Construction and optimization of efficient glucose–xylose co-fermenting yeast Yarrowia lipolytica for green and sustainable succinic acid production from lignocellulosic biomass†
Mianshen
Ge‡
ab,
Yuanyuan
Sha‡
ab,
Minrui
Lu
ab,
Yuwei
Zhang
ab,
Zhaoxian
Xu
ab,
Sitong
Chen
ab,
Ying
Ding
ab and
Mingjie
Jin
 *ab
*ab
aSchool of Environmental and Biological Engineering, Nanjing University of Science and Technology, Nanjing 210094, PR China. E-mail: jinmingjie@njust.edu.cn
bBiorefinery Research Institution, Nanjing University of Science and Technology, Nanjing 210094, PR China
First published on 22nd November 2024
Abstract
Xylose is the second most abundant carbohydrate present in nature, while its inefficient utilization severely restricts the economic viability of lignocellulosic biomass-based biorefinery. Herein, metabolic engineering strategies involving xylose metabolism and the succinic acid (SA) synthetic pathway were developed in Yarrowia lipolytica for the production of SA from lignocellulosic hydrolysate. First, the Ylsdh5 gene (succinate dehydrogenase subunit 5) was inactivated in Y. lipolytica BZ, which can grow on xylose as the sole carbon source, thereby obtaining a strain capable of synthesizing SA from xylose. Subsequently, the glucose–xylose assimilating rate and SA titers were further optimized by blocking the by-product pathway and enhancing the SA synthetic pathways. Then, with the overexpression of the crucial mitochondrial dicarboxylic acid transporter YlDic, the obtained SA producer Y. lipolytica BDic5 showed excellent xylose assimilation performance, which could utilize all the glucose and xylose in either pure culture or hydrolysate fermentation. Remarkably, BDic5 exhibited robust growth in 30% solid-loading of corn stover hydrolysate without hydrolysate detoxification or dilution, and the fermentation process did not require neutral pH maintenance. Finally, up to 105.42 g L−1 SA was produced from undetoxified lignocellulosic hydrolysate using the fed-batch strategy in a 3 L bioreactor, which was the highest SA titer achieved from lignocellulosic feedstock to date. Following downstream purification of the acidic fermentation broth, 61.75% of the total SA with purity of 92.81% was recovered. These promising results indicated that the recombinant strain exhibited great potential for bioconversion of lignocellulosic biomass into bio-SA, which demonstrated great prospects for industrial production.
1. Introduction
Succinic acid (SA) is among the top value-added platform chemicals that have received increasing attention due to its widespread application in the surfactant, biodegradable plastics, food additives, and pharmaceutical markets.1,2 Due to the enormous application values of SA, the worldwide market of SA exceeds 275 × 106 kg annually.3 Furthermore, it is anticipated that more recent industrial applications for 1,4-butanediol, poly-butylene succinate, alkyd resins, and plasticizers will further support the SA market's future expansion.4 Maleic anhydride, derived from petrochemical feedstock, is the primary substrate for SA in conventional chemical production. Despite the high efficiency of chemical synthesis methods, they still suffer from severe reaction conditions, intricate operation, and a high potential for environmental pollution.5 Microbial fermentation presents a promising technology for the sustainable production of bio-based chemicals and fuels due to numerous advantages, including high conversion rate, ease of product separation, environmental friendliness, etc. Biosynthesis has a broader range of raw materials and lower costs than chemical processing.6 Accordingly, researchers have concentrated on the utilization of low-cost alternative carbon sources to reduce production costs and achieve green and sustainable production of SA, thus improving the competitiveness of bio-based products.The development of low-cost and sustainable bio-based SA production processes using lignocellulosic biomass as raw materials is a major concern for its industrial production. Lignocellulosic biomass is a renewable raw material that is widely available and can be used to produce a variety of bio-based products.7,8 The primary components of lignocellulosic biomass are cellulose, hemicellulose, and lignin, which make up roughly 32–52, 16–33, and 9–32 weight percent (wt%), respectively, in different plant varieties.9,10 Lignocellulose is pretreated to destroy its resistant structure, and then, the released cellulose/hemicellulose is hydrolyzed to microbial fermentable monomeric sugars by enzymatic saccharification.11 One of the main challenges faced in subsequent microbial fermentation is that most microbial strains cannot efficiently metabolize lignocellulose degradation products.12 Lignocellulose-derived xylose, accounting for 30–40% of carbohydrates in lignocellulosic hydrolysate, frequently poses challenges to fermenting microbes when utilizing it as a carbon source.13 In addition, inhibitors in lignocellulosic hydrolysates usually prevent microbe strains from fermenting xylose to produce desired chemicals.14 Therefore, given the characteristics of lignocellulosic hydrolysate, efficient co-metabolism of glucose and xylose by engineered microorganisms in the presence of high concentrations of inhibitors is of great significance for sustainable and economical bio-chemical production.15
SA is an intermediate of the tricarboxylic acid cycle, and naturally present producers of SA are regarded as promising candidates for industrial SA.16 A variety of microorganisms, including Actinobacillus succinogenes, Anaerobiospirillum succiniciproducens, Basfia succiniciproducens, etc., have been reported to produce SA naturally.17 However, the industrial applications of these bacterial hosts are limited due to their potential pathogenicity, poor cell growth, and stress tolerance. In particular, the above-mentioned bacteria isolated from bovine rumen have multiple auxotrophies, and complex component media must be set up to promote their growth.18 An alternative strategy is to use genetically modified industrial microorganisms that do not naturally produce large amounts of SA, such as Escherichia coli, Corynebacterium glutamicum, etc. Through combined mutagenesis and subsequent optimization, Gao et al. constructed an engineered E. coli strain, FMME-N-30, which could obtain 119.0 g L−1 succinate from glucose.19 Li et al. introduced the isomerase pathway and the Weimberg pathway into C. glutamicum to obtain a recombinant strain that could simultaneously use glucose and xylose to produce succinate.20 However, due to the toxicity of various inhibitors in the hydrolysates, the utilization of lignocellulosic hydrolysates by bacteria requires the adoption of high-density fermentation strategies or diluting the hydrolysates with the medium before inoculation. Additionally, the pH during bacterial fermentation needs to be maintained at neutral, which greatly increases the production and downstream purification costs. In contrast to using bacteria, numerous studies have shown that yeast could produce more than 100 g L−1 of organic acids, including succinic acid and malic acid, without requiring neutral pH maintenance owing to its acid-tolerance property.21,22 For lignocellulosic biorefinery, the yeast Y. lipolytica Hi-SA2-YlGsh2 was reported to achieve a high SA titer of 45.34 g L−1 from lignocellulosic hydrolysate without pH control.23 Hence, yeast is considered a preferred candidate for industrial production due to its safety and robustness, especially when considering the intricate features of lignocellulosic biorefinery.10
Yarrowia lipolytica, an unconventional yeast generally recognized as safe, has attracted increasing interest due to its unique characteristics, including ease of genetic manipulation, broad substrate adaptability, and potential applications in the biosynthesis of different bio-chemicals.24,25 In recent years, various carbon sources have been used in genetically modified Y. lipolytica to enhance SA production.22,26,27 Among them, Y. lipolytica PGC01003 achieved the highest ever SA titer of 209.7 g L−1 using crude glycerol as the sole carbon source with an in situ fibrous bed bioreactor derived from sugarcane bagasse. In addition, the genetic pathway-optimized Y. lipolytica strain PGC62-SYF-Mae could obtain an SA titer exceeding 100 g L−1 with glucose as the sole carbon source.28 When considering xylose as the carbon source, even though it has been reported that the engineered Y. lipolytica without xylose metabolic pathway could also utilize a small amount of xylose in the presence of glucose, it remained unclear whether xylose could be successfully converted into SA.23,29 Researchers have also introduced xylose metabolism pathways in Y. lipolytica to generate recombinant strains PSA02004PP, which can only produce SA at a maximum titer of 22.3 g L−1 with a low yield of 0.15 g g−1 using xylose as the sole carbon resource. Besides, a low SA titer of 5.6 g L−1 was detected when sugarcane bagasse-derived hydrolysate was employed.30 Until now, despite many efforts that have been made to produce SA from xylose, these engineered Y. lipolytica yeasts are still unable to produce sufficient bio-SA to meet industrial demands employing lignocellulose as a carbon source.
Herein, an engineered Y. lipolytica yeast capable of biosynthesizing SA using xylose as the sole carbon source was first constructed (Fig. 1). Afterward, tailored strategies were developed for simulating the proportion of glucose and xylose components contained in lignocellulosic hydrolysate, which allowed the engineered strain to completely co-ferment high-concentration mixed sugars. Finally, an efficient fermentation process was developed for SA production from undetoxified corn stover (CS) hydrolysate. In general, this study demonstrated that glucose and xylose in actual lignocellulosic hydrolysate could be completely consumed by engineered Y. lipolytica to produce SA, and the developed process proposed based on this robust strain could reduce the overall production cost, offering a green and sustainable alternative for large-scale industrial production of bio-SA.
2. Materials and methods
2.1 Strains, media, and culture conditions
The xylose-utilization strain Y. lipolytica BZ used in this study originated from Y. lipolytica Po1f-ΔKu70 (disruption of the Ku70 gene) with overexpression of endogenous xylulokinase (XK), and heterologous expression of D-xylose reductase (XR) and D-xylitol dehydrogenase (XDH) from Scheffersomyces stipites. Each gene mentioned above was flanked with promoter PTEFin and the terminator TXPR, and these three fragments were then inserted into restriction sites of Mss I and EcoR I of plasmid 26s rDNA-HUH. The verified correct plasmid was linearized and integrated into the genome of Y. lipolytica Po1f-ΔKu70 to generate Y. lipolytica BZ for further genetic modification. All the strains and plasmids constructed in this study are listed in Table 1. Escherichia coli Top 10 was cultured at 37 °C in Luria–Bertani media (5 g L−1 yeast extract, 10 g L−1 tryptone, 10 g L−1 NaCl) supplemented with ampicillin (50 mg L−1) or kanamycin (10 mg L−1) as needed for plasmid construction and propagation. All the Y. lipolytica strains were cultured at 30 °C with shaking at 220 rpm. YPG20 medium (10 g L−1 yeast extract, 20 g L−1 tryptone, 20 g L−1 glycerol) was used for the routine culture of Y. lipolytica after the URA3 selection marker had been recycled. YPGX medium (10 g L−1 yeast extract, 20 g L−1 tryptone, 10 g L−1 glycerol, 20 g L−1 xylose) was used for the routine culture of engineered Y. lipolytica. YNBG plates (20 g L−1 glycerol, 1.7 g L−1 YNB without amino acids, 5 g L−1 NH4SO4, 22 g L−1 agar) were used for screening transformants. The URA3 marker was removed by counter-selection on YPG plates supplemented with 1.2 g L−1 5-fluoroorotic acid (5-FOA).31 For SA fermentation, the modified YPD medium (10 g L−1 yeast extract, 20 g L−1 tryptone, 40–80 g L−1 glucose), YPX medium (10 g L−1 yeast extract, 20 g L−1 tryptone, 20–40 g L−1 xylose), or YPDX medium (10 g L−1 yeast extract, 20 g L−1 tryptone, 40–80 g L−1 glucose, 20–40 g L−1 xylose) was used.| Strains and plasmids | Descriptions | Sources |
|---|---|---|
| a Randomly selected transformant with chromosomal iterative integration of related genes. | ||
| Strains | ||
| E. coli Top 10 | A host strain used for the amplification of constructed plasmids | Lab stock |
| BZ | MatA, xpr2-322, axp-2, leu2-270, ura3-302, Δku70::hisG, YlXk, SsXr, SsXdh | Lab stock |
| BS3302 | BZ ΔYlSdh5::hisG-URA3-hisG | This study |
| BAS4332 | BS3302 ΔYlAch1::hisG-URA3-hisG | This study |
| BPS2a10a | BAS4332 Δ26S rDNA::PTEFin-ScPck-TCYC, PTEFin-YlScs2-TXPR2 | This study |
| BPS2b10a | BAS4332 Δ26S rDNA::PTEFin-ScPck-TCYC, PTEFin-YlScs2-TXPR2 | This study |
| BPS2b14a | BAS4332 Δ26S rDNA::PTEFin-ScPck-TCYC, PTEFin-YlScs2-TXPR2 | This study |
| BDic5 | BPS2b14 ΔintE3::PTEFin-YlDic-TCYC | This study |
| BMae11 | BPS2b14 ΔintE3::PTEFin-YlMae-TCYC | This study |
| BDM14 | BPS2b14 ΔintE3::PTEFin-YlDic-TCYC, PTEFin-YlMae-TXPR2 | This study |
| YLGZ1-6 | BDic5 ΔA2::PTEFin-YlYht1-6-TCYC | This study |
| Plasmids | ||
| pUC-HUH | hisG-URA3-hisG (HUH) in pUC57 | Lab stock |
| 26s rDNA-HUH | hisG-URA3-hisG (HUH) in p-26s rDNA | Lab stock |
| pUC-intE3-HUH | IntE3 upstream and downstream homology arms inserted into pUC-HUH | This study |
| pUC-HUH-ΔSdh5 | YlSdh5 upstream and downstream homology arms inserted into pUC-HUH | This study |
| pUC-HUH-ΔAch1 | YlAch1 upstream and downstream homology arms inserted into pUC-HUH | This study |
| 26s rDNA-Pck-Scs | P TEFin -ScPck-TCYC and PTEFin-YlScs-TXPR2 cassettes in rDNA-HUH | This study |
| pUC-HUH-Dic | P TEFin -YlDic-TCYC cassette in pUC-intE3-HUH | This study |
| pUC-HUH-Mae | P TEFin -YlMae-TCYC cassette in pUC-intE3-HUH | This study |
| pUC-HUH-Dic-Mae | P TEFin -YlDic-TCYC and PTEFin-YlMae-TXPR2 cassettes in pUC-intE3-HUH | This study |
| pUC-HUH-Yht1-6 | P TEFin -YlYht1-6-TCYC cassette in pUC-intE3-HUH | This study |
2.2 Raw materials and pretreatment
The CS used in this study was purchased from a farm in Lianyungang, Jiangsu, China, and its main components were 30.33 wt% glucan and 19.36 wt% xylan. For the pretreatment of CS, an original pretreatment approach developed in our laboratory named DLCA(sa) (Densifying Lignocellulosic biomass with Sulfuric Acid followed by a regular steam Autoclave) was adopted. Pretreatment reagent dosage, process conditions, and subsequent enzymatic saccharification were applied as described in previous studies.32,33 Briefly, CS was mixed with sulfuric acid (0.075 g sulfuric acid per g dry biomass) and water (0.5 g water per g dry biomass), and then pelleted using a flat die pellet machine. Afterward, the obtained pellets were immediately autoclaved at 121 °C for 30 min to achieve DLCA(sa)-pretreated CS. Subsequently, the DLCA(sa)-CS was neutralized to a pH of 5.0 and hydrolyzed for 72 h with cellulase CTec3 HS at a dosage of 20 mg protein per g glucan using shake flasks at solid loadings of 20%, 25%, 30%, and 35% (w/w), respectively. For DLCA(ch)-pretreated CS, the whole process was similar to that for DLCA(sa) except that calcium hydroxide (0.15 g g−1 dry biomass) was used and the subsequent autoclaving time was extended to 60 min. The enzymatic saccharified liquid was harvested after centrifugation; details of the hydrolysate are listed in Table S1.†2.3 DNA manipulation
The synthesis of all primers and verification of DNA sequencing in this study were performed by Tsingke Biotechnology Co. Ltd (Nanjing, China). For deleting target genes YlSdh5 and YlAch1, the plasmid consisting of a 2000 bp upper homologous arm, hisG-URA3-hisG (HUH) cassette, and 2000 bp down homologous arm was used to disrupt the related genes using the URA-Blaster protocol,34 and the DNA fragments flanking both sides of the deletion site were amplified using the designed primers (Table S2†) for DNA sequencing verification. The integrative expression plasmid pUC-HUH, combined with different integration sites reported previously,35 was used for overexpression of the endogenous genes including YlScs, YlDic, and YlMae (amplified from the genomic DNA of Y. lipolytica), and the heterologous gene ScPck (amplified from the genomic DNA of S. cerevisiae) under the control of promoter PTEFin and terminator TCYC or TXPR2. All plasmids were obtained from the vector backbone and the corresponding fragment by one-step cloning employing the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The constructed plasmids were linearized and transformed into Y. lipolytica competent cells using the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Orange, CA) as previously described.31 The positive transformations were selected on YNBG plates, and the diagnostic PCR using the genomic DNA of the clones as a template was carried out for identification. All PCR reactions were performed using PrimerSTAR Max DNA Polymerase (TaKaRa, Beijing, China) or KOD-plus-Neo DNA polymerase (Toyobo, Osaka, Japan). By counter-selecting on YPG plates containing 1.2 g L−1 5-FOA, the URA3 selection marker was recycled to engineer subsequent strains.2.4 Testing sugar consumption of engineered strains in shake flasks
All the engineered yeast was fermented separately in shake flasks using YPD, YPX, or YPDX to test its capacity for sugar utilization. First, the engineered strains preserved in glycerol tubes at −80 °C were incubated overnight in test tubes with 5 mL of YPG to restore their growth activity. Then, a portion of the microbial solution in the test tube was transferred into 50 mL shake flasks containing 10 mL of YPGX medium as seed culture. Finally, yeast cells from the seed culture were harvested by centrifugation and inoculated into 250 mL shaking flasks that contained 50 mL of testing medium with an initial OD600 of 1.0 to initiate SA fermentation. Samples were extracted every 24 h (sampling intervals were shortened when necessary) during fermentation to determine cell growth, glucose and xylose consumption, and SA titer. It should be emphasized that the pH of the fermentation carried out in the shake flasks was not adjusted, and all the experiments were conducted in triplicate.2.5 Batch and fed-batch fermentation with DLCA(sa)-pretreated CS as feedstock
To test the fermentation performances of Y. lipolytica strains on real lignocellulosic hydrolysate, DLCA(sa)-CS was used as feedstock. Y. lipolytica BDic5 was cultured in YPGX medium as the fermentation seed, and cells were harvested and inoculated into hydrolysate containing 10 g L−1 yeast extract and 20 g L−1 tryptone with an OD600 value of 10.0 to initiate fermentation in shake flasks. The required pH was manually adjusted every 24 h under sterile conditions using 10 M NaOH. For hydrolysate fermentation, all the experiments were performed in duplicate.To achieve higher SA titers, fed-batch fermentation was carried out in a 3 L bioreactor (Bailun Bio, Shanghai, China). Y. lipolytica BDic5 was incubated overnight in the test tube with 5 mL of YPGX medium to restore growth activity. 1 mL of the culture was inoculated into 50 mL of YPGX medium in 250 mL shake flasks, and the cells were harvested and inoculated into DLCA(sa)-CS hydrolysate as seed culture at 30 °C and 220 rpm for 48–60 h. The seed culture was then inoculated into the bioreactor to start fed-batch fermentation. The fermentation was carried out at 30 °C, 800 rpm, and 1 vvm. By observing the online monitoring pH sensor, the pH of the fermentation system was manually adjusted to 6.5 with 10 M NaOH solution every 24 h. Antifoam agent Antifoam 204 (Sigma-Aldrich, A6426) was added to the bioreactor as required. When the residual xylose in the bioreactor dropped below 10 g L−1, approximately 100–120 mL of concentrated hydrolysate (hydrolysate after rotary steaming, which contained 505.6 g L−1 glucose and 284.1 g L−1 xylose) was supplemented.
2.6 SA recovery
The modified direct crystallization method was employed for SA recovery from the fermentation broth.36 Briefly, the broth was first centrifuged at 8000 rpm for 10 min to remove yeast cells and insoluble impurities. Afterward, the obtained supernatant was mixed directly with 8% (w/v) activated carbon and placed in a shaker at 250 rpm for 2 h for decolorization. In order to ensure complete decolorization, this step was performed in duplicate. The clarified filtrate was harvested by vacuum filtration, and the pH of that was adjusted to 2.0 using 35% (v/v) dilute hydrochloric acid. Subsequently, the decolorized filtrate was subjected to vacuum distillation and concentrated until crystals could be observed. Then, the concentrated filtrate was stored at 2–4 °C (>5 h) for SA crystallization. Finally, the SA crystals were collected after vacuum filtration, and the resulting filtrate was further subjected to the above steps for recovering residual SA.2.7 Quantitative analysis
An HPLC system (Agilent, 1260 Infinity II, USA), equipped with an Aminex HPX-87H column and a refractive index detector, was used to measure the concentrations of SA, residual sugars, and by-products after the sample supernatant had been appropriately diluted and filtered through a 0.45 μm syringe filter. The optical density of the fermentation broth at 600 nm (OD600) was measured using a TU-1810 spectrophotometer (PERSEE, Beijing, China) to track the cell growth of Y. lipolytica. Samples were eluted with 5 mM H2SO4 with a flow rate of 0.6 mL min−1 and the column temperature was set at 65 °C.3. Results and discussion
3.1 Establishment of the xylose metabolic pathway for SA production in Y. lipolytica
Succinic acid, as an intermediate of the tricarboxylic acid (TCA) cycle in cells, is catalytically depleted by the succinate dehydrogenase (SDH) complex in mitochondria. The SDH complex consists of five subunits, of which a soluble mitochondrial protein encoded by the subunits YlSdh5 is essential for SDH-dependent respiration.37 Therefore, the URA-Blaster method was used to knock out the Sdh5 gene in Y. lipolytica BZ, and the resulting mutant BS3302 was employed for further testing. The BS3302 strain with deletion of the YlSdh5 gene was able to grow on xylose as the sole carbon source, and succinic acid production could be detected in the fermentation broth. After being cultivated on YPX20 medium, the recombinant strain BS3302 quickly consumed 5.08 g of xylose at 24 h, resulting in a very low SA titer of 0.62 g L−1 (Fig. 2a). However, a large amount of the by-product acetic acid (1.70 g L−1) was detected in the supernatant, and the combined production of these two organic acids contributed to a rapid decrease in the pH of the whole fermentation system. Acetic acid is a common by-product produced during microbial fermentation, and its excessive accumulation may contribute to the loss of carbon sources and potentially prevent microbial growth.38 The maximum OD600 value obtained was 8.56 at 24 h, followed by a rapid decline to 3.83 at 96 h, which indicated that the excess acetic acid produced during the fermentation severely inhibited the growth of BS3302. The introduction of the xylose pathway resulted in 1.07 g L−1 SA with a yield of 0.19 g g−1. However, the acetic acid titer reached 2.55 g L−1 at the end of fermentation, which was significantly higher than that of the target product SA, proving that more carbon flux flowed into the biosynthesis of acetic acid due to the imbalance between glycolysis and the TCA cycle. Fig. 2b shows the time course profiles for glucose assimilation. The strain quickly ingested glucose and reached peak cell biomass (OD600 = 7.01) within 48 h, which is consistent with xylose metabolism data. At the same time, the fermentation process also produced a large amount of acetic acid, which caused the cell biomass of the strain to drop rapidly when it reached its peak. The recombinant strain was able to produce 1.11 g L−1 SA from glucose with 0.26 g g−1 yield. After incubation with glucose or xylose as the sole carbon source, co-fermentation experiments using glucose and xylose were performed (Fig. 2c). Similarly, consistent with its use of glucose as the sole carbon source, the strain rapidly consumed glucose and produced large amounts of acetic acid within 48 h, which severely inhibited the cell growth. The co-fermentation of glucose and xylose resulted in the maximum OD600 value of 7.15 at 48 h, and only an SA titer of 1.09 g L−1 was detected at 96 h. At the end of fermentation, the production of acetic acid reached 3.75 g L−1, which was 3.4 times that of SA, suggesting that elimination of the by-product metabolic flux was essential for enhancing SA accumulation. | ||
| Fig. 2 Fermentation performance of Y. lipolytica BS3302 (a, b, c) and BAS4332 (d, e, f) strains in media with different carbon sources. | ||
As described in the previous study, acetic acid accumulation was detrimental to SA production and cell growth in Sdh5-deleted Y. lipolytica, while these negative effects could be substantially mitigated by knocking out the vital gene YlAch, which encodes acetyl-CoA hydrolase.38 On the basis of BS3302, the YlAch gene was further knocked out to generate BAS4332, which was then cultivated in different fermentation media. As anticipated, the recombinant strain BAS4332 grew robustly when it was cultivated on 20 g L−1 xylose since there was a significant decrease in the acetic acid level (241 mg L−1). As shown in Fig. 2d, the xylose (22.21 g L−1) was completely utilized within 72 h, resulting in an SA titer of 6.9 g L−1 with a yield of 0.31 g g−1. The maximum OD600 obtained was 26.78 at 48 h, which then decreased slightly at 72 h due to the xylose being completely consumed. The xylose assimilation performance, SA yield, and cell growth of strain BAS4332 were greatly enhanced compared with that of BS3302. This phenomenon suggested that the elimination of the acetic acid pathway was essential for SA biosynthesis from xylose. When glucose was used as the sole carbon source (Fig. 2e), BAS4332 completely exhausted 40 g L−1 glucose within 120 h, yielding a maximum cell OD600 and SA titer of 24.97 and 15.3 g L−1, respectively. During the co-fermentation with 40 g L−1 glucose and 20 g L−1 xylose, glucose was completely metabolized within 120 h, while 10.13 g L−1 of xylose was consumed at the end of fermentation (Fig. 2f). Benefiting from the removal of the acetic acid overflow and the recovery of strain growth, Y. lipolytica BAS4332 successfully achieved xylose co-fermenting after large amounts of glucose were consumed. The co-fermentation of glucose and xylose resulted in the maximum OD600 value of 27.05 with an SA titer of 18.78 g L−1 at 168 h. Under this condition, the overall yield of BAS4332 using glucose and xylose as carbon resources reached 0.38 g g−1. Although the actual cumulative concentrations of glucose and xylose exceeded 60 g L−1, there was no lag phase in the initial fermentation stage. Further increasing the concentration of mixed sugars would cause a metabolic burden on the strain, thus the process showed a significant lag phase during the first 24 h of fermentation (Fig. S1†). The co-fermentation of glucose and xylose mixture resulted in the SA titer of 23.33 (60 g L−1 glucose and 30 g L−1 xylose) and 27.14 g L−1 (80 g L−1 glucose and 40 g L−1 xylose), respectively. However, although the cell growth of strain BAS4332 was partly recovered when acetic acid overflow was reduced, low SA yield and residual sugar (especially xylose) after fermentation remained resolved. In order to clearly understand the ability of the strain to assimilate xylose in the presence of glucose, subsequent experiments will need to be conducted using mixed sugars.
3.2 Optimizing metabolic pathways to enhance SA biosynthesis
As previously described, the YlSdh and YlAch double-deficient Y. lipolytica strain produced by-products at the end of fermentation, demonstrating the presence of metabolic flux inhibition from acetyl-CoA.38 To address this issue and further direct the carbon flow to SA, the key genes ScPck (encoding phosphoenolpyruvate carboxylase from S. cerevisiae) and YlScs2 (encoding succinyl-CoA synthase subunit 2) were overexpressed. The expression cassettes of the two genes were randomly integrated into multiple cloning (26s rDNA) sites in the genome of Y. lipolytica by homologous recombination.Due to the random integration of functional genes into yeast chromosomes, the resulting transformants obtained may have different gene copy numbers, thus exhibiting distinct metabolic characteristics. The sugar consumption and SA production of randomly selected transformants (genes integration has been verified by PCR) at 144 h fermentation are depicted in Fig. 3a. Combined with the experimental results, strains BPS2b10 (which exhibited the highest SA titer with a slightly lower conversion yield), BPS2b14 (characterized by a relatively lower SA titer but the highest conversion yield), and BPS2a10 (with an SA titer similar to that of BPS2b14 but a higher conversion yield compared to BPS2b10) have attracted attention. In the co-fermentation experiment with 80 g L−1 glucose and 40 g L−1 xylose, the fermentation conditions of three strains with glucose depletion as the time node are shown in Fig. 3b–d. During the fermentation, the highest value (30.8) of cell OD600 was achieved in BPS2a10 at 120 h, followed by BPS2b10 (30.68) at 120 h and BPS2b14 (30.22) at 144 h. In shake flask experiments, approximately 81.96, 84.67, and 86.05 g L−1 of glucose was consumed by BPS2a10, BPS2b10, and BPS2b14, respectively. BPS2b10 exhibited the highest xylose consumption (13.55 g L−1), followed by BPS2a10 (10.78 g L−1), which was relatively higher than that of BPS2b14 (8.64 g L−1). Although strain BPS2b14 had a comparatively lower xylose consumption, its SA titer (48.8 g L−1) and overall conversion yield (0.52 g g−1) were the highest among the three strains (Table 2). Hence, the strain BPS2b14 was selected for further genetic modification.
3.3 Transporter engineering enhances sugar assimilation and SA release
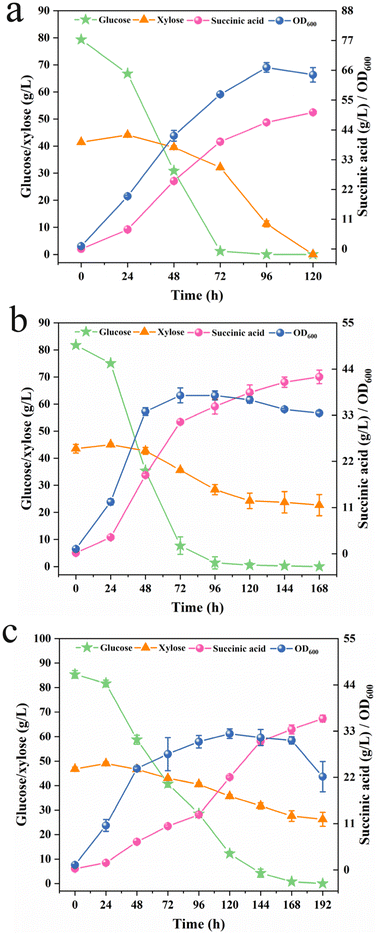
In the presence of glucose, the xylose assimilation was subjected to carbon catabolite repression, and the xylose uptake was permitted only after glucose was largely assimilated.30 One of the most important processes in sugar metabolism is the phosphorylation of hexoses, which is accomplished by specific kinases in the hexokinase gene family, primarily glucokinase and hexokinase.39 Previous studies have shown that deletion of the Hxk (hexokinase) gene in Saccharomyces cerevisiae reduced the maximum rate of glucose consumption, yet overexpression of the Hxk in Y. lipolytica increased biomass yield.40 Therefore, attempts were made to overexpress hexose phosphorylation-related proteins to obtain strains with rapid glucose metabolism ability. However, overexpression of YlGlk (YALI0E15488g) and YlHxk (YALI0B22308g) genes in BPS2b14 showed virtually no effect on Sdh-negative Y. lipolytica (data not shown).From the perspective of final product transport, timely secretion of the product minimized cytotoxicity caused by the accumulation of compounds in cells, and also avoided negative feedback inhibition to a certain extent, thereby further improving the production efficiency of target products.41 According to Jiang et al.,28 overexpression of mitochondrial dicarboxylic acid transporter YlDic1 (YALI0B03344g) or C4-dicarboxylic acid transporter YlMae (YALI0E24167g) in Sdh-negative Y. lipolytica could efficiently improve the SA titer. Encouragingly, the YlDic overexpressing strain BDic5 exhibited significantly improved sugar assimilation capacity (Fig. 4a). When inoculated in YPD80X40 medium, the recombinant strain BDic5 could consume glucose rapidly within approximately 72 h, concomitant with the complete depletion of xylose in 120 h. The sugar consumption rate of BDic5 was greatly accelerated compared with that of parent strain BPS2b14, and no residual xylose was detected after fermentation. Finally, it resulted in a maximum cell OD600 of 67.03 at 96 h, an SA titer of 50.48 g L−1 with an overall yield of 0.42 g g−1. Furthermore, the fermentation performance of BDic5 was tested by fed-batch fermentation in shake flasks, which yielded 102.44 and 73.89 g L−1 SA from the glucose–xylose mixture or pure xylose, respectively (Fig. S2†). In addition, the strain BMae3 (overexpressing YlMae in BPS2b14) also exhibited a higher glucose utilization rate. It could consume 80 g L−1 glucose in approximately 96 h, while xylose failed to be completely utilized even after extending the fermentation time to 168 h (Fig. 4b). In contrast, overexpression of the key gene YlDic significantly increased the sugar assimilation capacity of Y. lipolytica BDic5, particularly in xylose uptake. It could be inferred that overexpression of the mitochondrial dicarboxylic acid transporter responsible for transporting SA from mitochondria to cytoplasm could effectively alleviate the toxicity of SA to cells and alleviate the metabolic pressure of yeast to a certain extent, thereby promoting the bioconversion of sugars to SA. However, no apparent effect of co-expression of the two genes (YlDic and YlMae) on the improvement of sugar assimilation and SA biosynthesis was observed (Fig. 4c). Notably, although the sugar–SA conversion yield of BDic5 decreased compared to that of BPS2b14, its sugar assimilation ability was significantly improved. Moreover, there was no fermentation lag phase exhibited at higher sugar concentration, which was beneficial for the economy of lignocellulosic biorefinery. To further increase the rate of glucose assimilation, an attempt was made to overexpress glucose transporters in engineered strains. However, overexpression of glucose transporters (YlYht1-6) from Y. lipolytica in BDic5 failed to enhance the glucose assimilation rate (Fig. S3†).
3.4 Efficient SA production from undetoxified DLCA(sa)-pretreated corn stover in shake flasks
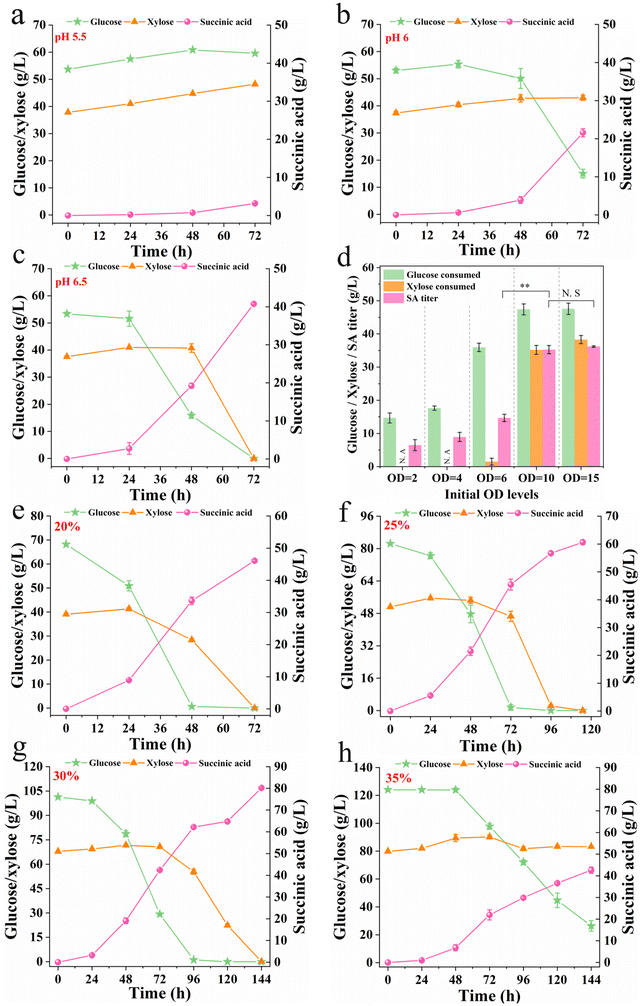
Since the strain BDic5 possessed superior glucose and xylose assimilation properties, it was further used for the production of SA from lignocellulosic biomass. Differing from pure culture fermentation, the implementation of BDic5 in the actual hydrolysate that contained various inhibitors to produce SA was full of indeterminacy. First of all, BDic5 was inoculated into DLCA(sa)-pretreated and DLCA(ch)-pretreated hydrolysates. It was found that the former hydrolysate was able to be utilized for SA biosynthesis while the latter significantly impeded the growth of the strain (data not shown). Afterward, the pH and initial inoculation levels were investigated to determine optimal conditions for SA production. The experimental results showed that the strain was almost completely inhibited under the condition of pH 5.5, and a lag period of nearly 72 h was required to recover the cell growth (Fig. 5a). At initial pH 6, the growth of the strain was partially recovered, but a relatively long growth lag period was still presented under this condition (Fig. 5b). When the initial pH was increased to 6.5, the fermentation performance of the strain almost recovered to that of pure culture fermentation, and the glucose and xylose in the system could be completely consumed within 72 h (Fig. 5c). Then, the inoculation levels of Y. lipolytica BDic5 were carried out under the optimized condition of pH 6.5. As shown in Fig. 5d, when the initial inoculation OD600 value was 2 or 4, the strain could only produce a low titer of SA after 72 h of fermentation. Further increasing the initial OD600 value to 6.0, the glucose and xylose were still not utilized after 72 h of fermentation. However, when the initial inoculation level increased to 10, the strain could resume normal performance and uptake all the sugars within 72 h. Further increasing the inoculum level had little effect on the fermentation performance. In order to reduce the lag phase of fermentation, an inoculation level of 10.0 was used for subsequent experiments. Finally, the fermentation performance of BDic5 was tested in the hydrolysate with different solid loadings under the optimum conditions (pH 6.5, initial inoculation OD600 of 10.0).As shown in Fig. 5, the engineered strain BDic5 survived well in 20–35% solid loading of corn stover hydrolysate and could successfully convert sugar into SA. When incubated in the 25% solid loading of corn stover hydrolysate, BDic5 was able to consume all the sugars (67.2 g L−1 glucose and 39.08 g L−1 xylose) within 72 h to produce an SA titer of 46.07 g L−1 with overall yield of 0.43 g g−1 (Fig. 5e). When the solid loading increased to 25% and 30%, higher solid loadings of feedstock produced more fermentable sugars for production of SA. From the experimental results, it can be seen that a total of 133.68 g L−1 (82.45 g L−1 glucose and 51.23 g L−1 xylose) and 169.14 g L−1 (101.34 g L−1 glucose and 67.8 g L−1 xylose) of fermentable sugars was produced under 25% and 30% solid loadings, respectively. At these sugar concentrations, the fermentation performance of strain BDic5 was not inhibited. After fermentation, BDic5 successfully consumed all the sugars contained in the system to produce 60.62 and 80.24 g L−1 SA under 25% and 30% solid loading, respectively (Fig. 5f and g). The conversion ratios of sugar to SA were 0.45 and 0.47 g g−1, respectively. A further increase in solid loading to 35% had a negative impact on both cell growth and SA biosynthesis (Fig. 5h). Under such high solid loading circumstances, the elevated initial sugar concentrations and increased inhibitor contents exhibit significant inhibitory effects, resulting in a more prolonged lag phase (48 h) and decreased SA productivity. Nevertheless, despite such a harsh environment, the strain still showed slow sugar assimilation and SA production, indicating that the engineered strain constructed in this paper has a strong ability to acclimate and tolerate changes in the external environment of the hydrolysate. Overall, the robust strain Y. lipolytica BDic5 exhibited non-weak performance in the hydrolysate when compared to pure culture fermentation, indicating its promising potential for application in the industrial production of bio-SA from lignocellulosic biomass.
3.5 Fed-batch fermentation with hydrolysate in the 3 L bioreactor
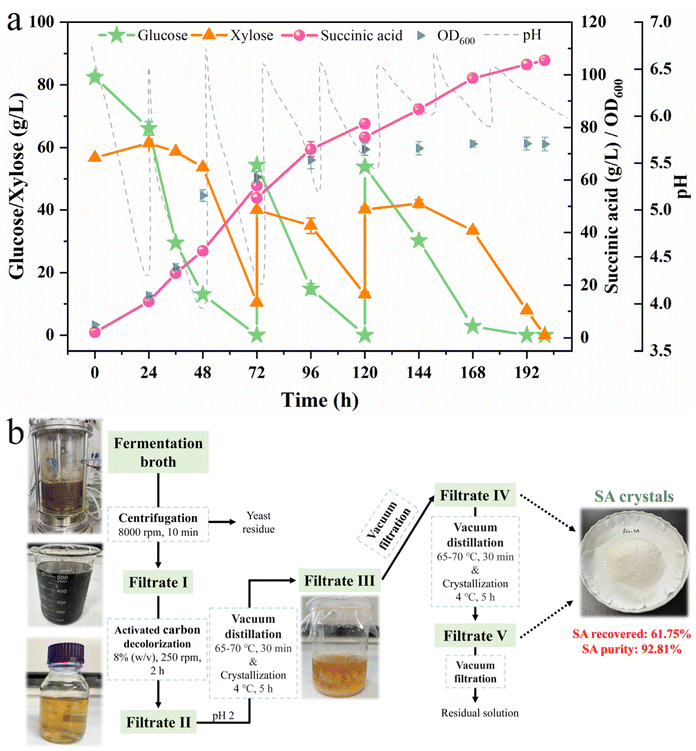
Compared with the strain BS3302 with YlSdh5 inactivation, BDic5 significantly improved the sugar assimilation efficiency in high concentrations of glucose–xylose mixture for efficient SA production. By conducting a batch fermentation test using actual hydrolysate with different solid loading, the strain showed excellent performance in lignocellulosic biorefinery. Higher solid loading of feedstock means that more fermentable sugars can be produced to be utilized by microorganisms for the production of desired products. Therefore, it is of great significance to obtain the hydrolysate with the high substrate solid loading for the production of SA without affecting the actual fermentation performance of the engineered strain. Hence, 30% solid loading DLCA(sa)-CS hydrolysate was selected in fed-batch fermentation. To further improve SA production by the strain BDic5 using undetoxified hydrolysates, fed-batch fermentation using concentrated hydrolysate as a carbon source was implemented in the 3 L bioreactor. Through optimization, the fermentation conditions were set as follows: stirring speed of 800 rpm, airflow rate of 1 vvm, and pH (6.5) adjustment every 24 h.As shown in Fig. 6a, since the seed culture of BDic5 was carried out in the same hydrolysate, the adapted strain (the initial OD600 after inoculation was 4.93) could rapidly consume glucose and xylose in the bioreactor and produce 57.85 g L−1 SA within 72 h. After 200 h of cultivation with two times feeding, all the glucose (190.52 g L−1) and xylose (113.51 g L−1) contained in the medium were completely assimilated by strain BDic5, and the final SA titer reached 105.42 g L−1. To our knowledge, this is the highest fermentative SA production achieved from lignocellulosic biomass so far (Table 3). The overall SA yield and average productivity were 0.35 g g−1 total sugars and 0.53 g L−1 h−1, respectively. Due to its great acidity and inhibitor tolerance, Y. lipolytica is an excellent microbial host for the production of bio-SA. In contrast to bacteria, which need to maintain a neutral pH condition throughout the process, the pH of Y. lipolytica fermentation is only adjusted at intervals to create suitable conditions for meeting the growth of yeast cells in harsh environments. Therefore, the cost of alkali reagent and subsequent SA purification will be further reduced. Moreover, the lignocellulosic hydrolysate with high solid loading can be directly utilized by the engineered strain BDic5 without further detoxification or additional dilution with complex media (Table 3). These results further confirmed that the engineered strain Y. lipolytica BDic5 has great potential for green and sustainable bio-SA production from lignocellulosic biomass. Even though more than 100 g L−1 SA was produced from lignocellulosic hydrolysate by Y. lipolytica BDic5, the yield and productivity of SA remained relatively low. Hence, further metabolic modification of the recombinant strain should be performed to enhance SA production efficiency.
| Strains | Subtract | Fermentation strategy | Xylose consumed (g L−1) | Xylose residue | Succinic acid | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Titer (g L−1) | Yield (g g−1) | Productivity (g L−1 h−1) | ||||||
| a Estimates based on graphical data from related papers. N. A: data not available. | ||||||||
| C. glutamicum K5 | Corn stover | Batch, high-density fermentation (OD600 = 30), detoxification of hydrolysate (washed by tap water during CASA pretreatment), Mg2(OH)2CO3 were used to prevent acidification | 16.5 | Yes | 64.16 | 0.76 | 1.07 | 20 |
| C. glutamicum CGS5 | Corn stalk | Batch, high-density fermentation (OD600 = 150), CGXIIB medium with diluted hydrolysate, neutralization by 4MgCO3·Mg(OH)2·5H2O | 30.1 | No | 98.6 | 0.87 | 4.29 | 42 |
| A. succinogenes 130Z | Corn stover | Batch, hydrolysate was diluted at 56% (v/v), the pH was maintained at 6.8 via Na2CO3 | 55.4 | No | 42.8 | 0.74 | 1.27 | 43 |
| E. coli DC1515 | Corn stalk | Batch, hydrolysate (after detoxification) supplied with yeast extract, MgCO3 was added as the pH buffer | N. A | N. A | 38.6 | 0.39 | 0.8 | 44 |
| E. coli SD121 | Corn stalk | Fed-batch, hydrolysates were diluted about 2-fold, the pH was maintained at 7.0 (aerobic phase) and 6.7 (anaerobic phase) | 18–20a | Yes | 57.8 | 0.87 | 0.96 | 45 |
| Y. lipolytica PSA02004 | Sugarcane bagasse | Batch, hydrolysate supplied with 2% corn steep liquor, the pH was maintained at 6.0 | 10.2 | Yes | 33.2 | 0.58 | 0.33 | 29 |
| Y. lipolytica Hi-SA2-YlGsh2 | Corncobs | Fed-batch, 20%–50% (v/v) hydrolysates were added to the culture medium, without pH control | 6.2 | Yes | 45.34 | 0.71 | 1.42 | 23 |
| Y. lipolytica BDic5 | Corn stover | Batch, high solid loading hydrolysate supplied with 1% yeast extract and 2% peptone, the pH was adjusted to 6.5 every 24 h | 67.8 | No | 80.24 | 0.47 | 0.56 | This study |
| Y. lipolytica BDic5 | Corn stover | Fed-batch, high solid loading hydrolysate supplied with 1% yeast extract and 2% peptone, the pH was adjusted to 6.5 every 24 h | 113.51 | No | 105.42 | 0.35 | 0.53 | This study |
The fermentative bio-SA was extracted and purified from the acidic broth (pH = 5.98) of the hydrolysate fermentation (Fig. 6b). Following the modified direct crystallization method, 61.75% of the total SA with purity of 92.81% (Fig. S4†) was recovered from the fermentation broth. In general, the total SA recovery obtained by direct crystallization methods was similar to the result reported by Alexandri et al., which used the B. succiniciproducens fermentation broth.36 Otherwise, the purity of the recovered SA was similar to that obtained from the acidic broth of the fed-batch SA fermentation (93.5%) using glucose as the substrate.22 This indicated that such a high purity of the SA product was triggered by the lower production of inhibitors during DLC pretreatment and the complete utilization of the major sugars in the hydrolysate. However, the purification process needs to be further optimized to improve SA recovery from SA fermentation broth of hydrolysate.
4. Conclusion
In this study, an engineered Y. lipolytica strain with superior glucose–xylose utilization efficiency was constructed to produce SA from lignocellulosic biomass. Through blocking by-products and channeling metabolic flow toward SA biosynthesis, the strain BPS2b14 capable of producing SA from high concentrations of the glucose–xylose mixture was initially constructed, yet the rate of sugar consumption remained low. By further overexpressing mitochondrial dicarboxylic acid transporter YlDic, an engineered strain BDic5 was obtained, which could produce 105.42 g L−1 SA from DLCA(sa)-CS hydrolysate in fed-batch fermentation. This is the highest SA titer achieved from lignocellulosic feedstock to date, and there was no xylose residue after fermentation. The lower broth pH value and higher SA purity make the entire lignocellulosic biorefinery more economical. This demonstrates that the strain constructed in this study contributes to the development of green and sustainable bio-SA from lignocellulosic biomass.Author contributions
Mianshen Ge designed and performed the experiments, analyzed the data, and drafted the manuscript. Yuanyuan Sha participated in the experimental design and revised the manuscript. Minrui Lu and Yuwei Zhang participated in the experiments. Zhaoxian Xu, Sitong Chen, and Ying Ding revised the manuscript. Mingjie Jin managed the project, supervised the work, and revised and proofread the manuscript.Data availability
The data that support the findings of this study are available from the corresponding author, Mingjie Jin, upon reasonable request.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the “National Key R&D Program of China” (No. 2021YFC2101301).References
- C. Pateraki, S. J. Andersen, D. Ladakis, A. Koutinas and K. Rabaey, Green Chem., 2019, 21, 2401–2411 RSC.
- E. Mancini, S. S. Mansouri, K. V. Gernaey, J. Luo and M. Pinelo, Crit. Rev. Environ. Sci. Technol., 2020, 50, 1829–1873 CrossRef CAS.
- D. Silva and E. Bogel-Łukasik, Green Chem., 2017, 19, 4048–4060 RSC.
- E. Mancini, R. Dickson, S. Fabbri, I. A. Udugama, H. I. Ullah, S. Vishwanath, K. V. Gernaey, J. Luo, M. Pinelo and S. S. Mansouri, Chem. Eng. Res. Des., 2022, 179, 401–414 CrossRef CAS.
- X. Liu, G. Zhao, S. Sun, C. Fan, X. Feng and P. Xiong, Front. Bioeng. Biotechnol., 2022, 10, 843887 CrossRef PubMed.
- S. Zhou, M. Zhang, L. Zhu, X. Zhao, J. Chen, W. Chen and C. Chang, Biotechnol. Biofuels Bioprod., 2023, 16, 1–17 CrossRef CAS PubMed.
- G. P. Robertson, S. K. Hamilton, B. L. Barham, B. E. Dale, R. C. Izaurralde, R. D. Jackson, D. A. Landis, S. M. Swinton, K. D. Thelen and J. M. Tiedje, Science, 2017, 356, eaal2324 CrossRef PubMed.
- V. Z. Ong and T. Y. Wu, Renewable Sustainable Energy Rev., 2020, 132, 109924 CrossRef CAS.
- M. Ge, Y. Liu, J. Zhou and M. Jin, Bioresour. Technol., 2022, 362, 127762 CrossRef CAS PubMed.
- S. Chen, Z. Xu, B. Ding, Y. Zhang, S. Liu, C. Cai, M. Li, B. E. Dale and M. Jin, Sci. Adv., 2023, 9, eadd8835 CrossRef CAS PubMed.
- Q. Zhao, Z. Zhang, Z. Liu, H. Liang, L. Gao, J. Zhao, G. Liu and Y. Qu, Chem. Eng. J., 2024, 480, 148272 CrossRef CAS.
- Z. Zhao, M. Xian, M. Liu and G. Zhao, Biotechnol. Biofuels, 2020, 13, 1–12 CrossRef PubMed.
- S. R. Kim, Y.-C. Park, Y.-S. Jin and J.-H. Seo, Biotechnol. Adv., 2013, 31, 851–861 CrossRef CAS PubMed.
- T. W. Jeffries, I. V. Grigoriev, J. Grimwood, J. M. Laplaza, A. Aerts, A. Salamov, J. Schmutz, E. Lindquist, P. Dehal and H. Shapiro, Nat. Biotechnol., 2007, 25, 319–326 CrossRef CAS PubMed.
- Y. Chen, J. Ind. Microbiol. Biotechnol., 2011, 38, 581–597 CrossRef CAS PubMed.
- J. W. Lee, J. Yi, T. Y. Kim, S. Choi, J. H. Ahn, H. Song, M.-H. Lee and S. Y. Lee, Metab. Eng., 2016, 38, 409–417 CrossRef CAS PubMed.
- H. Song and S. Y. Lee, Enzyme Microb. Technol., 2006, 39, 352–361 CrossRef CAS.
- J. J. Beauprez, M. De Mey and W. K. Soetaert, Process Biochem., 2010, 45, 1103–1114 CrossRef CAS.
- C. Gao, W. Tang, L. Guo, G. Hu, J. Liu, L. Liu and X. Chen, Syst. Microbiol. Biomanuf., 2022, 2, 331–344 CrossRef CAS.
- K. Li, C. Li, X.-Q. Zhao, C.-G. Liu and F.-W. Bai, Bioresour. Technol., 2023, 378, 128991 CrossRef CAS PubMed.
- Y. Xi, H. Xu, T. Zhan, Y. Qin, F. Fan and X. Zhang, Metab. Eng., 2023, 75, 170–180 CrossRef CAS PubMed.
- Z. Cui, Y. Zhong, Z. Sun, Z. Jiang, J. Deng, Q. Wang, J. Nielsen, J. Hou and Q. Qi, Nat. Commun., 2023, 14, 8480 CrossRef CAS PubMed.
- Y. Zhong, J. Gu, C. Shang, J. Deng, Y. Liu, Z. Cui, X. Lu and Q. Qi, Bioresour. Technol., 2024, 131166 CrossRef CAS PubMed.
- J.-H. Mou, W. Yan, Z.-H. Qin, M. A. Haque, Y.-H. Miao, F.-X. Xin, X. Wang, P. Fickers and C. S. K. Lin, Chem. Eng. J., 2024, 481, 148408 CrossRef CAS.
- E. Stylianou, J. M. Carvajal-Arroyo, D. Ladakis, C. S. K. Lin, V. Eßmann, S. Dörr, J. Marbach, K. Rabaey, A. Koutinas and C. Pateraki, Chem. Eng. J., 2023, 466, 142877 CrossRef CAS.
- V. Narisetty, A. A. Prabhu, R. R. Bommareddy, R. Cox, D. Agrawal, A. Misra, M. A. Haider, A. Bhatnagar, A. Pandey and V. Kumar, ACS Sustainable Chem. Eng., 2022, 10, 10858–10869 CrossRef PubMed.
- S. Zhang, F. Guo, Q. Yang, Y. Jiang, S. Yang, J. Ma, F. Xin, T. Hasunuma, A. Kondo and W. Zhang, Green Chem., 2023, 25, 183–195 RSC.
- Z. Jiang, Z. Cui, Z. Zhu, Y. Liu, Y.-J. Tang, J. Hou and Q. Qi, Biotechnol. Biofuels, 2021, 14, 1–10 CrossRef PubMed.
- K. L. Ong, C. Li, X. Li, Y. Zhang, J. Xu and C. S. K. Lin, Biochem. Eng. J., 2019, 148, 108–115 CrossRef CAS.
- A. A. Prabhu, R. Ledesma-Amaro, C. S. K. Lin, F. Coulon, V. K. Thakur and V. Kumar, Biotechnol. Biofuels, 2020, 13, 1–15 CrossRef PubMed.
- Q. Guo, Y. W. Li, F. Yan, K. Li, Y. T. Wang, C. Ye, T. Q. Shi and H. Huang, Biotechnol. Bioeng., 2022, 119, 2819–2830 CrossRef CAS PubMed.
- X. Yuan, X. Chen, G. Shen, S. Chen, J. Yu, R. Zhai, Z. Xu and M. Jin, Renewable Energy, 2022, 182, 377–389 CrossRef CAS.
- X. Chen, X. Yuan, S. Chen, J. Yu, R. Zhai, Z. Xu and M. Jin, Green Chem., 2021, 23, 4828–4839 RSC.
- Y. Ma, W. Li, J. Mai, J. Wang, Y. Wei, R. Ledesma-Amaro and X.-J. Ji, Green Chem., 2021, 23, 780–787 RSC.
- C. Holkenbrink, M. I. Dam, K. R. Kildegaard, J. Beder, J. Dahlin, D. Doménech Belda and I. Borodina, Biotechnol. J., 2018, 13, 1700543 CrossRef PubMed.
- M. Alexandri, A. Vlysidis, H. Papapostolou, O. Tverezovskaya, V. Tverezovskiy, I. K. Kookos and A. Koutinas, Sep. Purif. Technol., 2019, 209, 666–675 CrossRef CAS.
- B. Moosavi, E. A. Berry, X.-L. Zhu, W.-C. Yang and G.-F. Yang, Cell. Mol. Life Sci., 2019, 76, 4023–4042 CrossRef CAS PubMed.
- Z. Cui, C. Gao, J. Li, J. Hou, C. S. K. Lin and Q. Qi, Metab. Eng., 2017, 42, 126–133 CrossRef CAS PubMed.
- Z. Lazar, T. Dulermo, C. Neuvéglise, A.-M. Crutz-Le Coq and J.-M. Nicaud, Metab. Eng., 2014, 26, 89–99 CrossRef CAS PubMed.
- S. Qiang, J. Wang, X. C. Xiong, Y. L. Qu, L. Liu, C. Y. Hu and Y. H. Meng, Front. Microbiol., 2020, 11, 1346 CrossRef PubMed.
- S. A. van der Hoek and I. Borodina, Curr. Opin. Biotechnol., 2020, 66, 186–194 CrossRef CAS PubMed.
- Y. Mao, G. Li, Z. Chang, R. Tao, Z. Cui, Z. Wang, Y.-J. Tang, T. Chen and X. Zhao, Biotechnol. Biofuels, 2018, 11, 1–17 CrossRef CAS PubMed.
- D. Salvachúa, A. Mohagheghi, H. Smith, M. F. Bradfield, W. Nicol, B. A. Black, M. J. Biddy, N. Dowe and G. T. Beckham, Biotechnol. Biofuels, 2016, 9, 1–15 CrossRef PubMed.
- D. Wu, Q. Li, D. Wang and Y. Dong, Appl. Biochem. Biotechnol., 2013, 170, 1942–1949 CrossRef CAS PubMed.
- D. Wang, Q. Li, M. Yang, Y. Zhang, Z. Su and J. Xing, Process Biochem., 2011, 46, 365–371 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc04189e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |