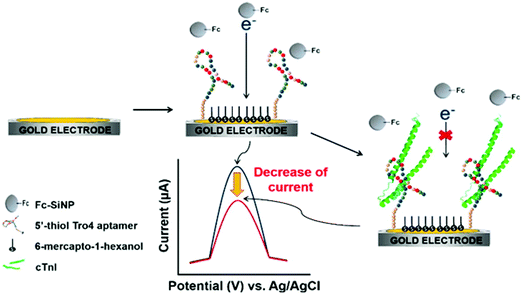 | ||
| Fig. 1 Schematic illustration of voltammetric detection of cTnI. Upon immobilization of the cTnI aptamer on the surface of a gold working electrode, cTnI is added. The cTnI concentration was quantified by monitoring the decrease in SWV signal resulting from the hindered approaching of Fc–SiNPs. Reproduced from ref. 90 with permission. | ||
The multiplexed detection of protein biomarkers for CVDs offers new opportunities for early diagnosis and efficient treatment. For troponins, a flexible disposable electrochemical biosensor comprising vertically oriented zinc oxide (ZnO) nanostructures was developed for rapid and simultaneous screening of cTnI and cTnT in a POC sensor format. Simultaneous detection was achieved through sensor platform design consisting of ZnO nanostructured electrode arrays with anti-troponin antibodies immobilized on their surface via dithiobis(succinimidyl propionate) (DSP). Using electrochemical impedance spectroscopy, linear calibration plots between 0.1 and 105 pg mL−1 for both analytes were achieved with a LOD value of 1 pg mL−1 in human serum.92
C-Reactive protein (CRP) is an acute-phase protein widely accepted as a biomarker for cardiovascular disease and inflammation, with a cut-off value of 3.0 mg L−1 in serum as an indication of high risk.100 A connection between high CRP levels and atrial fibrillation (AF) has also been proposed.94 Various electrochemical biosensors have been reported for CRP determination at low concentration (see Table 1). An illustrative example is the impedimetric immunosensor for CRP analysis in blood serum prepared using a glassy carbon electrode (GCE) coated with graphene quantum dots (GQD) fabricated from graphene oxide, and immobilization of the capture antibody onto polyethylene glycol (PEG)-thiol HS-C11-(EG)3-OCH2-COOH. The GQDs/GCE electrochemical platform exhibited high conductivity and detected variations in the charge transfer resistance related to CRP concentration in the 0.5–70 nM linear range, with a LOD value of 176 pM. The method was validated by analysis of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted serum samples with results in agreement with those obtained using an ELISA kit.94 A CRP electrochemical aptasensor was also prepared by assembling RNA aptamers specific to CRP on the surface of AuNPs via Au–S bond and using immunoprobes fabricated by immobilization of anti-CRP antibodies onto silica microspheres coated with gold nanoparticles and functionalized with Zn2+ ions (Zn2+/anti-CRP/AuNPs@SiMSs) as labels.95 The electrochemical reduction of metal ions by SWV was employed to quantify CRP with high sensitivity owing to the large amount of signal moieties loaded on the immunoprobes. A linear range from 5 pg mL−1 to 125 ng mL−1 CRP and a LOD of 1.7 pg mL−1 were obtained. The biosensor was applied to real serum samples. Our group reported the multiplexed determination of CRP and another cardiac biomarker, amino-terminal pro-B-type natriuretic peptide (NT-proBNP), using dual screen-printed carbon electrodes. Importantly, the clinically relevant concentrations of these biomarkers in serum differed by more than three orders of magnitude.10 MBs functionalized with carboxylic groups were used to covalently immobilize the specific capture antibodies. Biomarker determination was performed using indirect competitive and sandwich-type configurations for NT-proBNP and CRP, respectively, with HRP-labeled tracers. The dual immunosensor allowed simultaneous independent amperometric measurement of each analyte at Eapp = −0.1 V vs. Ag using the H2O2/TMB system, providing low LODs of 0.47 ng mL−1 for both biomarkers. This method was applied to analyze an international standard for CRP serum spiked with NT-proBNP standards.
10 diluted serum samples with results in agreement with those obtained using an ELISA kit.94 A CRP electrochemical aptasensor was also prepared by assembling RNA aptamers specific to CRP on the surface of AuNPs via Au–S bond and using immunoprobes fabricated by immobilization of anti-CRP antibodies onto silica microspheres coated with gold nanoparticles and functionalized with Zn2+ ions (Zn2+/anti-CRP/AuNPs@SiMSs) as labels.95 The electrochemical reduction of metal ions by SWV was employed to quantify CRP with high sensitivity owing to the large amount of signal moieties loaded on the immunoprobes. A linear range from 5 pg mL−1 to 125 ng mL−1 CRP and a LOD of 1.7 pg mL−1 were obtained. The biosensor was applied to real serum samples. Our group reported the multiplexed determination of CRP and another cardiac biomarker, amino-terminal pro-B-type natriuretic peptide (NT-proBNP), using dual screen-printed carbon electrodes. Importantly, the clinically relevant concentrations of these biomarkers in serum differed by more than three orders of magnitude.10 MBs functionalized with carboxylic groups were used to covalently immobilize the specific capture antibodies. Biomarker determination was performed using indirect competitive and sandwich-type configurations for NT-proBNP and CRP, respectively, with HRP-labeled tracers. The dual immunosensor allowed simultaneous independent amperometric measurement of each analyte at Eapp = −0.1 V vs. Ag using the H2O2/TMB system, providing low LODs of 0.47 ng mL−1 for both biomarkers. This method was applied to analyze an international standard for CRP serum spiked with NT-proBNP standards.
Heme-containing protein cardiac myoglobin (Mb) is among the biomarkers that show an increase after acute miocardial infarction (AMI). The small size of Mb (molecular weight = 17.8 kDa) facilitates its rapid release into circulation within 1–3 h of symptoms.101 A sensitive electrochemical aptasensor for Mb-specific recognition has been reported by Zhang et al.96 The method involved using meso-tetra(4-carboxyphenyl) porphyrin (TCPP)-functionalized graphene-conjugated AuNPs to immobilize human Mb-binding aptamer. TCPP is an anion porphyrin that can bind graphene through π–π stacking, improving the solubility and stability of this nanomaterial and acting as a catalyst to enhance electrochemical reactions. The Mb-binding aptamer was dual modified, with thiol at the 5′-end to allow immobilization on the AuNPs, and ferrocene at the 3′-end to allow electrochemical transduction (Fig. 2). In the absence of Mb, ferrocene exhibited a stronger signal, while in the presence of Mb, the aptamer on the electrode surface recognized the target specifically and its conformation changed. Simultaneously, ferrocene was far from the electrode surface and the electron transfer rate was hindered, leading to a decrease in the ferrocene signal. Interestingly, this method provided a very low LOD of 6.7 × 10−12 M (S/N = 3), which was attributed to the large amount of binding sites provided by AuNPs and the high electrocatalytic activity of these nanoparticles and graphene. The aptasensor was applied to perform recovery experiments using the standard additions method in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted serum spiked with Mb.
100 diluted serum spiked with Mb.
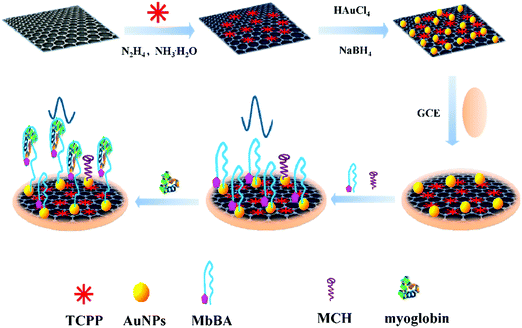 | ||
| Fig. 2 Synthesis of TCPP–graphene/AuNP nanocomposites and an electrochemical aptasensor for myoglobin detection. Reproduced from ref. 96 with permission. | ||
AXL has a potential role in the pathophysiology of heart failure (HF). The serum AXL concentration was higher in HF than in control patients with an established cut-off value of 71 ng mL−1.102 Elevated serum AXL levels are also associated with inflammatory biomarkers in acute coronary syndrome.97 The first electrochemical immunosensor for AXL was prepared using disposable immuno-platforms involving screen-printed carbon electrodes (SPCEs) modified with electropolymerized poly(pyrrole propionic acid) (pPPA), a conducting polymer containing several carboxylic moieties for covalent binding of biomolecules, which allows efficient immobilization of anti-AXL antibodies (Fig. 3). A sandwich-type immunoassay was designed involving enzyme-amplified electrochemical transduction. The developed HRP–Strept–biotin–anti-AXL–AXL–anti-AXL–pPPA/SPCE immunoplatform was successfully employed for AXL determination in human sera from patients with heart diseases. Interestingly, only two-fold dilution of serum with blocking buffer solution was required as sample treatment.103
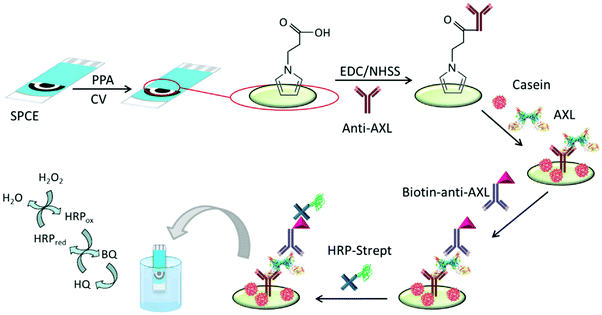 | ||
| Fig. 3 Schematic illustration of the steps involved in construction of the amperometric immunosensor for AXL involving pPPA-modified SPCEs and the covalent immobilization of anti-AXL. Reproduced from ref. 103 with permission. | ||
Serum levels of BNP, a neurohormone secreted from the cardiac ventricles as a response to ventricular volume expansion, pressure overload, and resultant wall tension,104 and NT-proBNP, the more stable amino terminal prohormone fragment, can be used to diagnose congestive heart failure and are strong indicators of future cardiac events in patients with and without symptomatic HF. An illustrative example of an electrochemical immunosensor for BNP determination in human serum is that reported by Serafín et al.30 This immunosensor was prepared by immobilization of capture antibodies onto gold nanoparticle (AuNP)-grafted-SPCEs through aryl diazonium salt chemistry using 4-aminothiophenol (AuNPs–S-Phe–SPCE). The nanostructured electrochemical platform afforded an ordered layer of AuNPs on SPCEs, providing high conductivity and improved stability of immobilized biomolecules. A sandwich-type immunoassay was implemented using an HRP-labeled detector antibody and amperometric transduction with the H2O2/HQ system. A linear range between 0.014 and 15 ng mL−1 BNP and a LOD of 4 pg mL−1, 100 times lower than the established cut-off value in serum for heart failure (HF) diagnosis, were achieved. The immunosensor was successfully applied to the analysis of human serum from HF patients with only 10-fold sample dilution as treatment.
Lipoprotein (a) (Lp(a)) is a complex polymorphic apolipoprotein, for which elevated levels are likely in the causal pathway for atherosclerotic cardiovascular diseases and aortic valve calcification.99 Although the concentration of this lipoprotein in serum can vary between 0 and 200 mg dL−1, concentrations of Lp(a) exceeding 30 mg dL−1 might increase the risk of thrombus formation. The integrated sandwich-type amperometric immunosensor involving covalent immobilization of anti-Lp(a) capture antibodies on the surface of N-[Nα,Nα-bis(carboxymethyl)-lysine]-12-mercaptododecamine (HS-NTA)-modified SPCEs is an example of an electrochemical biosensor for Lp(a) determination (Fig. 4). The current measured at −0.10 V vs. Ag pseudoreference electrode upon addition of TMB showed a linear range between 0.02 and 10 μg mL−1 and a LOD of 8 ng mL−1.9 The immunosensor was validated by analyzing both spiked serum samples and a reference serum with certified Lp(a) content.
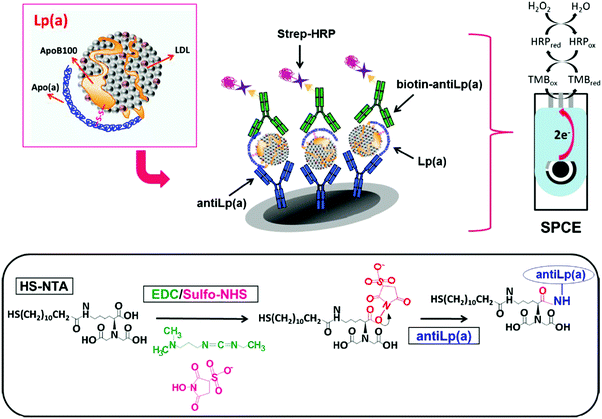 | ||
| Fig. 4 Schematic illustration of methodology involved in the development of a disposable Lp(a) sandwich-integrated immunosensor. Electrode and enzymatic reactions involved in the amperometric detection of H2O2 with TMB at the SPCE are also shown. Reproduced from ref. 9 with permission. | ||
Cancer is a multi-factorial disease in which various biomarkers might be involved. Demand for rapid, accurate, and cost-effective cancer detection has increasingly growing, with electrochemical biosensors currently among the most suitable strategies for early detection. Among electrochemical sensing bioplatforms, antibody-based biosensors have been used profusely for detecting various circulating tumor markers in serum. Furthermore, in recent years, the number of aptasensors developed with this purpose has grown.105Table 1 summarizes the characteristics of several electrochemical biosensors prepared for the detection of relatively unexplored or emerging cancer biomarkers, which have been applied to real serum samples. Some biosensors considered of special novelty or relevance are discussed below.
Human endoglin (CD105) is a 180 kDa homodimeric hypoxia-inducible cell transmembrane type III glycoprotein, densely expressed on the surface of angiogenic proliferating endothelial cells. Stronger CD105 expression has been found in a wide range of endothelial tumors, including colon, breast, brain, lung, prostate, and cervical cancers, compared with paired healthy tissues, suggesting the possible involvement of CD105 in tumor angiogenesis.106 Furthermore, increased levels of CD105 in biological fluids from affected patients might be used as an indicator for disease progression and metastasis risk. CD105 circulating levels have also been found to be altered in response to chemotherapy. An amperometric immunosensor for the determination of CD105, designed to comply with the sensitivity and accuracy requirements of clinical practice, has been reported recently.25 This immunosensing platform was implemented on disposable electrodes modified with pPPA and involved a sandwich configuration using a biotinylated detector antibody labeled with poly-HRP–streptavidin for signal amplification. Amperometric detection of H2O2 in the presence of hydroquinone (HQ) was employed as analytical readout. The resulting immunosensor provided a linear range between 0.18 and 20 ng mL−1, which was adequate for the determination of CD105 in serum, with a LOD of 140 pg mL−1. The usefulness of the immunosensor was tested by analyzing human serum samples collected from healthy individuals and patients of colorectal, breast, and lung cancer. The results were successfully compared with those provided by an ELISA kit.
MicroRNAs (miRNAs) are an important class of short (18–24 nucleotides) single-stranded and non-protein encoding RNAs involved in various biological processes, such as cell differentiation, proliferation, and apoptosis. The aberrant expression of miRNAs is generally associated with serious diseases, such as cancers, cardiovascular diseases, and neurological disorders. Therefore, several miRNAs are emerging as novel biomarkers for cancer diagnosis, therapy, and prognosis. An illustrative example of electrochemical determination of miRNAs is that developed for miRNA-21, a potential cancer biomarker upregulated in breast cancer cells that acts as a non-specific oncogene. Two single-stranded (ss) DNA probes, thiol terminated probe 1 and biotin conjugated probe 2, were designed to hybridize with the target miRNA-21. AuNPs were used to modify a pencil graphite electrode (PGE) for SH-P1 immobilization by Au–S bonds. Part of the miRNA-21 target was first hybridized with SH-P1, and then the other part of the target was hybridized with biotin-P2. Streptavidin–alkaline phosphatase conjugate was immobilized on biotin-P2 and electrochemical detection was performed upon α-naphthyl phosphate addition.107 Another interesting electrochemical biosensor for miRNA-21 was that prepared using Fe3O4/CeO2@Au magnetite nanoparticles, conjugated with detector probe (S1), as a nanocatalyst and catalytic hairpin assembly for signal amplification. In this configuration, target miRNA-21 hybridized with hairpin H2 to form H2-T ds DNA, which further opened the hairpin H1 for H1–H2 dsDNA formation. Simultaneously, Fe3O4/CeO2@Au-S1 hybridized with a single-stranded (ss) fragment of H1–H2 dsDNA to produce long dsDNA and absorb a large amount of methylene blue (MB) electroactive substance. This also acted as a nanocatalyst for the reduction of MB, which amplified the electrochemical signal. This strategy allowed the proposed biosensor to provide a wide linear range between 1 fM and 1 nM, with a LOD of 0.33 fM (defined as S/N = 3). This biosensor was successfully applied to human serum samples.108
Vascular endothelial growth factor (VEGF) is a key regulator of vascular formation and an important protein biomarker in cancer angiogenesis.109 Qureshi et al.110 developed a capacitive aptamer–antibody-based sandwich assay for the detection of VEGF in human serum. Fig. 5 shows that the assay involved capture through two epitopes, one with the anti-VEGF aptamer and the other with the antibody. The capacitive sensor was functionalized with anti-VEGF aptamer, which captured the VEGF protein, followed by sandwiching with antibody-conjugated magnetic beads (Abs–MB). Measurements by EIS provide a detection range between 5 pg mL−1 and 1 ng mL−1 VEGF in human serum.
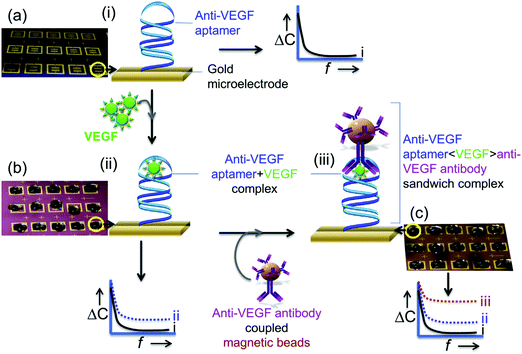 | ||
| Fig. 5 Schematic illustration of an apta-immunosensor constructed for VEGF determination through changes in capacitance against the applied AC electrical frequency: (a) sensor surfaces functionalized with aptamers as blank surfaces; (b) the aptasensor incubated with 0.1× serum with different VEGF concentrations; and (c) the sandwiching of the aptasensor with MB-Abs. Reproduced from ref. 110 with permission. | ||
The dual determination of VEGF and epidermal growth factor receptor (EGFR) was reported using gold screen-printed electrodes (Au-SPE) modified with a molecularly imprinted polymer (MIP) and amplified through antibody-conjugated nano-liposomes. The biosensor was prepared by modifying the Au-SPE with 3,3′-dithiodipropionic acid di(N-hydroxy-succinimide ester) (DTSP) via self-assembly. The target proteins were then covalently attached to the modified SPE. The corresponding MIPs were synthesized by polymerization of acrylamide and N,N′-methylene-bis(acrylamide) monomers around the EGFR and VEGF templates. Electrochemical signals were produced by loading nano-liposomes with Cd(II) and Cu(II) cations and decorating with antibodies specific for EGFR and VEGF. Potentiometric striping analysis was utilized for sensitive determination of the respective cations. The LOD values for EGFR and VEGF were 0.01 and 0.005 pg mL−1, with linear ranges of 0.05–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 0.01–7000 pg mL−1, respectively. This biosensor was successfully used for specific detection of EGFR and VEGF in real serum samples.111
000 and 0.01–7000 pg mL−1, respectively. This biosensor was successfully used for specific detection of EGFR and VEGF in real serum samples.111
Human epidermal growth factor receptor 2 (HER2) is a protein overexpressed in some breast cancer tumors, known as HER2-positive breast cancers, which tend to grow and spread more aggressively. Sensitive detection of low HER2 levels in serum is vital for early diagnosis and clinical management of breast cancer. Owing to its importance, various methods have been developed to assess HER2 status at the protein, RNA, or DNA levels. Tabasi et al.112 reported an electrochemical aptasensor for HER2 using a GCE modified with reduced graphene oxide (rGO) and chitosan (Chit) with a high fraction of amine groups to immobilize the aptamer. Using DPV and MB as an electrochemical probe, a detection limit of 0.21 ng mL−1 was obtained. A sandwich-type immunoassay for HER2 with silver signal enhancements was prepared by designing a platform composed of anti-HER2/3-aminopropyl-trimethoxy-silane (APTMS)–Fe3O4 bioconjugate immobilized on a GCE and labeling with AuNPs self-assembled with thiolated antibodies in the presence of hydrazine (anti-HER2/hydrazine@AuNPs/APTMS–Fe3O4. The chemical reduction of Ag(I) on AgNPs caused by hydrazine provoked significant enhancement of the signal related to HER2 measured by stripping DPV. The effectiveness of this protocol was evaluated by determining the level of this tumor marker in serum samples from breast cancer patients.113
Estrogen receptor alpha (ERα) is a nuclear hormone receptor and transcription factor that regulates the expression of genes affecting cell proliferation and differentiation. ERα is overexpressed in a high percentage of breast cancer cases and, therefore, the presence of high levels of ERα in breast epithelium may indicate an increased risk of breast cancer. A fully disposable microfluidic device (μFED) consisting of an array of eight electrodes was constructed and applied to ERα detection in undiluted calf serum. The DNA sequences where ERα binds specifically, known as estrogen response elements, were covalently immobilized via EDC on SCPCEs modified with PDDA. GSH–AuNP and paramagnetic particles (MP) heavily decorated with anti-ERα antibody and HRP (MP–Ab–HRP) were used to efficiently capture the analyte from the sample solution. The formed ERα–MP–Ab–HRP bioconjugate was injected into the μFED and incubated with the DNA-modified electrodes, followed by amperometric detection at −0.2 V vs. Ag|AgCl using the H2O2/HQ system. The achieved LOD was 10.0 fg mL−1.114
Among the various platelet-derived growth factors (PDGFs), PDGF-BB has been found to be directly involved in various cell processes, including tumor growth and progression, and, therefore, serves as an indicator of tumor angiogenesis.115 Fang et al.116 proposed an aptasensing configuration for PDGF-BB determination using a GCE modified with AuNPs and MoS2 to immobilize a large amount of capture aptamers (Apt1), and a tracer consisting of AuNP-labeled ferrocenyl hexanethiol conjugated with a detector aptamer (Fc–AuNPs–Apt2). Using the sandwich configuration and dual signal amplification, a wide linear response in the range of 0.001–10 nM and a LOD of 0.3 pM were obtained. The same group prepared another aptasensor using layered molybdenum selenide–graphene (MoSe2–Gr) composites modified with AuNPs and Exonuclease III (Exo III) to aid signal amplification. MoSe2 is electrocatalytically active, but its relatively low electronic conductivity can be improved by fabricating selenide with a sheet structure and combining it with graphene sheets, resulting in a good substrate for sensor preparation. Furthermore, the addition of AuNPs facilitated the immobilization of biomolecules. Exo III has specific exo-deoxyribonuclease activity toward duplex DNAs in the direction from the 3′ to 5′ termini, but limited activity on duplex DNAs with more than four mismatched terminal bases at the 3′ ends. The aptamer and complementary DNA (cDNA) sequences were designed with four thymine bases on the 3′ ends, such that, when the aptamer associated with the target protein, the duplex DNA was digested by Exo III and released cDNA, which hybridized with signal DNA to perform a new cleavage process. However, as shown in Fig. 6, this process did not occur in the absence of target protein because aptamer hybridization with cDNA inhibited Exo III-assisted nucleotide cleavage. The electrochemical DPV response was obtained using HRP and the H2O2/HQ system, achieving a wide detection range of 0.1 pM to 1 nM and a LOD of 20 fM.117
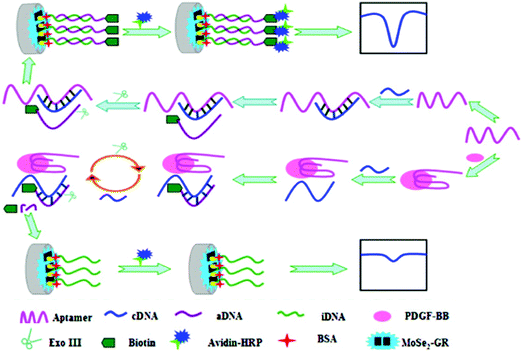 | ||
| Fig. 6 Schematic illustration of aptasensor construction and the fundamentals of PGDF-BB detection using Exonuclease III-assisted signal amplification. Reproduced from ref. 117 with permission. | ||
Aydin et al.118 recently reported a label-free immunosensor for detecting colorectal cancer biomarker p53 protein using an ITO electrode modified with a star-shaped poly(glycidylmethacrylate) (StarPGMA) polymer. This material contains a large number of epoxy ends to link covalently anti-p53 antibodies, providing a high sensitivity in EIS measurements using Fe(CN)63−/4− as redox probe. A 7 fg mL−1 LOD and a linear detection range between 0.02 pg mL−1 and 4 pg mL−1 were achieved. The immunosensor was used to analyze 1000-fold diluted serum samples supplied by a hospital and spiked with known amounts of p53.
Biomarkers for inflammatory and autoimmune diseases are also important in the field of electrochemical biodetection. Notably, interest in cytokine determination is growing owing to their demonstrated relationship with inflammation or disease progression. Such an example is the multifunctional TGF-β1 (transforming growth factor β1) cytokine, whose levels increase in patients with various types of diseases related to inflammatory processes. An electrochemical immunosensor for the determination of TGF-β1 was prepared using multi-walled carbon nanotube (MWCNT)-modified SPCEs. MWCNTs were functionalized by copper(I) catalyzed azide–alkyne cycloaddition (“click” chemistry) to allow covalent immobilization of immunoreagents without altering their configurations and preserving their biological activity. Alkyne-functionalized IgGs were also prepared and used to assemble IgG–alkyne–azide–MWCNT conjugates as scaffolds for immunosensor preparation. After a blocking step with casein, anti-TGF-β1 was immobilized and the target cytokine was sandwiched with biotinylated anti-TGF labeled with poly-HRP labeled streptavidin. The affinity reaction was monitored amperometrically at −0.20 V using the H2O2/HQ system. The calibration plot for TGF-β1 exhibited a range of linearity between 5 and 200 pg mL−1, with a LOD of 1.3 pg mL−1. The immunosensor was applied to the analysis of spiked human serum.119
Also belonging to the cytokine family, tumor necrosis factor α (TNF-α) is a proinflammatory cytokine involved in various biological processes. TNF-α levels have been correlated with rheumatoid arthritis (RA) among other inflammatory diseases. An enzyme-free sandwich-type electrochemical aptasensor was reported for TNF-α detection. The aptasensor involved a SPE modified with a AuNPs/graphene/chitosan nanocomposite, which was proposed as a platform for aptamer immobilization. Furthermore, bimetallic Ag@Pt core–shell nanoparticle-functionalized graphene nanosheets (Ag@Pt–GRs) acted as labels for signal amplification. The modified electrode provided a large surface area, allowing a large aptamer loading, and exhibited electrocatalytic activity toward the analytical signal yielded by catechol oxidation. This electrochemical aptasensor showed a dynamic range from 5.0 pg mL−1 to 70 pg mL−1 with a LOD of 1.64 pg mL−1, and was applied to the analysis of human serum.120 An aptasensor with enhanced performance for the determination of TNF-α was prepared using a hybridization chain reaction (HCR) and target recycling with host–guest interactions for signal probe collection. Fig. 7 shows that a bare GCE was modified with single-walled carbon nanohorns (SWCNHs) and electrodeposited AuNPs for further immobilization of S1 (capture DNA probe) and S2 (TNF-α binding aptamer) probes and blocking with hexanethiol (HT). The resulting electrode was incubated with MB and auxiliary (H1 and H2) probes. Separately, cucurbituril 7 (CB) was deposited onto an AuNPs@chit modified-GCE. For TNF-α determination, the electrode with DNA nanowires was incubated in cytokine solutions containing RecJf exonuclease. After TNF-α was linked to the corresponding aptamer, recycling was initiated, resulting in dissociation of the intercalated MB, while the RecJf exonuclease facilitated further recycling of TNF-α to obtain more dissociated DNA nanowires. With the assistance of duplex-specific nuclease (DSN), the dissociated DNA nanowires with MB in solution released free MB, which was selectively captured by CB in the CB/AuNPs@chit/GCE to produce the electrochemical signal. A wide linear range from 0.001 to 100 ng mL−1 and a LOD of 0.5 pg mL−1 were obtained. The aptasensor was applied to clinical real serum and spiked serum samples.121
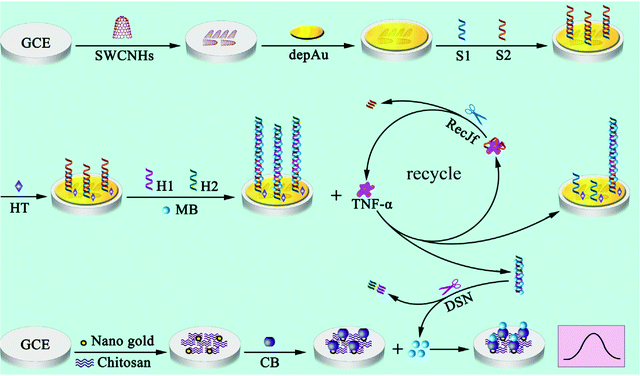 | ||
| Fig. 7 Schematic illustration of the HCR and target recycling enhanced aptasensor for TNF-α with host–guest interactions for signal probe collection. Reproduced from ref. 121 with permission. | ||
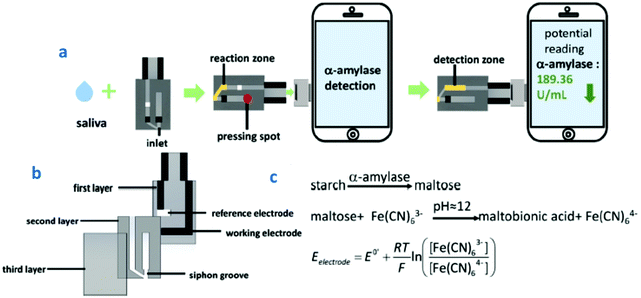 | ||
| Fig. 8 (a) Schematic illustration of HSA detection using a smartphone-based sensor; (b) the three layers of the sensor; and (c) the reaction steps involved in HSA detection. Reproduced and adapted from ref. 124 with permission. | ||
Cortisol is a steroid hormone secreted from the adrenal cortex of the kidney that plays an important role in increasing blood pressure and glucose levels. Cortisol production is known to be a biological response associated with stress. A strong correlation exists between salivary and blood cortisol levels, making the quantification of salivary cortisol an important diagnostic tool. An interesting example of biosensors for salivary determination of cortisol at the POC level is a label-free paper-based electrical chip in which anti-cortisol antibody is immobilized on top of gold microelectrodes using a DTSP SAM. A signal amplification strategy using poly(styrene)-block-poly(acrylic acid) polymer and a graphene nanoplatelet suspension coated on filter paper was implemented. Chip integration with a printed circuit board provided an electrical connection and wireless transmission/reception of the electrical signals using MATLAB.125
Regarding detection of a cancer biomarker, an important example is the determination of cytokine interleukin 8 (IL-8). IL-8 is a multifunctional pro-inflammatory cytokine produced in response to inflammatory stimulation. Furthermore, an increase in IL-8 concentration is associated with melanoma, breast, renal, gastric, ovarian, pancreatic, and colorectal cancers.126 IL-8 is also an important diagnostic biomarker in oral cancer. The simultaneous determination of protein IL-8 and its messenger RNA (IL-8 mRNA) in human saliva has been accomplished using two electrochemical magnetoimmunosensors.16 Functionalized MBs were used to immobilize a specific antibody against IL-8 protein and a specific hairpin DNA sequence for IL-8 mRNA, respectively (Fig. 9). Amperometric detection at disposable dual screen-printed carbon electrodes provided LODs of 0.21 nM for IL-8 mRNA and 72.4 pg mL−1 for IL-8 protein, far below the clinically established cut-off of 600 pg mL−1, in undiluted saliva. This magnetoimmunosensor was used to determine the endogenous content of IL-8 protein in saliva samples from seven healthy individuals, with results statistically in agreement with those provided by a commercial ELISA kit.
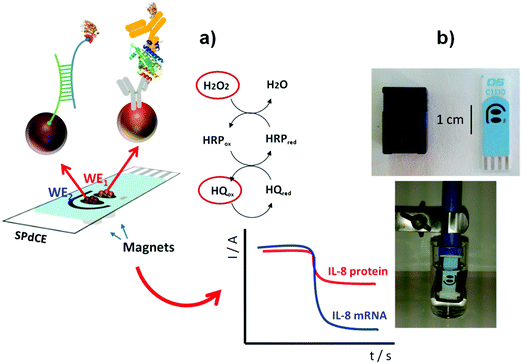 | ||
| Fig. 9 (a) Schematic illustration of dual MB-based biosensor for simultaneous determination of IL-8 protein and IL-8 mRNA. (b) Pictures of a SPdCE and the homemade magnet-holding block (top), and MBs on the SPdCE assembled on the magnet-holding block (bottom). Reproduced and adapted from ref. 16 with permission. | ||
A label-free impedimetric immunosensor has been reported for IL-8 detection in human serum and saliva using a 6-phosphonohexanoic acid (PHA)-modified ITO electrode.127 Good analytical performance with a wide linear range from 0.02 to 3 pg mL−1 and a low LOD of 6 fg mL−1 was achieved. The same authors prepared another impedimetric immunosensor for IL-8 using a disposable ITO electrode decorated with a conductive composite consisting of Super P carbon black, polyvinylidene fluoride, and star polymer (SPGMA) material.128 Anti-IL-8 antibodies were covalently bound to the polymer epoxy groups and, under optimum conditions, a wide linear range of 0.01–3 pg mL−1 and LOD of 3.3 fg mL−1 were obtained. In both cases, the immunosensors were applied to determine the endogenous cytokine in saliva. Recently, Ma et al. reported the preparation of an electrochemical scaffold for DNA species related to Oral Cancer Overexpressed 1 (ORAOV1) in saliva.129 ORAOVI 1 is a candidate proto-oncogene located on 11q13 that plays a functional role in the tumorigenesis of various oral cancers in humans.130 The biosensing process relied on a hybridization recognition followed by homogeneous Exonuclease III (Exo III)-aided target recycling amplification. As discussed above, Exo III is an exodeoxy-ribonuclease that catalyzes the stepwise removal of mononucleotides from 3′-hydroxyl termini of duplex DNA with blunt or recessed 3′-termini, providing an excellent amplification strategy without requiring any specific recognition sequence. Fig. 10(a) shows that the presence of the target DNA initiated homogeneous enzymatic cleavage of the ferrocene-labeled probe DNA (Fc-P1) to remove mononucleotides in a stepwise manner, leading to a highly efficient 1:N target-responsive recycling mechanism and the consumption of multiple Fc-P1 copies. The Fc-P1 consumed was proportional to the amount of target DNA, while residual Fc-P1 was able to hybridize with the methylene blue-labeled hairpin DNA (MB-PP1) on the electrode, resulting in a dual electrochemical signal. The ratiometric readout of both responses led to high sensitivity and eliminated interferences in addition to false-negative and false-positive signals. Notably, the method gave high sensitivity with a linear range of 0.02 pM to 2 nM and a LOD of 12.8 fM for the target DNA. The method was validated in saliva obtained from different individuals after centrifugation.
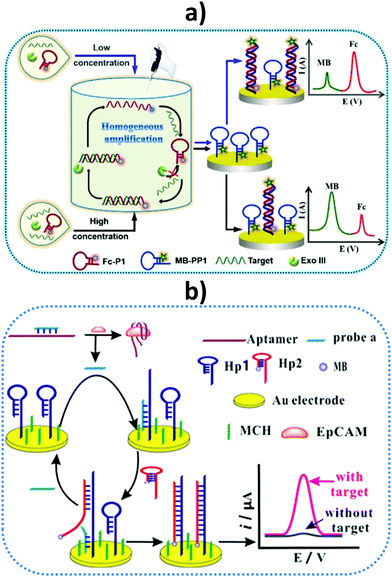 | ||
| Fig. 10 Schematic illustrations of (a) a homogeneous Exo III-assisted target recycling amplification and dual-signal ratiometric electrochemical DNA biosensor, and (b) a sensor design and amplification strategy for the determination of epithelial cell adhesion molecule (EpCAM) and EpCAM-driven toehold-mediated DNA recycling amplification. MB, methylene blue; Fc, ferrocene; MCH, 6-mercapto-1-hexanol. Reproduced from (a) ref. 129 and (b) ref. 131 with permission. | ||
Another interesting method relies on the development of an electrochemical aptasensor for the determination of epithelial cell adhesion molecule (EpCAM). EpCAM is a transmembrane glycoprotein expressed at low levels in most healthy epithelial tissues, but overexpressed in human colon cancer and various carcinomas. Chen et al. designed a strategy for detecting this biomarker by combining the specific binding of EpCAM to the aptamer and EpCAM-driven toehold-mediated DNA recycling amplification.131Fig. 10(b) shows that, upon adding the target analyte, probe (a) was liberated from the complex and hybridized with the toehold domain of Hp1, exposing another toehold domain of Hp1 for further hybridization with Hp2 to form the duplex that brought electroactive reporter MB close to the electrode. Spiked saliva, among other biological samples, was used to demonstrate the usefulness of the aptasensor.131 A microfluidic paper-based electrochemical DNA biosensor was constructed for detecting epidermal growth factor receptor (EGFR) mutations in saliva of patients suffering non-small cell lung cancer (NSCLC). A gold surface was deposited onto the paper working electrode (PWE) and the capture probe was copolymerized with pyrrole (Py) onto the AuNPs/PWE by cyclic voltammetry. The target DNA was mixed with the HRP-labeled detection probe and the electrochemical response was obtained by DPV upon addition of H2O2 and MB.132 Another oral cancer biomarker, the Cytokeratin 19 Fragment (CYFRA-21-1) antigen, is known to be over-secreted in saliva, with levels reaching around tens ng mL−1, and is also correlated with CK19mRNA expression in oral squamous cell carcinoma.133 Using hafnia-modified ITO electrodes, which provided high current changes in CV at different analyte concentrations, a disposable immunosensor was reported for antigen detection involving immobilization of the specific antibody via APTES. The immunosensor was applied to real saliva samples from oral cancer patients, with results validated against colorimetric ELISA.134
Human fetuin A (HFA) is a relevant biomarker for pancreatic and liver cancers, inflammatory processes, and obesity and related diseases. SPCEs modified with grafted p-aminobenzoic acid and streptavidin were used as scaffolds for the preparation of an electrochemical immunosensor for HFA determination. Biotinylated capture antibodies were immobilized and a sandwich assay configuration was implemented using magnetic MWCNTs conjugated with HRP and anti-HFA antibodies as the detection labels for signal amplification. HFA determination was accomplished between 20 and 2000 pg mL−1, with a LOD value of 16 pg mL−1. The usefulness of the immunosensor was demonstrated by HFA analysis in saliva with minimal sample treatment.23 The same group also prepared a MBs-based immunosensor for human interleukin-6 (IL-6) detection. IL-6 is a pleiotropic cytokine encoded in humans by the IL-6 gene, which has a leading role in the inflammatory response, and constitutes a suitable biomarker for prostate cancer or head and neck squamous cell carcinoma (HNSCC).135 Interestingly, elevated IL-6 concentrations in saliva were also found in patients with oral neoplastic and preneoplastic lesions and, therefore, IL-6 can be used as a biomarker for the early detection and screening of tongue cancer.136 The reported strategy involved covalent immobilization of anti-IL-6 antibodies onto carboxyl-functionalized magnetic microparticles and a sandwich-type immunoassay with signal amplification using poly-HRP–streptavidin conjugates. Important features to note are the low LOD of 0.39 pg mL−1 and the high storage stability of the anti-IL-6–MBs immunoconjugates (36 days). This immunosensor was successfully applied to determination of the endogenous content of IL-6 in three different saliva samples corresponding to a periodontitis patient, a smoker volunteer, and a non-smoker volunteer, with results agreeing statistically with those obtained by a commercial ELISA kit.137
Cytokine TNF-α is involved in vascular dysfunction and cardiac diseases, such as heart failure (HF).138 The pathophysiology of HF is exceedingly complex, and HF patients are also characterized by systemic inflammation, highlighted by raised circulating levels of several inflammatory cytokines such as TNF-α, with the increase correlated to the degree of disease severity.139 The TNF-α concentrations in saliva reflect those in serum, making this cytokine an ideal HF-related salivary biomarker. A fully integrated electrochemical biosensor platform for TNF-α was reported using eight gold working microelectrodes, where the specific capture anti-TNF-α antibodies were immobilized through functionalization with carboxyl diazonium. Electrochemical impedance spectroscopy was used to determine the cytokine in the linear range of 1–100 pg mL−1, which were the critical concentrations for patients suffering HF.140 Recently, the same team described an immunosensor for TNF-α involving the immobilization of antibodies onto gold electrodes modified with 4-carboxymethylaniline (CMA) and a sandwich-type configuration with amperometric detection. A linear range of 1–30 pg mL−1 and a LOD of 1 pg mL−1 were reported.141 Both immunosensors were applied to TNF-α determination in saliva from healthy individuals after dilution and spiking.
Our group proposed using dual SPCEs modified with 4-carboxyphenyl-functionalized double-walled carbon nanotubes as scaffolds for the preparation of electrochemical immunosensors for simultaneous determination of TNF-α and interleukin-1β (IL-1β) cytokines. Interestingly, both proteins can be used as biomarkers to predict side effects of cancer therapy, such as inflammatory oral mucositis.142 Furthermore, determination of the salivary contents of these cytokines in patients with leucoplakia and oral cancer is of great interest and they have recently been considered as novel biomarkers for the detection of periodontal diseases.143 The dual configuration involved the oriented immobilization of capture antibodies onto HOOC-Phe-DWCNTs/SPCEs, making use of commercial polymeric coating Mix&Go. Sandwich-type immunoassays were implemented with amperometric signal amplification using poly-HRP streptavidin conjugates and the H2O2/HQ system. The linear ranges extended to 1–200 pg mL−1 and 0.5–100 pg mL−1 for TNF-α and IL-1β, respectively, which were adequate for cytokine determinations in clinical samples. The LODs were 0.85 pg mL−1 (TNF-α) and 0.38 pg mL−1 (IL-1β). This dual immunosensor was applied to the simultaneous determination of both proteins in real saliva samples from smoker and non-smoker male and female volunteers, with the results in agreement with those obtained using an ELISA kit.19
Saliva is also the sample of choice for detecting AMI, which is among the most immediately life-threatening types of acute coronary syndromes. Following myocardial damage, the troponin complex is broken and proteins, including cTnI, are released into the bloodstream, appearing in saliva at concentrations 0–100-fold lower than in serum. A label-free aptasensor was prepared with nitrogen-doped reduced graphene oxide (N-prGO) modified with 1-pyrenecarboxylic acid (py-COOH) and poly(ethylene glycol)-modified pyrene (py-PEG), allowing covalent integration of the Tro4 aptamer and DPV quantification of cTnI. The fundamental details of this biosensor are shown in Fig. 11. With a linear range of 0.001–100 ng mL−1 and LOD value of 1 pg mL−1, the aptasensor exhibited the high sensitivity required for application to real saliva samples from AMI-diagnosed patients.144
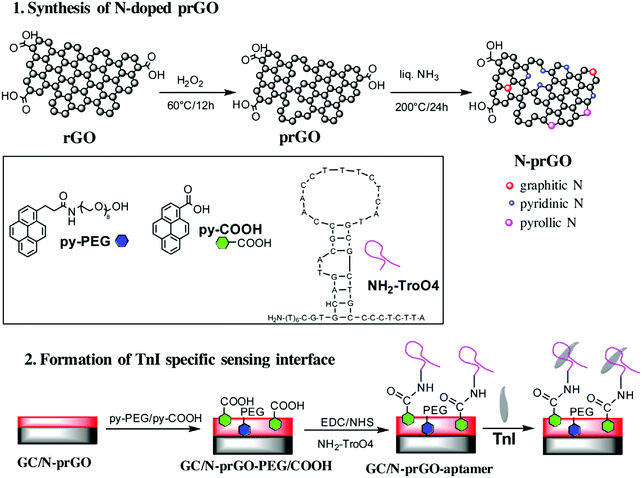 | ||
| Fig. 11 Schematic illustration of the synthetic strategy for N-prGO and the preparation of an immunosensor for cTnI. Reproduced from ref. 144 with permission. | ||
In addition to the types of biosensor discussed above, enzyme biosensors have also been used for the determination of some biomarkers in saliva. Specifically, monitoring lactate in saliva is important for diverse biomedical and fitness monitoring activities. Wang and coworkers designed a wearable saliva metabolite biosensor by integrating a printable lactate oxidase electrode on a mouthguard to detect the peroxide product, which provided real-time information.145 Amperometric measurements of lactate were performed in connection with a Prussian blue (PB)–poly-o-phenylenediamine system, where PB acted as an artificial peroxidase, allowing a low potential to be used for selective H2O2 detection. Based on a similar design, a disposable biosensor for analyzing lactate in saliva was constructed by Palleschi et al.146 The developed biosensor was used to analyze real saliva samples after treatment with tris(2-carboxyethyl)phosphine hydrochloride (TCEP) for mucoprotein precipitation.
A recently identified biomarker for bladder cancer, Apo-A1 protein, is present at high levels in urine at the early stage of this neoplasm.150 An electrochemical immunoassay platform on ITO glass strips has been recently reported for the determination of Apo-A1. ITO electrodes were modified with avidin to immobilize biotinylated-anti-Apo-A1 and a sandwich-type immunoassay was implemented using a detector antibody labeled with alkaline phosphatase. The electrochemical readout was obtained by dropping L-ascorbic acid-2-phosphate (AAP) and measuring the oxidation of L-ascorbic acid obtained after the enzyme reaction. Interestingly, chronocoulometry was used as the electrochemical technique using data recorded at 50 s from the chronocoulograms recorded at +0.45 V. The developed method allowed Apo-A1 determination in urine over a wide range of concentrations between 1 pM and 100 nM.151
Systemic lupus erythematosus (SLE) is an autoimmune disease affecting multiple organs including kidneys.152 Vascular cell adhesion molecule-1 (VCAM-1) is a biomarker useful as an indicator of renal disease in SCL because its levels, correlated to higher renal activity, are significantly higher in lupus nephritis. A label-free impedimetric immunosensor was developed by Selvam et al. for VCAM-1 detection.153 The immunosensor consisted of a sandwich-type configuration, providing a calibration plot in a dynamic range between 8 fg mL−1 and 80 pg mL−1, and was validated with a blinded cohort of 12 patient urine samples after a 5000-fold dilution. The semiquantitative results were compared successfully with those obtained by an ELISA test.
The use of whole blood for direct analysis is an emerging trend in biosensing research. Blood is a particularly complex mixture composed of proteins, glucose, inorganic salts, hormones, and other substances. Significant advances have been made in biomarker determination in whole blood.154 An important example is the development of an immunosensor for the determination of α-fetoprotein (AFP), an important biomarker for the early diagnosis of liver cancer. This biosensor involved the modification of a GCE surface with heparin (Hep), γ-polyglutamic acid (PGA), and polypyrrole (PPy) (Hep–PGA–PPy) nanoparticles. Combining the inherent conductivity of PPy and the biocompatibility of Hep, the Hep–PGA–PPy nanoparticles improved not only the antibiofouling properties of the surface, but also the electrochemical properties of the immunosensor. The capture antibody was immobilized on the modified GCE surface and, after conjugation to AFP antigen, DPV measurements using Fe(CN)64− as the electrochemical probe allowed AFP detection in a linear range of 0.1–100 ng mL−1 with a LOD of 0.099 ng mL−1. Furthermore, AFP detection in five human blood samples showed satisfactory accuracy.155
Spinal muscular atrophy (SMA), cystic fibrosis (CF), and Duchenne muscular dystrophy (DMD) are well-known progressive hereditary disorders associated with increased morbidity and mortality. Rapid detection of biomarkers for these diseases in newborns offers new opportunities for early diagnosis and effective treatment. A disposable carbon nanofiber-based electrochemical immunosensor for the simultaneous detection of survival motor neuron 1 (SMN1), cystic fibrosis transmembrane conductance regulator (CFTR), and DMD proteins has been reported. The electrode array was functionalized by electroreduction of carboxyphenyl diazonium salt and the corresponding antibodies were covalently immobilized. LODs of 0.9, 0.7, and 0.74 pg mL−1 were achieved for CFTR, DMD, and SMN1, respectively. High recoveries were obtained when the immunosensor was applied to analyze whole blood samples from volunteers. The samples were subjected to a single freeze–thaw cycle, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 diluted with 10× RIPA buffer to lyse the red blood cells, diluted 1
40 diluted with 10× RIPA buffer to lyse the red blood cells, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS buffer (pH 7.4), and finally spiked with SMN1, CFTR, and DMD, using only a few drops of blood.156
100 in PBS buffer (pH 7.4), and finally spiked with SMN1, CFTR, and DMD, using only a few drops of blood.156
4.2. Electrochemical biosensing in cells
The main electrochemical strategies described so far for biosensing in cells (mainly cancer cells owing to their great relevance) include determination in extracted genomic material (mainly total RNA or genomic DNA), raw cell lysates, and whole cells through specific extracellular protein receptors. These methods are summarized in Table 2.| Electrode | Type of biosensor/format | Bioreceptor/s | Biomarker (disease) | Cell/type of sample (lysate amount, μg) | Electrochemical technique | L.R.b | LODb | Assay time | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Abbreviations: AuNPs: Au nanoparticles; CDH-17: cadherin-17; cDNA: complementary DNA; ERα: estrogen receptor α; FGFR4: fibroblast growth factor receptor 4; HCR: hybridization chain reaction; HER2: human epidermal growth factor receptor 2; IL-13sRα2: interleukin-13 α2 receptor; MBs: magnetic microbeads; 5-mC: 5-methyl-cytosine; PR: progesterone receptor; rGO–CMC: reduced graphene oxide–carboxymethylcellulose hybrid nanomaterial; RNAt: total RNA; SPCE: screen-printed carbon electrode.a After the solid supports (MBs or AuNPs-SPCEs) were modified with the capture bioreceptor and blocked.b Results for standards and synthetic target nucleic acids (DNAs or miRNAs). | |||||||||
| Determination in genomic material extracted from cells | |||||||||
| rGO-CMC-modified SPCE | DNA biosensor | Hairpin DNA probe | Wild-type TP53 (cancer) | MCF-10A, MCF-7 and SK-BR-7/cDNA (50 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.01–0.1 μM | 2.9 nM (29 fmol) | 45 mina | 13 |
| SPCE | Affinity biosensor onto Chitin-MBs | p19 protein (capture) and biotinylated synthetic RNA probe (detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (500 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.14–10.0 nM | 40 pM (0.4 fmol) | 105 mina | 164 |
| SPCE | Affinity biosensor onto Strep–MBs | Biotinylated synthetic RNA probe (capture) and p19 protein (detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (500 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 1.4–10.0 nM | 0.42 nM (4.2 fmol) | 45 min | 165 |
| SPCE | Affinity biosensor onto ProtG-MBs | Anti-RNA/DNA antibody (capture) and biotinylated synthetic DNA probe (detector) | miRNA-205 (tumor suppressor) | MCF-10A, MCF-7/RNAt (50 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 8.2–250 pM | 2.4 pM (60 amol) | 120 mina | 167 |
| SPCE | HCR-sandwich hybridization DNA biosensor onto Strep–MBs | Synthetic DNA probes (biotinylated: capture and unmodified: detector) and 2 additional biotinylated DNA probes (for HCR) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (500 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.2–5.0 nM | 60 pM (1.5 fmol) | 45 mina | 61 |
| SPCE | Affinity biosensor onto Strep–MBs | Biotinylated synthetic DNA probe (capture) and anti-RNA/DNA antibody (detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (250 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 1.0–100 pM | 0.4 pM (10 amol) | 30 mina | 168 |
| AuNPs-SPCE | Competitive hybridization RNA/RNA biosensor | Thiolated synthetic RNA probe (capture) and biotinylated RNA probe (detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (50 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.1–25 pM | 25 fM (0.25 attomol) | 75 mina | 22 |
| SPCE | Competitive hybridization DNA/RNA biosensor onto Strep–MBs | Biotinylated synthetic DNA probes (capture and detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (1000 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.7–10.0 nM | 0.2 nM (5 fmol) | 120 mina | 166 |
| AuNPs-SPCE | Affinity sensor | Thiolated synthetic RNA probe (capture) and p19 protein (detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (50 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.5–50 pM | 142 fM (1.42 attomol) | 60 mina | 34 |
| AuNPs-SPCE | Affinity sensor | Thiolated synthetic DNA probe (capture) and anti-RNA/DNA antibody (detector) | miRNA-21 (oncogen) | MCF-10A, MCF-7/RNAt (10 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.096–25 pM | 29 fM (0.29 attomol) | 90 mina | 35 |
| SPCE | DNA sensor onto Strep–MBs | Biotinylated synthetic DNA probe (capture) and anti-5-mC antibody (detector) | 5-mCs in DNA (glioblastoma) | U87 and HeLa/genomic DNA (100 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 90–2500 pM | 26 pM (0.6 fmol) | 60 mina | 27 |
| SPCE | Immuno-DNA sensor onto HOOC-MBs | Anti-5-mC antibody (capture) and biotinylated synthetic DNA probe (detector) | 5-mCs in DNA (glioblastoma) | U87 and HeLa/genomic DNA (100 ng) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 3.9–500 pM | 1.2 pM (30 amol) | 45 mina | 28 |
| Determination in cellular lysates | |||||||||
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and biotinylated detector antibodies | PR (breast cancer) | MCF-7, MDA-MB-436 and SK-BR-3/raw cell lysate (2.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 73–1500 pg mL−1 | 22 pg mL−1 | 130 mina | 172 |
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and biotinylated detector antibodies | ERα (breast cancer) | MCF-7, MDA-MB-436, SK-BR-3 and BxPC3/raw cell lysate (2.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 63–2000 pg mL−1 | 19 pg mL−1 | 130 mina | 53 |
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and HRP-conjugated detector antibodies | Human p53 | BxPC3, MCF-7, KM12SM, SW620, KM12C, MDA-MB-436 and SW480/raw cell lysate (2.0 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 5–150 ng mL−1 | 1.29 ng mL−1 | 30 mina | 173 |
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and biotinylated detector antibodies | FGFR4 (cancer) | MCF-7, MDA-MB-436, SK-BR-3, KM12C, KM12SM, SW480, SW620, BxPC3/raw cell lysate (2.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 160.6–7500 pg mL−1 | 48.2 pg mL−1pg mL−1 | 15 mina | 17 |
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and HRP-conjugated detector antibodies | CDH-17 (gastric, hepatocellular, and colorectal tumors) | SW480, SW620, KM12C and KM12SM/raw cell lysate (0.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 4.76–1000 ng mL−1 | 1.43 ng mL−1 | 15 mina | 32 |
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and biotinylated detector antibodies | E-cadherin (cancer) | MCF-7, SK-BR-3, MDA-MB-436, SW480, SW620, KM12C and KM12SM/raw cell lysate (0.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.50–25 ng mL−1 | 0.16 ng mL−1 | 75 mina | 33 |
| Determination in whole cells | |||||||||
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and HRP-conjugated detector antibodies | HER2 (breast cancer) | MDA-MB-436, MCF-7 and SK-BR-3/whole and lysed cells (2.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.1–32.0 ng mL−1 | 26 pg mL−1 | 60 mina | 11 |
| SPCE | Sandwich immunosensor onto HOOC-MBs | Specific capture and biotinylated detector antibodies | IL-13Rα2 | SW480, SW620, KM12C and KM12SM/whole and lysed cells (0.1–2.5 μg) | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 3.9–100 ng mL−1 | 1.2 ng mL−1 | 75 mina | 31 |
Esteban-Fernández de Ávila et al.13 constructed disposable electrochemical DNA sensors for the detection of a specific target DNA sequence within the p53 tumor suppressor gene (TP53). The electrochemical platforms consisted of SPCEs functionalized with a water-soluble reduced graphene oxide–carboxymethylcellulose hybrid nanomaterial. An amino-terminated hairpin specific DNA capture probe was covalently immobilized through carbodiimide chemistry onto the nanostructured platforms. Using Strep–HRP conjugate as an electrochemical indicator, the hybridization reaction was monitored by recording the amperometric responses obtained upon adding 3,3′,5,5′-tetramethylbenzidine (or TMB) as redox mediator and H2O2 as enzyme substrate, leading to a typical “on–off” change resulting from the markedly increased distance between the attached enzymatic conjugate and the electrode surface, which hindered electron transfer. The synthetic target sequence corresponded to a region of the wild type TP53 gene. The electrochemical bioplatform allowed a LOD of 2.9 fmol to be achieved for the target sequence, and showed a 15 day storage stability at 4 °C, attractive non-fouling properties in untreated human serum and saliva samples, and complete discrimination between perfectly matched and single-base mismatched DNA (a mutant genotype containing a single nucleotide change at codon 175, leading to cancer-triggering deactivation of this tumor suppressor protein). Importantly, the developed method was applied to analyze the endogenous TP53 status in total RNA (previous synthesis of cDNA by reverse transcriptase) extracted from different human breast cell lines, namely, one epithelial nontumorigenic (MCF-10A) and two cancer cell lines (MCF-7 and SK-BR-3). The obtained results agreed with the TP53 gene status reported for the three cell lines, confirming the usefulness of the constructed bioplatform for this purpose.
Several electroanalytical bioplatforms have been reported for PCR-free determination of miRNAs in total RNA extracted from cells. Owing to the relatively simple handling and integration into a portable device, short assay time, and excellent analytical performance, the strategies based on using commercial bioreceptors with a high affinity for RNA duplexes,48 competitive hybridization,22,168 or easily implemented amplification methods, such as the hybridization chain reaction (HCR),61 are highlighted in this section. These strategies involve specific DNA or RNA probes coupled to novel commercial bioreceptors used for selective capturing or labeling of the corresponding hybrids. The bioreceptors included viral protein p19, which is highly selective for RNA/RNA homohybrids of short length,34,164,165 or commercial antibodies (anti-RNA/DNA) with high specificity towards DNA/RNA heterohybrids.35,166,167 The methods were all implemented either on the surface of MBs coupled with disposable electrodes to perform electrochemical transduction,61,164–168 or directly as integrated configurations on the surface of commercial AuNPs nanostructured disposable electrodes.22,34,35 The methodologies exhibited attractive analytical characteristics for the determination of target miRNAs (Table 2) and were successfully applied to the reliable determination of miRNAs endogenous content in total RNA samples (amounts of 10–1000 ng) extracted from non-cancerous human breast epithelial (MCF-10A) and human breast cancer (MCF-7) cells without previous amplification or preconcentration of the genetic material. Importantly, in contrast with qRT-PCR, these electrochemical bioplatforms allow direct determination of the target miRNA in raw RNAt extracted without reverse transcription into cDNA or internal controls, such as house-keeping genes, to alleviate the variability of PCR amplification,169 which implies significantly lower costs and shorter analysis times.
Pingarrón and coworkers recently developed two PCR-free amperometric biosensing strategies to detect the presence of 5-mCs in the promoter region of the O6-methylguanine-DNA methyltransferase (MGMT) tumor suppressor gene without conventional methylated DNA bisulfite or amplification pretreatments.27,28 One strategy (DNA sensor, strategy 1 in Fig. 12(a)) involved immobilization of a biotinylated DNA capture probe, specific to the methylated sequence to be detected, on the surface of streptavidin-modified micromagnetic particles (Strep–MBs). Methylations in the captured DNA were detected using a 5-mC specific antibody (anti-5-mC), which was recognized with a secondary HRP-conjugated antibody.27 The second strategy (immuno-DNA sensor, strategy 2 in Fig. 12) implied capturing of methylated DNA with anti-5-mC antibodies that were covalently immobilized on modified HOOC-MBs. The methylated DNA was selectively detected with a specific biotinylated DNA probe conjugated with a commercial Strep–HRP polymer.28 In both strategies, the resulting magnetic bioconjugates were magnetically captured on the surface of the screen-printed electrodes and amperometric transduction was conducted through the H2O2/HQ system. The biosensing platforms showed good measurement reproducibility and a wide linear range, allowing determination of the methylated sequence of the MGMT promoter region at the picomolar level (see data in Table 2). Furthermore, these platforms were applied to determination of the methylation status in the promoter region of the MGMT gene using 100 ng of genomic DNA extracted and fragmented with ultrasound from U87 and HeLa cells. Fig. 12(b) shows that amperometric responses significantly different to the blank were only obtained for DNA extracted from U87 cells. These results were consistent with specific hypermethylation of the MGMT promoter in these human glioblastoma cells. As methylation in MGMT promoter is considered a useful predictor of malignant glioma patient response to the action of alkylating agents, these pioneering applications confirmed the usefulness of the developed bioplatforms for therapeutic action.170,171
 | ||
| Fig. 12 (a) Schematic illustration of strategies developed for the PCR-free determination of 5-mCs in DNA (DNA sensor, 1, and immuno-DNA sensor, 2), and (b) amperometric responses provided by the DNA sensor in the absence of target DNA (blank) and in the presence of 100 ng of genomic DNA extracted and fragmented with ultrasound from U87 and HeLa cells. Error bars are three times the standard deviation of three replicates. Reproduced and adapted from ref. 27 and 28 with permission. | ||
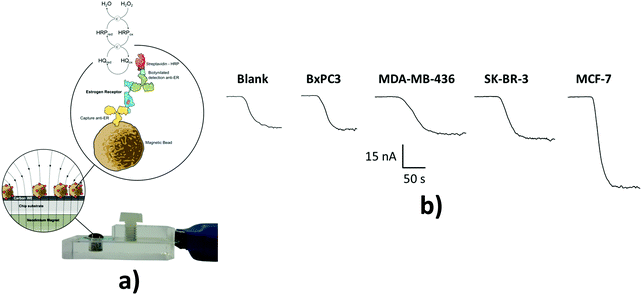 | ||
| Fig. 13 (a) Schematic illustration of MB-based sandwich immunosensor developed for ERα determination. (b) Amperometric responses recorded in the absence (blank) and presence of 2.5 μg of raw lysates from the corresponding cells. Reproduced and adapted from ref. 53 with permission. | ||
 | ||
| Fig. 14 Photomicrographs obtained for the sandwich immunoassays conducted with cell suspensions (1.0 × 106 cells mL−1) on MBs unmodified and modified with the capture antibody (CAb). Reproduced and adapted from ref. 31 with permission. | ||
4.3. Electrochemical biosensing in solid biopsies (fresh and paraffin-embedded tissues)
Electrochemical biosensors have also been used for the simple, reliable, and accurate determination of biomarkers at different molecular levels in fresh,61,164,165,167,168 and formalin-fixed paraffin-embedded (FFPE)18,27,28,32,33 human tissues. The developed biosensors were used to determine miRNAs in RNAt extracted from fresh and FFPE tissues of patients diagnosed with breast cancer,61,164,165,167,168 5-mCs in denatured genomic DNA extracted from paraffin-embedded brain tumor tissues from patients diagnosed with glioblastoma,27,28 and protein biomarkers related to metastatic processes proposed as oncogene (CDH-1732) and tumor suppressors (E-cadherin33) directly in protein extracts from paraffin-embedded colon cancer tissues with different metastatic grades. Fig. 15 shows fundamental details of the immunosensor developed for the determination of CDH-17 and amperometric traces recorded in tumor (T) and paired healthy (N) tissue extracts from five patients diagnosed with colorectal cancer at different stages. The assay time for the determination in the tissue samples was between 1532 and 12018,167 min. Determinations of miRNAs, 5-mCs, and protein markers were feasible with raw RNAt amounts between 100 and 1000 ng RNAt, 100 ng genomic DNA, and just 0.5 μg of tissue protein extract, respectively. Interestingly, electrochemical bioplatforms developed for the determination of miRNAs exhibited similar analytical performance in both fresh-frozen and FFPE breast tissue samples. The results obtained with FFPE tissue samples are of great interest owing to the benefits of using this type of sample in clinical research for the diagnosis and therapeutic follow-up of a wide range of cancers and other human pathologies (such as inflammation and immune-related diseases). As FFPE is the standard tissue processing method in pathology departments for cancer diagnosis, the possibility of analyzing the vast amount of available samples housed in hospitals, clinics, and research facilities should allow rapid advanced research into disease diagnostics, outcomes, and therapies involving different candidate biomarkers.18 | ||
| Fig. 15 (a) Schematic illustration of the immunosensor developed for CDH-17 determination. Illustrative examples of amperograms recorded with the immunosensor of 0.5 μg of protein extracts obtained from a tumor (T) and paired healthy tissue (N), and CDH-17 concentrations obtained in the analyzed tissue extracts. Error bars are three times the standard deviation of three replicates. Reproduced and adapted from ref. 32 with permission. | ||
5. Electrochemical multiplexing in challenging samples
Multiplexing with electrochemical platforms has been achieved using multielectrode arrays or barcode configurations (which involve a single electrode platform and labels with differentiated electrochemical behavior for each analyte).57 Similar to electrochemical platforms designed for the determination of a single analyte, the required sensitivity and selectivity have been achieved in multideterminations through appropriate use of single or hybrid nanomaterials as surface electrode modifiers or tags,57 or in connection with isothermal nucleic acid amplification strategies.24,63 A rational design of platforms involving surface chemistry through diazonium salt grafting has also been exploited to develop multiplexing immunoplatforms with excellent analytical performance.15,174Electrochemical bioplatforms with excellent analytical characteristics have been reported for the multidetermination of biomarkers of the same or different molecular levels, allowing matching of the clinically relevant concentration ranges of target analytes. Yáñez-Sedeño et al.57 recently reviewed the applications of electrochemical immunosensors to the multidetermination of protein biomarkers with relevance in cancer, cardiovascular, infectious, and autoimmune diseases, metabolic disorders, inflammation processes, and apoptosis in liquid biopsies (mainly serum, but also plasma and saliva), tumor cells,172,175 and cell culture supernatants.8 None of the reported bioplatforms showed “cross-talk” between the adjacent transduction elements and used different redox tags with sufficiently separated detection potentials. Furthermore, in general, while configurations based on multi-electrode arrays used disposable electrodes, those based on barcode configurations involved conventional electrodes (mostly GCE and more scarcely AuE). Importantly, some of these bioplatforms were integrated into chips176,177 or microfluidic devices,178–180 and even fabricated on paper substrates.181–183 Interestingly, some of the reported strategies were able to simultaneously determine serum biomarkers with clinically relevant cut-off concentrations differing in three orders of magnitude, such as CRP and NT-proBNP with recommended clinical thresholds in serum/plasma samples of 1000 and 1 ng mL−1, respectively,10 overcoming the limitations of conventional methodologies for performing these challenging determinations. Furthermore, different immunoassay formats (sandwich and indirect competitive immunoassays) could be used for each biomarker on the same platform.10Fig. 16 shows representative examples of multiplexed electrochemical immunoplatforms for the determination of relevant biomarkers in saliva and serum using multielectrode arrays or barcode configurations.
 | ||
| Fig. 16 Multiplexed electrochemical sandwich immunoplatforms using (a) multielectrode arrays or (b) barcode configurations. Simultaneous amperometric determination of TNF and IL-1β at a SPdCE modified with 4-carboxyphenyl-functionalized double-walled carbon nanotubes (HOOC-Phe-DWCNTs) and polymeric coating Mix&Go to immobilize the capture antibodies in an oriented manner (a). Schematic illustration of the bioplatform developed for the simultaneous SWV determination of IL-6 and IL-7 using polystyrene spheres (PS) containing Cd2+ or ferrocene (Fc) as tags (b, left). SWV traces recorded for simultaneous determination of increasing concentrations of IL-6 and IL-17 standards (b, right; from (a) to (e): 5, 50, 100, 500, 1000 pg mL−1). Reproduced and adapted from (a) ref. 19 and (b) ref. 184 with permission. | ||
To our knowledge, the use of electrochemical platforms for multiplexed determination of biomarkers of genetic nature or different molecular levels has yet to be reviewed. Table 3 summarizes the main analytical characteristics of these multiplexed bioplatforms reported to date.
| Electrode | Type of biosensor/format | Bioreceptor/s | Biomarker (disease) | Sample | Electrochemical technique | L.R.b | LODb | Assay timea | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Abbreviations: 5-hmC: 5-hydroxymethylcytosine; 5-mC: 5-methylcytosine; HSIL: squamous intraepithelial lesions; HCR: hybridization chain reaction; IL-1β: interleukin-1β; IL-8: interleukin-8; MBs: magnetic beads; LAMP: loop-mediated amplification; SPdCE: screen-printed dual carbon electrode.a After the solid supports (MBs or electrode array) were modified with the capture bioreceptor and blocked.b Results for protein standards and synthetic target nucleic acids (DNAs or miRNAs). | |||||||||
| SPdCE | Sandwich immunosensors (global level), immuno-DNA sensors (gene-specific level) onto MBs | Anti-5-mC, anti-5-hmC and specific DNA probes | 5-mC, 5-hmC (cancer) | Serum and genomic DNA extracted from cells and paraffin-embedded tissues | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | Global: 14–2500 pg 5-mC positive control, (0.04–0.55)% 5-hmC (per 100 ng ssDNA) | Global: 4.0 pg 5-mC positive control, 0.004% 5-hmC (per 100 ng ssDNA) | 45 min (global), 90 min (gene-specific level) | 185 |
| Gene-specific: 4.0-250 pM (5-mC), 1.44-100 pM (5-hmC) | Gene-specific: 1.2 pM (30 amol, 5-mC), 0.43 pM (11 amol, 5-hmC) | ||||||||
| SPdCE | Direct hybridization combined with LAMP onto MBs | Specific DNA probes | HPV16, HPV18 (cancer) | DNA extracted from cervival smears | Chronomperometry (Eapp = −0.30 V vs. Ag pseudoreference electrode) | 0.6–50.0 ng DNA | 0.1 ng DNA | 75 min (+LAMP) | 63 |
| SPdCE | Direct hybridization and selective recognition by viral protein p19-MBs | p19 and specific RNA probes | miRNA-21, miRNA-205 (cancer) | RNAt extracted from breast cancer cells, tissues | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 2.0–10.0 nM (both miRNAs) | 0.6 nM (6 fmol, both miRNAs) | 120 min | 186 |
| Screen-printed carbon electrode array of 8 electrodes | Direct hybridization combined with HCR onto MBs | Specific DNA probes | miRNA-21, miRNA-31, miRNA-let7a (cancer) | RNAt extracted from cancer cells and cervical smears fom patients diagnosed with HSIL | Chronomperometry (Eapp = −0.30 V vs. Ag pseudoreference electrode) | 1.2–100 pM (miRNA-21) | 0.66 nM (33 amol, miRNA-21) | 120 min | 24 |
| 16 integrated Au electrode arrays | Direct hybridization (IL-8 mRNA), sandwich immunosensor (IL-8 protein) onto the Au array modified with conducting polymer and streptavidin modified dendrimer nanoparticles | Hairpin specific probe (IL-8 mRNA), antibodies (IL-8 protein) | IL-8 mRNA, IL-8 protein (cancer) | Saliva samples from oral cancer patients | Amperometry (Eapp = −0.20 V vs. Au pseudoreference electrode) | 5 fM–50 pM (IL-8 mRNA), 10–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 pg mL−1 (IL-8 protein) 500 pg mL−1 (IL-8 protein) |
3.9 fM (IL-8 mRNA), 7.4 pg mL−1 (IL-8 protein) | ∼10 min | 187 |
| 16 integrated Au electrode arrays | DNA dendrimer and polypyrrole | IL-8 mRNA, IL-8 protein, IL-1β protein (cancer) | — | Amperometry (Eapp = −0.20 V vs. Au pseudoreference electrode) | 10 aM–10 pM (IL-8 mRNA), 200–4000 fg mL−1 (IL-8 protein) 100–4000 fg mL−1-(IL-1β protein) | 10 aM (IL-8 mRNA), 200 fg mL−1 (IL-8 protein), 100 fg mL−1 (IL-1β protein) | 60 min | 188 | |
| SPdCE | Direct hybridization (IL-8 mRNA), sandwich immunosensor (IL-8 protein) onto MBs | Hairpin specific probe (IL-8 mRNA), antibodies (IL-8 protein) | IL-8 mRNA, IL-8 protein (cancer) | Undiluted saliva samples | Amperometry (Eapp = −0.20 V vs. Ag pseudoreference electrode) | 0.32–7.5 nM (IL-8 mRNA), 87.9–5000 pg mL−1 (IL-8 protein) | 0.10 nM (IL-8 mRNA), 26.4 pg mL−1 (IL-8 protein) | 70 min (IL-8 mRNA) 105 min (IL-8 protein) | 16 |
The multidetermination of biomarkers of genetic nature was conducted with bioplatforms using modified MBs and electrochemical detection at screen-printed dual carbon electrodes or at integrated carbon or Au electrode arrays. MB-based bioplatforms were satisfactorily applied to the simultaneous determination of 5-mC and 5-hmC, both at global and gene specific levels in the promoter region of two different tumor suppressor genes (RASSF1A and MGMT)185 and the most oncogenic high-risk of human papillomavirus (HPV) types, HPV16 and HPV18, in DNA extracted from cell lines and cervical smears from women suffering the most severe grade of squamous intraepithelial lesions (HSIL).63 Furthermore, other MB-based nucleic acid biosensors were used for simultaneous determination of two different miRNAs (miRNA-21 and miRNA-205) in RNAt extracted from breast cancer cells and tissues,186 or three miRNAs (miRNA-21, miRNA-31 and miRNA-let7a) in RNAt extracted from cancer cells and cervical precancerous lesions of women diagnosed with HSIL.24 The same bioplatforms mentioned for multiplexed determination of miRNA-21 and miRNA-205186 were also useful for evaluating the potential action and mechanism of a new drug as an antineoplastic agent for breast cancer stem cells. The obtained results (unpublished) showed that the new drug modulated the expression of both miRNAs, reducing the oncogenic (miRNA-21) and increasing the tumor suppressor (miRNA-205) expression in RNAt extracted from breast adenocarcinoma cells (MCF-7) at non-cytotoxic doses. Notably, to achieve the sensitivity required for the target biomarkers, some of the developed strategies combined electrochemical detection with isothermal amplification methods of nucleic acids other than target DNA, such as loop-mediated amplification (LAMP)63 or hybridization chain reaction (HCR),24 for easy implementation in routine and decentralized analyses.
Of particular relevance is the simultaneous determination of biomarkers of different molecular levels in a single assay to provide highly accurate diagnostic tools.2 For example, electrochemical biosensing platforms developed both in integrated formats187,188 and MB-based approaches16 were proposed for the simultaneous determination of protein IL-8 and its messenger RNA (IL-8 mRNA), which are biomarkers associated with salivary oral cancer. These bioplatforms were able to determine with great sensitivity and selectivity both biomarkers in saliva supernatants in just 10 min187 and directly in untreated human saliva samples.16
6. Key aspects, current trends, and future perspectives
Important achievements in recent years in this fast-growing field have made single and multiple electrochemical biosensing extremely promising for improving the reliability and speed of diagnostics and therapy monitoring, leading to more rapid clinical decision-making and corresponding reductions in patient stress and healthcare costs. The illustrative works discussed in this review show the unique opportunities offered by electrochemical biosensors for the determination of analytes at different molecular levels in particularly challenging samples using simple protocols.Requirements associated with the determination of different analytes, in terms of nature and concentration, and different sample types, make the development of a universal platform capable of monitoring any molecular analyte difficult. However, knowledge derived from successful examples reported in the last few years should guide researchers toward the key aspects to be considered for the design of novel strategies suitable for fulfilling the demands of particular applications.
Key aspects involved in the development of electrochemical biosensors for challenging clinical applications include the type of electrode substrate, bioreceptor, bioassay format, and electrochemical technique, and the strategies used for bioreceptor immobilization and signal amplification. In this context, SPEs offer suitable performance for this type of applications; sandwich formats in both immuno- and nucleic acid sensors are, in general, easier to implement and leading to more selective methodologies; electrochemical techniques that provide higher sensitivity are DPV and amperometry; strategies allowing the stable and oriented immobilization of large bioreceptor loadings (for instance, using diazonium salts on integrated substrates or MBs as solid supports) are preferred; and amplification strategies based on multienzyme labeling bioreagents should be more easily transferable to marketable devices. Regarding the samples for analysis, the use of electrode surfaces with antifouling properties is adequate for the determination of certain analytes in particular biofluids, although it cannot be extrapolated beforehand to determine other analytes or samples. In this context, after appropriate selection of the bioreceptor and bioassay format, methodologies involving the use of magnetic particles as solid supports seem easy to translate to the determination of different analytes within clinical ranges and in complex samples (biofluids, whole cells, and raw tissue and cellular extracts). Furthermore, switching-based electrochemical biosensors involving thiolated DNA, aptamer, and peptide probes self-assembled onto gold electrodes are particularly appealing for reagentless and real-time monitoring of relevant analytes in static or flowing biofluids, providing unprecedented simple and fast molecular monitoring in the clinical field. Biofouling, biocompatibility, stability, and calibration issues are among current problems prevented extension of their in vivo applications.
Despite the important achievements demonstrated, additional efforts are necessary for the determination of several (more than two) biomarkers through the design of new electrochemical probes that can produce more than two independent signals. The development of platforms suitable for the multidetermination of different molecular biomarkers or with large differences in threshold levels should be explored further. Owing to the great heterogeneity of cancer, the implementation of such platforms can serve as a basis for the detection of multiple types of cancer from a single blood sample, with the subsequent significant impact on early detection marking a turning point regarding improved treatments for cancer patients. Furthermore, the cost of these tests is expected to be lower than that of other diagnostic tests already in clinical use.
Despite the tremendous possibilities exhibited by electrochemical biosensors, requirements for the direct determination of target analytes in samples rich in proteins, or with extreme pH values (denaturing the biorecognition elements), or after prolonged incubation periods in these types of matrices, are clearly important challenges yet to be faced. Strategies based on efficient electrode modification with a wide range of antifouling (bio)materials (polymers, hydrogels, peptides, and thiolated monolayers) allow the preparation of biosensors for electrochemical determination of fouling analytes or in fouling samples exhibiting excellent performance. Nevertheless, in most cases, non-fouling strategies have been tested only on a particular sample matrix. Therefore, future efforts must be focused on evaluating these strategies in a wide range of matrices (blood, serum, urine, saliva) to extend the range of applications. The active pursuit of new strategies and/or materials with unique or improved properties will lead to highly stable electrochemical biosensing systems for continuous on-body determination or in vivo monitoring of important analytes in various bodily fluids with different pH ranges and compositions.
Additionally, a thorough clinical validation of electrochemical biosensors using minimally treated real samples and an exhaustive comparison with other current methodologies are needed. Efforts to guarantee the appropriate functionality of the biosensors after transportation and during storage, and further work on their integration in microfluidic devices to achieve automation and decrease the analysis time, will also influence their future development, stimulate interest from industry, and ensure their transition from research laboratories to the real-world use. Furthermore, the identification and clinical validation of new biomarkers and reliable signatures, and applications in little or yet-to-be explored clinical samples, are issues to be addressed to further push the limits of electrochemical bioplatforms toward new challenging applications in clinical diagnosis, prognosis, and therapeutic action of relevant diseases. However, the unique merits and practical superiority demonstrated by electrochemical biosensing platforms compared with other available technologies in terms of versatility, simplicity, cost, and portability, have provided significant motivation for the relentless exploration of other urgently demanded practical applications and their introduction to the market as ideal minimal-handling, affordable, portable, and easy-to-use quantitative devices.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from CTQ2015-70023-R and CTQ2015-64402-C2-1-R (Spanish Ministerio de Economía y Competitividad Research Projects) and S2013/MT3029 (NANOAVANSENS Program from the Comunidad de Madrid) are gratefully acknowledged.References
- S. E. Ilyin, S. M. Belkowski and C. R. Plata-Salamán, Trends Biotechnol., 2004, 2, 411 CrossRef PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi, X. Lou, J. Shi and C. Fan, Nat. Protoc., 2016, 11, 1244 CrossRef CAS PubMed.
- V. Kulasingam and E. P. Diamandis, Nat. Clin. Pract. Oncol., 2008, 5, 588 CrossRef CAS PubMed.
- C. Paoletti and D. F. Hayes, Annu. Rev. Med., 2014, 65, 95 CrossRef CAS PubMed.
- J. D. Cohen, L. Li, Y. Wang, C. Thoburn, B. Afsari, L. Danilova, C. Douville, A. A. Javed, F. Wong, A. Mattox, R. H. Hruban, C. L. Wolfgang, M. G. Goggins, M. Dal Molin, T.-L. Wang, R. Roden, A. P. Klein, J. Ptak, L. Dobbyn, J. Schaefer, N. Silliman, M. Popoli, J. T. Vogelstein, J. D. Browne, R. E. Schoen, R. E. Brand, J. Tie, P. Gibbs, H.-L. Wong, A. S. Mansfield, J. Jen, S. M. Hanash, M. Falconi, P. J. Allen, S. Zhou, C. Bettegowda, L. A. Diaz, C. Tomasetti, K. W. Kinzler, B. Vogelstein, A. M. Lennon and N. Papadopoulos, Science, 2018, 359, 926 CrossRef CAS PubMed.
- S. Campuzano, P. Yánez-Sedeño and J. M. Pingarrón, Sensors, 2017, 17, 866, DOI:10.3390/s17040866.
- S. Campuzano, M. Pedrero and J. M. Pingarrón, Sensors, 2017, 17, 1993, DOI:10.3390/s17091993.
- V. Escamilla-Gómez, D. Hernández-Santos, M. B. González-García, J. M. Pingarrón-Carrazón and A. Costa-García, Biosens. Bioelectron., 2009, 24, 2678 CrossRef PubMed.
- B. Esteban-Fernández de Ávila, S. Campuzano, M. Pedrero, J. P. Salvador, M. P. Marco and J. M. Pingarrón, Anal. Bioanal. Chem., 2014, 406, 5379 CrossRef PubMed.
- B. Esteban-Fernández de Ávila, V. Escamilla-Gómez, V. Lanzone, S. Campuzano, M. Pedrero, D. Compagnone and J. M. Pingarrón, Electroanalysis, 2014, 26, 254 CrossRef.
- U. Eletxigerra, J. Martinez-Perdiguero, S. Merino, R. Barderas, R. M. Torrente Rodríguez, R. Villalonga, J. M. Pingarrón and S. Campuzano, Biosens. Bioelectron., 2015, 70, 34 CrossRef CAS PubMed.
- I. Ojeda, M. Barrejón, L. M. Arellano, A. González-Cortés, P. Yáñez-Sedeño, F. Langa and J. M. Pingarrón, Biosens. Bioelectron., 2015, 74, 24 CrossRef CAS PubMed.
- B. Esteban-Fernández de Ávila, E. Araque, S. Campuzano, M. Pedrero, B. Dalkiran, R. Barderas, E. Kilic, R. Villalonga and J. M. Pingarrón, Anal. Chem., 2015, 87, 2290 CrossRef PubMed.
- M. Garranzo-Asensio, A. Guzmán-Aránguez, C. Povés, M. J. Fernández-Aceñero, R. M. Torrente-Rodríguez, V. Ruiz-Valdepeñas-Montiel, G. Domínguez, M. Villalba, J. M. Pingarrón, S. Campuzano and R. Barderas, Anal. Chem., 2016, 88, 12339 CrossRef CAS PubMed.
- G. Martínez-García, L. Agüí, P. Yáñez-Sedeño and J. M. Pingarrón, Electrochim. Acta, 2016, 202, 209 CrossRef.
- R. M. Torrente-Rodríguez, S. Campuzano, V. Ruiz-Valdepeñas Montiel, M. Gamella and J. M. Pingarrón, Biosens. Bioelectron., 2016, 77, 543 CrossRef PubMed.
- R. M. Torrente-Rodríguez, V. Ruiz-Valdepeñas Montiel, S. Campuzano, M. Pedrero, M. Farchado, E. Vargas, F. J. Manuel de Villena, M. Garranzo-Asensio, R. Barderas and J. M. Pingarrón, PLoS One, 2017, 12, e0175056 CrossRef PubMed.
- R. M. Torrente-Rodríguez, S. Campuzano, V. Ruiz-Valdepeñas Montiel, A. Sagrera, J. J. Domínguez-Cañete, E. Vargas, J. J. Montoya, R. Granados, J. M. Sánchez-Puelles and J. M. Pingarrón, J. Biotechnol. Biomed. Eng., 2016, 3, 1064 Search PubMed.
- E. Sánchez-Tirado, C. Salvo, A. González-Cortés, P. Yáñez-Sedeño, F. Langa and J. M. Pingarrón, Anal. Chim. Acta, 2017, 959, 66 CrossRef PubMed.
- E. Sánchez-Tirado, L. M. Arellano, A. González-Cortés, P. Yáñez-Sedeño, F. Langa and J. M. Pingarrón, Biosens. Bioelectron., 2017, 98, 240 CrossRef PubMed.
- E. Sánchez-Tirado, A. González-Cortés, M. Yudasaka, S. Iijima, F. Langa, P. Yáñez-Sedeño and J. M. Pingarrón, J. Electroanal. Chem., 2017, 793, 197 CrossRef.
- M. Zouari, S. Campuzano, J. M. Pingarrón and N. Raouafi, Biosens. Bioelectron., 2017, 91, 40 CrossRef CAS PubMed.
- E. Sánchez-Tirado, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Biosens. Bioelectron., 2018, 113, 88 CrossRef PubMed.
- L. Jirakova, R. Hrstka, S. Campuzano, J. M. Pingarrón and M. Bartosik, Electroanalysis DOI:10.1002/elan.201800573.
- E. Martínez-Periñán, E. Sánchez-Tirado, A. González-Cortés, R. Barderas, J. M. Sánchez-Puelles, L. Martínez-Santamaría, S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Electrochim. Acta, 2018, 292, 887 CrossRef.
- F. Mollarasouli, V. Serafín, S. Campuzano, P. Yáñez-Sedeño, J. M. Pingarrón and K. Asadpour-Zeynali, Anal. Chim. Acta, 2018, 1011, 28 CrossRef CAS PubMed.
- E. Povedano, E. Vargas, V. Ruiz-Valdepeñas Montiel, R. M. Torrente-Rodríguez, M. Pedrero, R. Barderas, P. San Segundo-Acosta, A. Peláez-García, M. Mendiola, D. Hardisson, S. Campuzano and J. M. Pingarrón, Sci. Rep., 2018, 8, 6418 CrossRef PubMed.
- E. Povedano, A. Valverde, V. Ruiz-Valdepeñas Montiel, M. Pedrero, P. Yáñez-Sedeño, R. Barderas, P. San Segundo-Acosta, A. Peláez-García, M. Mendiola, D. Hardisson, S. Campuzano and J. M. Pingarrón, Angew. Chem., Int. Ed., 2018, 57, 8194 CrossRef CAS PubMed.
- V. Serafín, G. Martínez-García, J. Aznar-Poveda, J. A. Lopez-Pastor, A. J. Garcia-Sanchez, J. Garcia-Haro, S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Anal. Chim. Acta, 2019, 1049, 65 CrossRef PubMed.
- V. Serafín, R. M. Torrente-Rodríguez, P. García de Frutos, M. Sabaté, S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Talanta, 2018, 179, 131 CrossRef PubMed.
- A. Valverde, E. Povedano, V. Ruiz-Valdepeñas Montiel, P. Yáñez-Sedeño, M. Garranzo, R. Barderas, S. Campuzano and J. M. Pingarrón, Biosens. Bioelectron., 2018, 117, 766 CrossRef CAS PubMed.
- A. Valverde, E. Povedano, V. Ruiz-Valdepeñas Montiel, P. Yáñez-Sedeño, M. Garranzo-Asensio, N. Rodríguez, G. Domínguez, R. Barderas, S. Campuzano and J. M. Pingarrón, Anal. Chem., 2018, 90, 11161 CrossRef CAS PubMed.
- C. Muñoz-San Martín, M. Pedrero, F. J. Manuel de Villena, M. Garranzo-Asensio, N. Rodríguez, G. Domínguez, R. Barderas, S. Campuzano and J. M. Pingarrón, Electroanalysis DOI:10.1002/elan.201800645.
- M. Zouari, S. Campuzano, J. M. Pingarrón and N. Raouafi, Electrochim. Acta, 2018, 262, 39 CrossRef CAS.
- M. Zouari, S. Campuzano, J. M. Pingarrón and N. Raouafi, ACS Omega, 2018, 3, 8923 CrossRef CAS.
- P. Yáñez-Sedeño, A. González-Cortés, L. Agüí and J. M. Pingarrón, Electroanalysis, 2016, 28, 1679 CrossRef.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Diagnostics, 2017, 7, 2, DOI:10.3390/diagnostics01000S.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Anal. Bioanal. Chem., 2018 DOI:10.1007/s00216-018-1273-6.
- Y. Y. Yu, Z. G. Chen, L. J. Shi, F. Yang, J. B. Pan, B. B. Zhang and D. P. Sun, Anal. Chem., 2014, 86, 8200 CrossRef CAS PubMed.
- P. Miao, B. Wang, F. Meng, J. Yin and Y. Tang, Bioconjugate Chem., 2015, 26, 602 CrossRef CAS PubMed.
- V. Ruiz-Valdepeñas Montiel, M. L. Gutiérrez, R. M. Torrente-Rodríguez, E. Povedano, E. Vargas, A. Julio Reviejo, R. Linacero, F. J. Gallego, S. Campuzano and J. M. Pingarrón, Anal. Chem., 2017, 89, 9474 CrossRef PubMed.
- V. Ruiz-Valdepeñas Montiel, E. Povedano, E. Vargas, R. M. Torrente-Rodríguez, M. Pedrero, A. J. Reviejo, S. Campuzano and J. M. Pingarrón, ACS Sens., 2018, 3, 211 CrossRef PubMed.
- P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Sensors, 2016, 16, 1585 CrossRef PubMed.
- M. F. Brugnera, R. Bundalian, T. Laube, E. Julián, M. Luquin, M. V. Boldrin-Zanoni and M. I. Pividori, Talanta, 2016, 153, 38 CrossRef CAS PubMed.
- P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Sensors, 2018, 18, 675 CrossRef PubMed.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, ChemElectroChem, 2019, 6, 60 CrossRef CAS.
- S. Campuzano, V. Salema, M. Moreno-Guzmán, M. Gamella, P. Yáñez-Sedeño, L. A. Fernández and J. M. Pingarrón, Biosens. Bioelectron., 2014, 52, 255 CrossRef CAS PubMed.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Electrochim. Acta, 2017, 230, 271 CrossRef CAS.
- S. Centi, S. Laschi, M. Frànek and M. Mascini, Anal. Chim. Acta, 2005, 538, 205 CrossRef CAS.
- E. Zacco, J. Adrian, R. Galve, M. P. Marco, S. Alegret and M. I. Pividori, Biosens. Bioelectron., 2007, 22, 2184 CrossRef CAS PubMed.
- F. Ricci, G. Volpe, L. Micheli and G. Palleschi, Anal. Chim. Acta, 2007, 605, 111 CrossRef CAS PubMed.
- B. Esteban-Fernández de Ávila, V. Escamilla-Gómez, S. Campuzano, M. Pedrero, J. P. Salvador, M. P. Marco and J. M. Pingarrón, Sens. Actuators, B, 2013, 188, 212 CrossRef.
- U. Eletxigerra, J. Martinez-Perdiguero, S. Merino, R. Barderas, V. Ruiz-Valdepeñas Montiel, R. Villalonga, J. M. Pingarrón and S. Campuzano, Sens. Biosensing Res., 2016, 7, 71 CrossRef.
- W. Ma, B. Situ, W. Lv, B. Li, X. Yin, P. Vadgama, L. Zheng and W. Wang, Biosens. Bioelectron., 2016, 80, 344 CrossRef CAS PubMed.
- V. Serafín, R. M. Torrente-Rodríguez, M. Batlle, P. García de Frutos, S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Microchim. Acta, 2017, 184, 4251 CrossRef.
- F. Arduini, L. Micheli, D. Moscone, G. Palleschi, S. Piermarini, F. Ricci and G. Volpe, TrAC, Trends Anal. Chem., 2016, 79, 114 CrossRef CAS.
- P. Yánez-Sedeño, S. Campuzano and J. M. Pingarrón, Sensors, 2017, 17, 965 CrossRef.
- B. P. Corgier, C. A. Marquette and L. J. Blum, J. Am. Chem. Soc., 2005, 127, 18328 CrossRef CAS.
- W. Zheng, R. van den Hurk, Y. Cao, R. Du, X. Sun, Y. Wang, M. T. McDermott and S. Evoy, Biosens. Bioelectron., 2016, 6, 8 CrossRef PubMed.
- M. J. Lobo-Castañón, Anal. Bioanal. Chem., 2016, 408, 8581 CrossRef PubMed.
- R. M. Torrente-Rodríguez, S. Campuzano, V. Ruiz-Valdepeñas Montiel, J. J. Montoya and J. M. Pingarrón, Biosens. Bioelectron., 2016, 86, 516 CrossRef PubMed.
- S. Barreda-García, R. Miranda-Castro, N. de-los-Santos-Álvarez, A. J. Miranda-Ordieres and M. J. Lobo-Castañón, Anal. Bioanal. Chem., 2016, 408, 8603 CrossRef.
- M. Bartosik, L. Jirakova, M. Anton, B. Vojtesek and R. Hrstka, Anal. Chim. Acta, 2018, 1042, 37 CrossRef CAS PubMed.
- C. Blaszykowski, S. Sheikh and M. Thompson, Chem. Soc. Rev., 2012, 41, 5599 RSC.
- Y. Wang, M. Cui, M. Jiao and X. Luo, Anal. Bioanal. Chem., 2018, 410, 5871 CrossRef CAS PubMed.
- B. L. Hanssen, S. Siraj and D. K. Y. Wong, Rev. Anal. Chem., 2016, 35, 1 CAS.
- M. Cui, Y. Wang, M. Jiao, S. Jayachandran, Y. Wu, X. Fan and X. Luo, ACS Sens., 2017, 2, 490 CrossRef CAS PubMed.
- V. Ruiz-Valdepeñas Montiel, J. R. Sempionatto, B. Esteban-Fernández de Ávila, A. Whitworth, S. Campuzano, J. M. Pingarrón and J. Wang, J. Am. Chem. Soc., 2018, 140, 14050 CrossRef PubMed.
- V. Ruiz-Valdepeñas Montiel, J. R. Sempionatto, S. Campuzano, J. M. Pingarrón, B. Esteban-Fernández de Ávila and J. Wang, Anal. Bioanal. Chem. DOI:10.1007/s00216-018-1528-2.
- N. Arroyo-Currás, J. Somerson, P. A. Vieira, K. L. Ploense, T. E. Kippine and K. W. Plaxco, PNAS, 2017, 114, 645 CrossRef PubMed.
- A. A. Lubin and K. W. Plaxco, Acc. Chem. Res., 2010, 43, 496 CrossRef CAS PubMed.
- K. W. Plaxco and H. T. Soh, Trends Biotechnol., 2011, 29, 1 CrossRef CAS PubMed.
- S. Ranallo, M. Rossetti, K. W. Plaxco, A. Vallée-Bélisle and F. Ricci, Angew. Chem., Int. Ed., 2015, 54, 13214 CrossRef CAS PubMed.
- C. Li, X. Hu, J. Lu, X. Mao, Y. Xiang, Y. Shu and G. Li, Chem. Sci., 2018, 9, 979 RSC.
- Q. Wang, F. Gao, J. Ni, X. Liao, X. Zhang and Z. Lin, Sci. Rep., 2016, 6, 22441 CrossRef CAS PubMed.
- A. J. Zaitouna and R. Y. Lai, Anal. Chim. Acta, 2014, 828, 85 CrossRef CAS PubMed.
- M. D. Mayer and R. Y. Lai, Talanta, 2018, 189, 585 CrossRef CAS PubMed.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi, X. Zuo and C. Fan, Angew. Chem., Int. Ed., 2015, 54, 2151 CrossRef CAS PubMed.
- H. Pei, N. Lu, Y. Wen, S. Song, Y. Liu, H. Yan and C. Fan, Adv. Mater., 2010, 22, 4754 CrossRef CAS PubMed.
- S. Dong, R. Zhao, J. Zhu, X. Lu, Y. Li, S. Qiu, L. Ji, X. Jiao, S. Song, C. Fan, R. Z. Hao and H. B. Song, ACS Appl. Mater. Interfaces, 2015, 5, 8834 CrossRef PubMed.
- X. Chen, G. Zhou, P. Song, J. Wang, J. Gao, J. Lu, C. Fan and X. Zuo, Anal. Chem., 2014, 86, 7337 CrossRef CAS PubMed.
- Y. Wen, H. Pei, Y. Wan, Y. Su, Q. Huang, S. Song and C. Fan, Anal. Chem., 2011, 83, 7418 CrossRef CAS PubMed.
- Y. L. Huang, S. Mo, Z. F. Gao, J. R. Chen, J. L. Lei, H. Q. Luo and N. B. Li, Microchim. Acta, 2017, 184, 2597 CrossRef CAS.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Sensors, 2017, 17, 2533, DOI:10.3390/s17112533.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, TrAC, Trends Anal. Chem., 2017, 86, 14 CrossRef CAS.
- S. Campuzano, P. Yañez-Sedeño and J. M. Pingarrón, Curr. Opin. Electrochem., 2018, 12, 81 CrossRef CAS.
- S. Campuzano and J. M. Pingarrón, Electroanalysis, 2018, 7, 1201 CrossRef.
- N. K. Bakirhan, G. Ozcelikay and S. A. Ozkan, J. Pharm. Biomed. Anal., 2018, 159, 406 CrossRef CAS PubMed.
- L. Farzin, M. Shamsipur, L. Samandari and S. Sheibani, J. Pharm. Biomed. Anal., 2018, 161, 344 CrossRef CAS PubMed.
- H. Jo, H. Gu, W. Jeon, H. Youn, J. Her, S. K. Kim, J. Lee, J. H. Shin and C. Ban, Anal. Chem., 2015, 87, 9869 CrossRef CAS PubMed.
- R. Akter, B. Jeong, Y. M. Lee, J. S. Choi and M. A. Rahman, Biosens. Bioelectron., 2017, 91, 637 CrossRef CAS PubMed.
- N. R. Shanmugam, S. Muthukumar, S. Chaudhry, J. Anguiano and S. Prasad, Biosens. Bioelectron., 2017, 89, 764 CrossRef PubMed.
- M. Abdorahim, M. Rabiee, S. N. Alhosseini, M. Tahriri, S. Yazdanpanah, S. H. Alavi and L. Tayebi, TrAC, Trends Anal. Chem., 2016, 82, 337–347 CrossRef CAS.
- G. Bing and X. Wang, Int. J. Electrochem. Sci., 2017, 12, 6304 CrossRef.
- J. Wang, J. Guo, J. Zhang, W. Zhang and Y. Zhang, Biosens. Bioelectron., 2017, 95, 100 CrossRef CAS PubMed.
- G. Zhang, Z. Liu, L. Wang and Y. Guo, Sensor, 2016, 16, 1803 CrossRef PubMed.
- Y. W. Liu, Q. F. Yang, P. Y. Zuo, C. L. Xiao, X. L. Chen and C. Y. Liu, Am. J. Med. Sci., 2015, 349, 124 CrossRef PubMed.
- J. Melin, G. Rundström, C. Peterson, J. Bakker, B. D. MacCraith, M. Read, O. Öhman and C. Jönsson, Anal. Biochem., 2011, 109, 7 CrossRef PubMed.
- A. Saeed and S. S. Virani, Front. Biosci. (Landmark Ed)., 2018, 23, 1099 DOI:10.2741/4635.
- T. Bryan, X. L. Luo, P. R. Bueno and J. J. Davis, Biosens. Bioelectron., 2013, 39, 94 CrossRef CAS PubMed.
- H. Y. Lee, J. S. C. Hoi, P. G. Uruprasath and B. L. Ee, Anal. Sci., 2015, 31, 699 CrossRef CAS PubMed.
- M. Batlle, P. Recarte-Oelz, E. Roig, M. A. Castel, M. Cardona, M. Farrero, B. Campos, M. J. Pulgarín, J. Ramírez, F. Pérez-Villa and P. García de Frutos, Int. J. Cardiol., 2014, 173, 402 CrossRef CAS PubMed.
- V. Serafín, R. M. Torrente-Rodríguez, M. Batlle, P. García de Frutos, S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Sens. Actuators, B, 2017, 240, 1251 CrossRef.
- V. Bhatia, P. Nayyar and S. Dhindsa, J. Postgrad. Med., 2003, 49, 182 CAS.
- R. Eivazzadeh-Keihan, P. Pashazadeh-Panahi, B. Baradaran, A. Maleki, M. Hejazi, A. Mokhtarzadeh and M. de la Guardia, TrAC, Trends Anal. Chem., 2018, 100, 103 CrossRef CAS.
- B. Bodey, S. E. Siegel and H. E. Kaiser, Anticancer Res., 1998, 18, 3621 CAS.
- J. Mandli, H. Mohammadi and A. Amine, Bioelectrochemistry, 2017, 116, 17 CrossRef CAS PubMed.
- S. Liu, Z. Yang, Y. Chang, Y. Chai and R. Yuan, Biosens. Bioelectron., 2018, 119, 170 CrossRef CAS PubMed.
- S. Dehghani, R. Nosrati, M. Yousefi, A. Nezami, F. Soltani, S. M. Taghdisi, K. Abnous, M. Alibolandi and M. Ramezani, Biosens. Bioelectron., 2018, 110, 23 CrossRef CAS PubMed.
- A. Qureshi, Y. Gurbuz and J. H. Niazi, Sens. Actuators, B, 2015, 209, 645 CrossRef CAS.
- M. Johari-Ahar, P. Karami, M. Ghanei, A. Afkhami and H. Bagheri, Biosens. Bioelectron., 2018, 107, 26 CrossRef CAS PubMed.
- A. Tabasi, A. Noorbakhsh and E. Sharifi, Biosens. Bioelectron., 2017, 95, 117 CrossRef CAS PubMed.
- M. Shamsipur, M. Emami, L. Farzin and R. Saberd, Biosens. Bioelectron., 2018, 103, 54 CrossRef CAS PubMed.
- C. V. Uliana, C. R. Peverari, A. S. Afonso, M. R. Cominetti and R. C. Faria, Biosens. Bioelectron., 2018, 99, 156 CrossRef CAS PubMed.
- N. Razmi, B. Baradaran, M. Hejazi, M. Hasanzadeh, J. Mosafer, A. Mokhtarzadeh and M. de la Guardia, Biosens. Bioelectron., 2018, 113, 58 CrossRef CAS PubMed.
- L.-X. Fang, K.-J. Huang and Y. Liu, Biosens. Bioelectron., 2015, 71, 171 CrossRef CAS PubMed.
- K.-J. Huang, H.-L. Shuai and J.-Z. Zhang, Biosens. Bioelectron., 2016, 77, 69 CrossRef CAS PubMed.
- E. B. Aydın and M. K. Sezgintürk, Anal. Biochem., 2018, 554, 44 CrossRef PubMed.
- E. Sánchez-Tirado, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Analyst, 2016, 141, 5730 RSC.
- M. Mazloum-Ardakani and L. Hosseinzadeh, J. Electrochem. Soc., 2016, 163, B119 CrossRef CAS.
- L. Liu, F. Liu, D. Jiang, G. Xiang, C. Liu, J. Yang and X. Pu, Sens. Actuators, B, 2016, 231, 680 CrossRef CAS.
- S. Mishra, D. Saadat, O. Kwon, Y. Lee, W.-S. Choi, J.-H. Kim and W.-H. Yeo, Biosens. Bioelectron., 2016, 81, 181 CrossRef CAS PubMed.
- B. Della Ventura, N. Sakač, R. Funari and R. Velotta, Talanta, 2017, 174, 52 CrossRef CAS PubMed.
- L. Zhang, W. Yang, Y. Yang, H. Liu and Z. Gu, Analyst, 2015, 140, 7399 RSC.
- M. S. Khan, S. K. Misra, Z. Wang, E. Daza, A. S. Schwartz-Duval, J. M. Kus, D. Pan and D. Pan, Anal. Chem., 2017, 89, 2107 CrossRef CAS PubMed.
- K. Rajkumar, G. Nandhini, R. Ramya, P. Rajashree, A. R. Kumar and S. N. Anandan, Oral Surg., Oral Med., Oral Pathol., Oral Radiol., 2014, 118, 309, DOI:10.1016/j.oooo.2014.04.008.
- M. Aydın, E. B. Aydın and M. K. Sezgintürk, Biosens. Bioelectron., 2018, 107, 1 CrossRef PubMed.
- M. Aydın, E. B. Aydın and M. K. Sezgintürk, Biosens. Bioelectron., 2018, 117, 720 CrossRef PubMed.
- R.-N. Ma, L.-L. Wang, H.-F. Wang, L.-P. Jia, W. Zhang, L. Shang, Q.-W. Xue, W.-L. Jia, Q.-Y. Liu and H.-S. Wang, Sens. Actuators, B, 2018, 269, 173 CrossRef CAS.
- L. Jiang, X. Zeng, Z. Wang, J. Ning, Y. Zhou, X. T. Liu and Q. M. Chen, Mol. Cancer, 2010, 9, 20 CrossRef PubMed.
- M. Chen, Y. Wang, H. Su, L. Mao, X. Jiang and X. Dai, Sens. Actuators, B, 2018, 255, 2910 CrossRef CAS.
- T. Tian, H. Liu, L. Li, J. Yu, S. Ge, X. Song and M. Yan, Sens. Actuators, B, 2017, 251, 440 CrossRef CAS.
- R. Malhotra, A. B. Urs, A. Chakravarti, S. Kumar, V. Gupta and B. Mahajan, Tumor Biol., 2016, 1 Search PubMed.
- S. Kumar, S. Kumar, S. Tiwari, S. Augustine, S. Srivastava, B. K. Yadav and B. D. Malhotra, Sens. Actuators, B, 2016, 235, 1 CrossRef CAS.
- R. Malhotra, V. Patel, J. P. Vaqué, J. S. Gutkind and J. F. Rusling, Anal. Chem., 2010, 82, 3118 CrossRef CAS PubMed.
- J. Liu and Y. Duan, Oral Oncol., 2012, 48, 569 CrossRef PubMed.
- I. Ojeda, M. Moreno-Guzmán, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Anal. Bioanal. Chem., 2014, 406, 6363 CrossRef CAS.
- S. D. Anker and S. von Haehling, Heart, 2004, 90, 464 CrossRef CAS PubMed.
- A. Yndestad, J. K. Damas, E. Oie, T. Ueland, L. Gullestad and P. Aukrust, Heart Failure Rev., 2006, 11, 83 CrossRef CAS PubMed.
- F. G. Bellagambi, A. Baraket, A. Longo, M. Vatteroni, N. Zine, J. Bausells, R. Fuoco, F. Di Francesco, P. Salvo, G. S. Karanasiou, D. I. Fotiadis, A. Menciassi and A. Errachid, Sens. Actuators, B, 2017, 251, 1026 CrossRef CAS.
- L. Barhoumi, A. Baraket, F. G. Bellagambi, G. S. Karanasiou, M. Ben Ali, D. I. Fotiadis, J. Bausells, N. Zine, M. Sigaud and A. Errachid, Sens. Actuators, B, 2018, 266, 477 CrossRef CAS.
- V. Brailo, V. Vucicevic-Boras, J. Lukac, D. Biocina-Lukenda, I. Zilic-Alajbeg, A. Milenovic and M. Balija, Med. Oral. Patol. Oral Cir. Bucal., 2012, 17, 10 CrossRef.
- F. I. Fernandes Gomes, M. G. Brito Aragão, F. C. Barroso Barbosa, M. Marques Bezerra, V. de P. Teixeira Pinto and H. Vasconcelos Chaves, J. Oral Maxillofac. Res., 2016, 7, e2 Search PubMed.
- F. Chekin, A. Vasilescu, R. Jijie, S. K. Singh, S. Kurungot, M. Iancu, G. Badea, R. Boukherroub and S. Szunerits, Sens. Actuators, B, 2018, 262, 180 CrossRef CAS.
- J. Kim, G. Valdés-Ramírez, A. J. Bandodkar, W. Jia, A. G. Martinez, J. Ramírez, P. Mercier and J. Wang, Analyst, 2014, 139, 1632 RSC.
- K. Petropoulos, S. Piermarini, S. Bernardini, G. Palleschi and D. Moscone, Sens. Actuators, B, 2016, 237, 8 CrossRef CAS.
- I. Ojeda, B. Garcinuño, M. Moreno-Guzmán, A. González-Cortés, M. Yudasaka, S. Iijima, F. Langa, P. Yáñez-Sedeño and J. M. Pingarrón, Anal. Chem., 2014, 86, 7749 CrossRef CAS.
- E. Sánchez-Tirado, G. Martínez-García, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Biosens. Bioelectron., 2017, 88, 9 CrossRef.
- D. A. Smith, L. J. Newbury, G. Drago, T. Bowen and J. E. Redman, Sens. Actuators, B, 2017, 253, 335 CrossRef CAS.
- H. Li, C. Li, H. Wu, T. Zhang, J. Wang, S. Wang and J. Chang, Proteome Sci., 2011, 9, 21 CrossRef CAS PubMed.
- S.-E. Kim, Y. J. Kim, S. Song, K.-N. Lee and W. K. Seong, Sens. Actuators, B, 2019, 278, 103 CrossRef CAS.
- S. V. Parikh, A. Alvarado, A. Malvar and B. H. Rovin, Semin. Nephrol., 2015, 35, 465 CrossRef.
- A. P. Selvam, A. Wangzhou, M. Jacobs, T. Wu, C. Mohan and S. Prasad, Future Sci. OA, 2017, 3, FSO224 CrossRef PubMed , eISSN 2056.
- M. Hasanzadeh and N. Shadjou, TrAC, Trends Anal. Chem., 2016, 80, 167 CrossRef CAS.
- T. Xu, B. Chi, J. Gao, M. Chu, W. Fan, M. Yi, H. Xu and C. Mao, Anal. Chim. Acta, 2017, 977, 36 CrossRef CAS PubMed.
- S. Eissa, N. Alshehri, M. Abduljabbar, A. M. A. Rahman, M. Dasouki, I. Y. Nizami, M. A. Al-Muhaizea and M. Zourob, Biosens. Bioelectron., 2018, 117, 84 CrossRef CAS PubMed.
- S. Singh, S. K. Tuteja, D. Sillu, A. Deep and C. R. Suri, Microchim. Acta, 2016, 183, 1729 CrossRef CAS.
- G. Yammouri, J. Mandli, H. Mohammadi and A. Amine, J. Electroanal. Chem., 2017, 806, 75 CrossRef CAS.
- A. R. Cardoso, F. T. C. Moreira, R. Fernandes and M. G. F. Sales, Biosens. Bioelectron., 2016, 80, 621 CrossRef CAS PubMed.
- A. Qureshi, Y. Gurbuz and J. H. Niazi, Sens. Actuators, B, 2015, 220, 1145 CrossRef CAS.
- M. Hasanzadeh, N. Razmi., A. Mokhtarzadeh, N. Shadjou and S. Mahboob, Int. J. Biol. Macromol., 2018, 108, 69 CrossRef CAS PubMed.
- M. Mazloum-Ardakani, L. Hosseinzadeh and A. Khoshroo, J. Electroanal. Chem., 2015, 757, 58 CrossRef CAS.
- Q. Chen, W. Hu, B. Shang, J. Wei, L. Chen, X. Guo, F. Ran, W. Chen, X. Ding, Y. Xu and Y. Wu, Microchim. Acta, 2018, 185, 202 CrossRef PubMed.
- S. Campuzano, R. M. Torrente-Rodríguez, E. López-Hernández, F. Conzuelo, R. Granados, J. M. Sánchez-Puelles and J. M. Pingarrón, Angew. Chem., Int. Ed., 2014, 53, 6168 CrossRef CAS PubMed.
- R. M. Torrente-Rodríguez, S. Campuzano, E. López-Hernández, R. Granados, J. M. Sánchez-Puelles and J. M. Pingarrón, Electroanalysis, 2014, 26, 2080 CrossRef.
- E. Vargas, E. Povedano, V. Ruiz-Valdepeñas Montiel, R. M. Torrente-Rodríguez, M. Zouari, J. J. Montoya, N. Raouaffi, S. Campuzano and J. M. Pingarrón, Sensors, 2018, 18, 863 CrossRef PubMed.
- R. M. Torrente-Rodríguez, V. Ruiz-Valdepeñas Montiel, S. Campuzano, M. Farchado-Dinia, R. Barderas, P. San Segundo-Acosta, J. J. Montoya and J. M. Pingarrón, ACS Sens., 2016, 1, 896 CrossRef.
- E. Vargas, R. M. Torrente-Rodríguez, V. Ruiz-Valdepeñas Montiel, E. Povedano, M. Pedrero, J. J. Montoya, S. Campuzano and J. M. Pingarrón, Int. J. Mol. Sci., 2017, 18, 2151 CrossRef PubMed.
- R. M. Graybill and R. C. Bailey, Anal. Chem., 2016, 88, 431 CrossRef CAS PubMed.
- A. N. Bhatt, R. Mathur, A. Farooque, A. Verma and B. S. Dwarakanath, Indian J. Med. Res., 2010, 132, 129 CAS.
- O. J. Switzeny, M. Christmann, M. Renovanz, A. Giese, C. Sommer and B. Kaina, Clin. Epigenet., 2016, 8, 49 CrossRef PubMed.
- U. Eletxigerra, J. Martinez-Perdiguero, S. Merino, R. Barderas, V. Ruiz-Valdepeñas Montiel, R. Villalonga, J. M. Pingarrón and S. Campuzano, Electroanalysis, 2016, 28, 1787 CrossRef CAS.
- M. Pedrero, F. Javier Manuel de Villena, C. Muñoz-San Martín, S. Campuzano, M. Garranzo-Asensio, R. Barderas and J. M. Pingarrón, Biosensors, 2016, 6, 56 CrossRef PubMed.
- M. Moreno-Guzmán, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Electroanalysis, 2012, 24, 1100 CrossRef.
- S. Zhou, Y. Wang and J. J. Zhu, ACS Appl. Mater. Interfaces, 2016, 8, 7674 CrossRef CAS PubMed.
- G.-J. Zhang, Z. H. H. Luo, M. J. Huang, J. J. Ang, T. G. Kang and H. Ji, Biosens. Bioelectron., 2011, 28, 459 CrossRef CAS.
- R. K. Gupta, R. Pandya, T. Sieffert, M. Meyyappan and J. E. Koehne, J. Electroanal. Chem., 2016, 773, 53 CrossRef CAS.
- F. Zhou, M. Lu, W. Wang, Z.-P. Bian, J.-R. Zhang and J.-J. Zhu, Clin. Chem., 2010, 56, 1701 CrossRef CAS PubMed.
- B. V. Chikkaveeraiah, V. Mania, V. Patel, J. S. Gutkind and J. F. Rusling, Biosens. Bioelectron., 2011, 26, 4477 CrossRef CAS PubMed.
- J. Tang, D. Tang, R. Niessner, G. Chen and D. Knopp, Anal. Chem., 2011, 83, 5407 CrossRef CAS PubMed.
- W. Li, L. Li, S. Ge, X. Song, L. Ge, M. Yan and J. Yu, Biosens. Bioelectron., 2014, 56, 167 CrossRef CAS PubMed.
- G. Sun, L. Zhang, Y. Zhang, H. Yang, C. Ma, S. Ge, M. Yan, J. Yu and X. Song, Biosens. Bioelectron., 2015, 71, 30 CrossRef CAS.
- C. Zhao and X. Liu, Biomicrofluidics, 2016, 10, 024119 CrossRef PubMed.
- T. Li, B. Shu, B. Jiang, L. Ding, H. Qi, M. Yang and F. Qu, Sens. Actuators, B, 2013, 186, 768 CrossRef CAS.
- E. Povedano, V. Ruiz-Valdepeñas Montiel, A. Valverde, F. Navarro-Villoslada, P. Yánez-Sedeño, M. Pedrero, A. Montero-Calle, R. Barderas, A. Peláez-García, M. Mendiola, D. Hardisson, J. Feliú, J. Camps, E. Rodríguez-Tomàs, J. Joven, M. Arenas, S. Campuzano and J. M. Pingarrón, ACS Sens. DOI:10.1021/acssensors.8b01339.
- R. M. Torrente-Rodríguez, S. Campuzano, E. López-Hernández, V. Ruiz-Valdepeñas Montiel, R. Barderas, R. Granados, J. M. Sánchez-Puelles and J. M. Pingarrón, Biosens. Bioelectron., 2015, 66, 385 CrossRef PubMed.
- F. Wei, P. Patel, W. Liao, K. Chaudhry, L. Zhang, M. Arellano-Garcia, S. Hu, D. Elashoff, H. Zhou, S. Shukla, F. Shah, C.-M. Ho and D. T. Wong, Clin. Cancer Res., 2009, 15 DOI:10.1158/1078-0432.CCR-09-0050.
- F. Wei, W. Liao, Z. Xu, Y. Yang, D. T. Wong and C.-M. Ho, Small, 2009, 5, 1784 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |



