Stereoselective coronas regulate the fate of chiral gold nanoparticles in vivo†
Didar
Baimanov‡
 ab,
Liming
Wang‡
ab,
Liming
Wang‡
 a,
Ke
Liu‡
ac,
Mengmeng
Pan
b,
Rui
Cai
a,
Hao
Yuan
a,
Wanxia
Huang
d,
Qingxi
Yuan
d,
Yunlong
Zhou
a,
Ke
Liu‡
ac,
Mengmeng
Pan
b,
Rui
Cai
a,
Hao
Yuan
a,
Wanxia
Huang
d,
Qingxi
Yuan
d,
Yunlong
Zhou
 *b,
Chunying
Chen
*b,
Chunying
Chen
 *aef and
Yuliang
Zhao
*aef
*aef and
Yuliang
Zhao
*aef
aCAS Key Laboratory for Biological Effects of Nanomaterials and Nanosafety, CAS Center for Excellence in Nanoscience, CAS-HKU Joint Laboratory of Metallomics on Health and Environment, National Center for Nanoscience and Technology of China and Institute of High Energy Physics, Chinese Academy of Sciences, Beijing 100190, P. R. China. E-mail: chenchy@nanoctr.cn; zhaoyl@nanoctr.cn
bEngineering Research Center of Clinical Functional Materials and Diagnosis &Treatment Devices of Zhejiang Province, Wenzhou Institute, University of Chinese Academy of Sciences, Wenzhou 325000, P. R. China. E-mail: zhouyl@ucas.ac.cn
cCollege of Materials and Chemistry & Chemical Engineering, Chengdu University of Technology, Sichuan province, Chengdu 610059, P. R. China
dBeijing Synchrotron Radiation Facility, Institute of High Energy Physics, Chinese Academy of Sciences, Beijing 100049, P. R. China
eGBA Research Innovation Institute for Nanotechnology, Guangzhou 510700, Guangdong, P. R. China
fResearch Unit of Nanoscience and Technology, Chinese Academy of Medical Sciences, Beijing 100730, P. R. China
First published on 22nd May 2023
Abstract
It is unknown how the identity provided by protein coronas on the surface of chiral nanoparticles determines their blood circulation, distribution, and clearance fates of the nanoparticles in vivo. Here, we attempt to investigate how the mirrored surface of gold nanoparticles with distinct chirality reshapes the coronal composition that mediates their subsequent clearance from blood and biodistribution. We found that chiral gold nanoparticles exhibited surface chirality-specific recognition for the coronal components, including the lipoproteins, complement components, and acute phase proteins, ultimately resulting in distinct cell uptake and tissue accumulation in vivo. We observed that these stereoselective behaviors were correlated to subgroups of the corona composition that could bind to low-density lipoprotein receptors. Therefore, this study reveals how chirality-specific protein compositions selectively recognize and interact with cell receptors for chirality-mediated tissue accumulation. This study will deepen our understanding of how chiral nanoparticles/nanomedicine/nanocarriers interact with biological systems to guide the efficient fabrication of target nanomedicines.
New conceptsOur findings support a new concept that surface chirality plays a crucial role in the biodistribution and cellular uptake of chiral nanoparticles via stereoselective formation of the protein corona. In this work, we found that the chirality property can be transferred from nanoparticles’ surface ligands to the adsorbed proteins, i.e., the protein corona. Chirality-specific protein coronas were observed in complement and acute phase proteins, whereas lipoproteins on chiral nanoparticles exhibited altered functionality, acting as dysopsonin or opsonin. On the surface of chiral gold nanoparticles, lipoproteins could enhance or reduce LDL receptor-mediated macrophage uptake. This study revealed the role of chirality in mediating protein corona formation and its effect on the blood clearance and biodistribution. Overall, this study demonstrated that the stereoselective coronas regulate the behavior and fate of chiral gold nanoparticles in vivo. We highlight the importance of surface chirality in the development of targeted nanomedicine and open a new avenue for the design of more efficient and effective nanomedicines. |
Introduction
Chirality is a symmetry property inherent in nature.1 It is well known that most biomolecules, e.g., proteins, nucleic acids, are chiral and attain stereospecific interactions in different biological processes. To approach the advantage of their chiral recognition by a living system, majority of chiral nanomaterials are obtained by the transfer of chirality from the small biomolecule to the nanomaterial.2–4 Besides the interaction of the metal core with a chiral molecule, chiroptical effects have been observed in a variety of inorganic nanostructures with chiral shapes.5,6 The principle called ‘chiral self-sorting’ is attracting much interest and could explain certain biological chirality-related processes.7 Based on this principle, the preference of biomolecules or/and chiral nanoparticles to specifically recognize and combine with molecules of the same chirality can explain the origin of ‘homochirality’.1 However, the interaction of biomolecules and nanoparticles with different chirality at the nano-bio interface has barely been investigated. It has been shown that the bioactivity of nanoparticles within a living system is determined by the orientation of the coronal proteins adopted on the surface of nanoparticles.8 Moreover, the kinds of rotation or translation of chirality from nanoparticles to the biomolecules, as determined by the direction of the spin, leading to varied in vivo outcomes remain unknown. Thus, investigation of the origin of ‘homochirality’ at the nano-bio interface remains a crucial task for the efficient development of targeted nanomedicines.Engineered nanoparticles are mainly fabricated to deliver drugs into a target cell in the desired tissue.9–11 It is crucial to understand how surface chemistry regulates cell recognition and the uptake of nanomaterials, as well as biological distribution, metabolism, and clearance fates in vivo, which is a fundamental question in the nanobiology and nanomedical fields. Combining the surface modifications of nanoparticles with adopted identities from adsorbed coronal proteins could further determine the nanoparticle interactions within the living system.12–14 Corona proteins undergo continuous dynamic exchanges throughout the transfer of biological fluids.15 Different protein sources could affect the protein corona compositions and lead to varied outcomes of nanoparticles.16 Therefore, tightly abundant coronal proteins are widely analyzed to predict nanoparticle behavior in vivo.17–19 Corona formation is dynamic, rapid, and strictly depends on the unique surface properties of the nanoparticles.20 In this aspect, recognition of the coronal protein subgroups by cell receptors due to the altered proteins functionality obtained on the nanoparticle surface should be considered to predict the cell and tissue accumulation.21 The relationship with neighboring proteins at the nano-bio interface was found to play a major role in the formation of protein corona and the regulation of nanoparticles' fate.22 Thus, the nanoparticle's shape, size, and solubility can be used as a tool to modulate a controllable targeted drug delivery system.23 However, due to dynamic nature of coronal fingerprints, the interactions between the nanoparticle-protein complexes and cell receptors need to be fully uncovered that can better predict nanoparticles' transport to desired tissues for the purpose of efficient drug delivery in vivo.
In this work, we aimed to investigate how the surface chirality mediates the formation of stereoselective coronal fingerprints that leads to a varied distribution of nanoparticles in vivo. Herein, we prepared two model chiral gold nanoparticles with an identical size, shape, and nature, but with opposite chirality. This study allowed us to perform a direct comparison of stereoselective coronal fingerprints with a single factor of variety, i.e., the surface chirality. Thus, we focused on trying to attain a systematic understanding of how surface chirality affects the formation of nanoparticles’ stereoselective protein coronas in serum, and how these alter serum protein adsorption, which could aid determining cell association, blood circulation, and tissue distribution. According to the findings, specific serum proteins exhibit stereoselective prevalence on chiral surfaces, which results in functional variations in their coronal fingerprints and discrepancies in the behavior of chiral gold nanoparticles, such as blood clearance and macrophage uptake rates. This work helps reveal how intrinsic chirality properties are transferred to the formed protein corona, such as the biological identity of chiral nanomaterials and their subsequent mediated biological fates and responses. This study also highlights a fundamental role of surface chirality in determining the interactions of nanoparticles with biomolecules. Our findings aim to lay a foundation for elucidating the routes and reasons for chirality-dependent tissue targeting.
Results and discussion
Synthesis and characterization
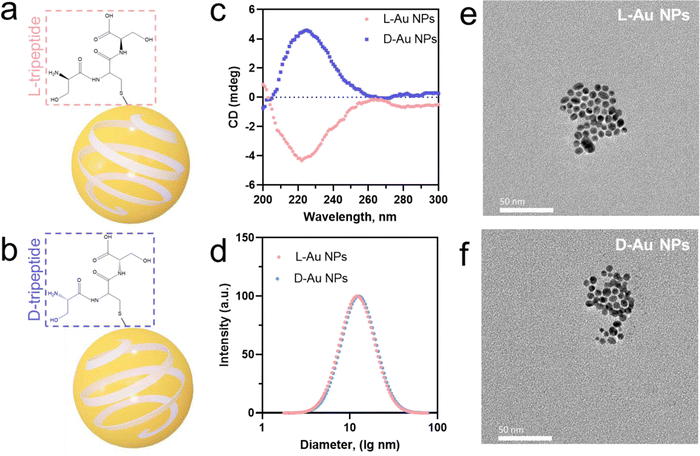
We prepared two chiral gold nanoparticles (Au NPs) with similar sizes and surface charges but opposite chirality and exposed them to human serum for studying their stereoselective coronal fingerprints. Herein, spherical nanoparticles with gold metal core and diameter under 5 nm were chosen as the model system. Chiral gold nanoparticles (Au NPs) were directly prepared by mixing a small molecule, i.e., tripeptide, as the protecting agent with sodium borohydride as the reducing agent.24 This method was used to develop small chiral Au NPs (Fig. 1(a) and (b)) through enantioselective interactions by transferring chirality from the ligands (L-Ser-L-Cys-L-Ser and D-Ser-D-Cys-D-Ser tripeptides) to chiral nanoparticles (L-chiral gold nanoparticles, L-Au NPs; and D-chiral gold nanoparticles, D-Au NPs). The circular dichroism (CD) spectra (Fig. 1(c)) and hydrodynamic diameter (Fig. 1(d)) showed the chiral formation in the fabricated gold nanoparticles with almost identical diameters but distinct chirality. Here, the L-tripeptide-stabilized chiral gold nanoparticles showed right circularly polarized light, while the D-tripeptide-stabilized ones showed an exactly inverted CD response4 at the same peak position. We observed spherical small-sized chiral gold nanoparticles with a similar size of about 5 nm by transmission electron microscopy (TEM, Fig. 1(e) and (f)). UV-Visible absorbance measurements of the chiral gold nanoparticles showed a band for surface plasmon peaks (λmax) at 520 nm (Fig. S1a, ESI†). According to ELS, the zeta-potential (ζ-potential) values were about −22.6 mV, indicating the colloidal stability of the chiral Au NPs (Fig. S1b, ESI†). The concentration of the chiral Au NPs was determined by inductively coupled plasma mass spectrometry (ICP-MS) measurements prior to the further experiments.22 The outline of the chiral nanoparticles used in this study in both cases was similar to a spherical shape with a core size of about 5 nm.Identifying the coronal proteins on the chiral gold nanoparticles
We selected chiral Au NPs with a high density of gold elements that could be directly used in a centrifugation method to separate the protein corona from serum.25 We obtained serum-coated chiral Au NPs by incubating them with 10% human serum26 to directly compare the formation of the protein corona on the chiral surface. The hydrodynamic diameters of both chiral gold nanoparticles were increased from ∼11.9 to ∼81.3 nm for L-Au NPs and from ∼12.3 to ∼111.2 nm for D-Au NPs after the formation of the protein corona (Fig. S2a, ESI†). Additionally, the adsorbed coronal proteins were quantified via a BCA protein assay, and the results showed that the adsorbed proteins on the chiral NPs were significantly different (Fig. S2b, ESI†), suggesting the subsequent interactions between the serum proteins and chiral surface. Negative stained TEM images further confirmed the proteins were adsorbed on the chiral nanoparticles (Fig. S3, ESI†). These findings demonstrate the formation of the serum protein corona on the surface of the chiral Au NPs.To assess the composition of the stereoselective coronal fingerprints, the isolated proteins of chiral gold nanoparticles were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In this study, the relative abundance of the identified proteins represented the average of three biological replications of each sample, which were used to obtain a normalized result. The data for the relative abundance and standard deviations of the identified proteins in the triplicate biological samples are presented in the Supplementary Tables (ESI†). Noticeably, we observed that the protein corona composition on the chiral gold nanoparticles was significantly different from that in the serum (Fig. S4, ESI†). As expected, albumin was the most abundant protein, followed by the other proteins, acute phase, lipoproteins, complement components, coagulation, and tissue leakage proteins in the serum (Fig. 2(a)). Compared to the serum, the relative abundance of lipoproteins, complement components, coagulation, and tissue leakage proteins increased, while albumin and the other proteins decreased in the coronal fingerprints of the chiral gold nanoparticles (Fig. 2(b) and (c)). Although the coronal identity on the chiral Au NPs was similar, we observed that the protein abundance was significantly altered, suggesting that the serum proteins could stereoselectively recognize the chiral surfaces. These findings emphasize the importance of our study to reveal how chiral surfaces could contribute to the recognition and distribution of nanomedicines in vivo.
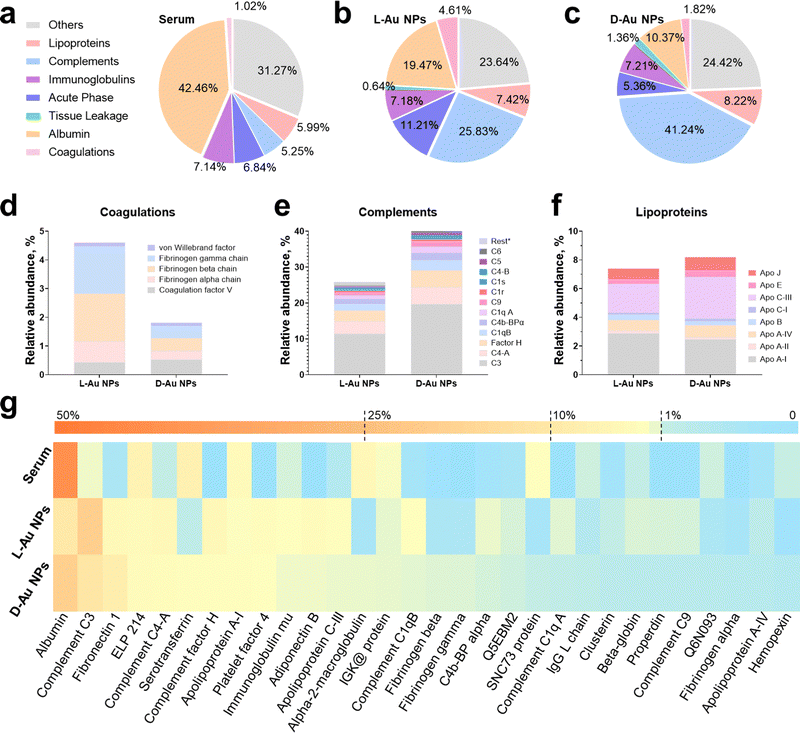 | ||
| Fig. 2 Identification of stereoselective protein coronas. Protein components of human serum (a), and protein corona fingerprints on L-Au NPs (b), and D-Au NPs (c) as determined by LC-MS/MS. Identified proteins are categorized into seven groups by biological functions. (d)–(f) Stereoselective protein families, including complements (d), coagulations (e), and lipoproteins (f). Coronal proteins were searched against the human database on the Uniprot website. (g) Heatmap of the most abundant proteins in human serum as well as on the corona of chiral gold nanoparticles. Heatmap shows the median abundance of protein groups (columns) on two chiral gold nanoparticles and in human serum. All data are shown as mean values for triplicate biological samples (n = 3) and standard deviations that are shown in the Supplementary Tables (ESI†). | ||
Comparing the stereoselectivity of the coronal proteins
To comprehensively reveal the effect of surface chirality on the corona formation, we first analyzed the coronal protein categories (Fig. 2(a)–(c)). The most abundant protein, serum albumin, decreased to ∼19% and 10% on L-Au NPs and D-Au NPs compared to serum (Fig. S5a, ESI†). The total abundance of coagulation (∼4.8% vs. ∼1.8%) and the acute phase (∼11.2% vs. ∼5.4%) proteins on the L-Au NPs were 2.5-fold and 2-fold higher than those on D-Au NPs. Oppositely, the total amount of lipoproteins (∼7.4% vs. ∼8.2%) and complement components (∼25.8% vs. ∼41.2%) were higher on the surface of D-Au NPs. Interestingly, we observed that the abundance of immunoglobulins and other proteins on the surface of the chiral Au NPs were similar (Fig. S5b, ESI†). Immunoglobulins showed a neglectable difference (<0.5%) on both chiral Au NPs, suggesting that the immunoglobulins might not exhibit any specific recognition for the chiral Au NPs. We thus confirmed that biomolecules can recognize the surface chirality of nanomaterials, leading to the possible alteration in their biological fates, such as phagocytosis and tissue accumulation in vivo.We next analyzed the stereoselective abundance of proteins adsorbed on the surface of nanoparticles at the single protein level. The coagulation family of proteins, such as fibrinogen (Fig. 2(d)), showed at least a 2-fold greater adsorption, while the acute phase family proteins, i.e., fibronectin (Fig. S5c, ESI†), showed a 1.8-fold higher adsorption onto L-Au NPs, suggesting that these proteins might exhibit distinct preferences to bind L-chiral nanomaterials. Similarly, almost all the components in the complement family proteins (Fig. 2(e)) showed a 1.2–2-fold higher abundance onto D-Au NPs, suggesting a higher preference for the D-chiral nanomaterials by these proteins. However, the lipoproteins (Fig. 2(f)) showed a slightly altered stereoselective recognition of the chiral surface. In detail, ApoC3, clusterin, and ApoE were slightly enhanced on the surface of D-Au NPs, while ApoA1 showed a slight preference to adsorb onto L-Au NPs. Among the other proteins, adiponectin (1.8-fold), beta-globulin (2-fold), and alpha-2 globin (2-fold) showed a higher abundance on D-Au NPs, while the abundance of serotransferrin (3.5-fold) was lower on D-Au NPs (Fig. S5d, ESI†). The 20 most abundant proteins are listed in Table S1 (ESI†). The top-ten most abundant proteins on the surface of L-Au NPs were: albumin, complement C3, fibronectin 1, epididymis luminal protein 214, complement C4-A, serotransferrin, complement factor H, apolipoprotein A-I, platelet factor 4, and immunoglobulin heavy constant mu. The top-ten most abundant proteins on the surface of D-Au NPs were: albumin, complement C3, complement C4-A, complement factor H, fibronectin, epididymis luminal protein 214, adiponectin B, platelet factor 4, apolipoprotein C-III, and complement C1q, constituting roughly 61% of the adsorbed proteins for both chiral gold nanoparticles. In summary, the amounts of albumin, serotransferrin, fibrinogen, and fibronectin were comparably higher on the surface of L-Au NPs, while most of the complement and lipoprotein family proteins showed enhanced adsorption on the surface of D-Au NPs. The most abundant coronal proteins are summarized in the heatmap in Fig. 2(g). It was shown that distinct proteins, such as serum albumin27 and transferrin,28 presented orientational and conformational changes during the adsorption onto the chiral surface of the gold nanoparticles, resulting in their altered uptake by cells. Moreover, it was shown that coagulation factor XII exhibited a chirality-specific binding affinity by altering the complex conformation and inducing enzymatic activation.29 Although the chiral surface modifications are not capable of preventing the shielding effects of coronal proteins, they might transfer the chirality from molecules to the nanoparticles’ surface to better realize chiral-dependent recognition by cell receptors.30 Overall, surface chirality could influence the interaction with individual serum proteins, altering their functionality and stability and vice versa, while the underlying mechanism of stereoselective corona recognition is not yet well understood. Taken together, these results suggested that surface chirality plays a significant role in controlling the formation of specific protein coronas.
Coronal fingerprint formation is a complex process; therefore, we would like to emphasize that at first, the surface properties of nanoparticles dictate the corona formations, while the coronal composition itself might impact its further evolution, which finally would determine the fate of nanomaterials. We aimed to investigate how proteins selectively recognize the mirrored surface of chiral Au NPs. Consequently, we analyzed the enrichment factor of coronal proteins (Table S2, ESI†) due to their presented significant stereoselectivity. Our results demonstrated that apart from binding to foreign objects in the blood, complement family proteins could selectively recognize the D-chiral NPs. Interestingly, ApoE proteins showed higher enrichment levels on D-chiral nanoparticles, suggesting a higher adsorption of this protein onto chiral gold nanoparticles. Therefore, we hypothesized that the complement components and lipoproteins could determine the stereoselectivity of chiral gold nanoparticles within a living system.
Distinct binding affinity of coronal proteins on chiral gold nanoparticles
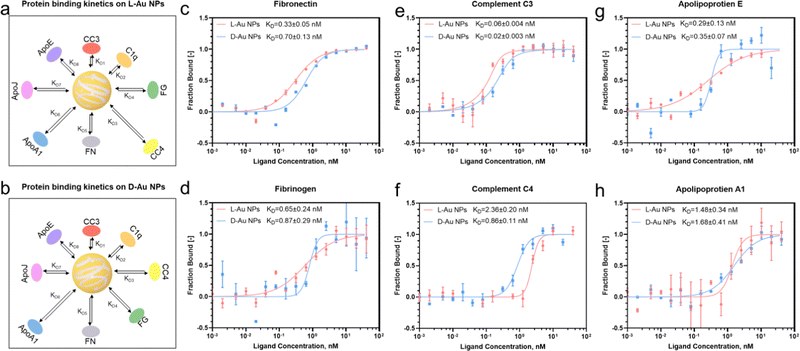
Next, we investigated the thermodynamic and kinetic aspects of adsorption, determining the final composition of the protein corona. It is well known that protein association and dissociation vary depending on the surface properties of nanoparticles.13,22 We performed microscale thermophoresis (MST) analysis to determine the affinity between selected proteins and chiral gold nanoparticles. Herein, we selected three complement proteins, three lipoproteins, fibronectin from the acute phase, and fibrinogen from coagulations due to their presented chirality-dependent selectivity. First, the coronal proteins' stereoselectivity, i.e., the preference of proteins to bind to the surface of distinct chiral Au NPs, were determined and their thermodynamic dissociation constants (KD) were schematically illustrated (Fig. 3(a) and (b)). Second, similar to our proteomics results, two coronal proteins, i.e., fibronectin and fibrinogen, showed stronger binding on the surface of L-Au NPs (Fig. 3(c) and (d)). Third, we observed that complement C3 and C4 proteins maintained a stronger binding affinity with D-Au NPs (Fig. 3(e) and (f)), supporting our previous conclusion. However, no obvious difference was noticed in the thermodynamic KD values of the complement C1q protein (Fig. S6a, ESI†) and lipoproteins (Fig. 3(g), (h) and Fig. S6b, ESI†) on the surface of the chiral gold nanoparticles. Our thermodynamic and kinetic adsorption experiments once again confirmed our hypothesis that individual biomolecules have chirality-specific preferences upon interactions with chiral nanoparticles. These findings revealed that stereoselective fingerprints determine the further fate of nanoparticles in vivo and might provide a clue to understanding the ‘homochirality’. However, we agree that an individual protein binding approach has limitations and may not reflect the corona formation in a complex system. For example, the lipoproteins here did not show a significant difference in binding affinity with chiral gold nanoparticles. It should be noted that coronal fingerprint formation and evolution are dynamic, therefore we considered that binding affinity alone cannot thoroughly address the adsorption and exchange rate of certain proteins in a complex biological fluid. However, we are confident that these individual protein association and dissociation rates could appropriately address the stereoselective behavior of the individual serum proteins to adsorb on the chiral surface by forming a hard protein corona.Determining the interaction between the stereoselective coronal fingerprint and macrophages
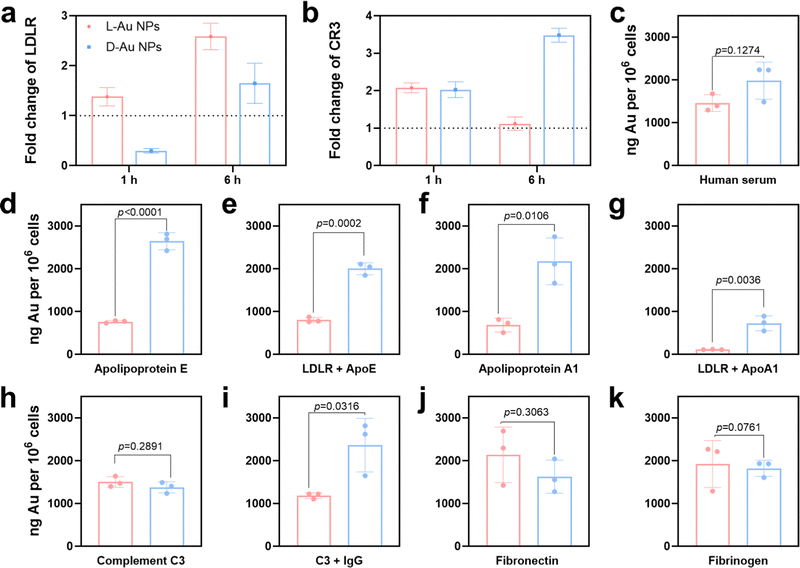
To predict the receptor-mediated cellular uptake of chiral Au NPs, we used a search tool for the retrieval of the interacting genes (STRING).31 We inputted the identities of the top 25 most absorbed coronal proteins on chiral Au NPs and the macrophage receptors possibly involved in cell uptake into the STRING website. A list of predicted protein–receptor interaction scores was returned, as listed in Table S3 (ESI†). It should be noted that the STRING score represents the confidence of the predicted interaction. We decided to investigate the interactions among coronal proteins with a score greater than 0.80 and those among eight receptors, i.e., low-density lipoprotein receptor (LDLR);32 complement receptor type 1 (CR1) and complement receptor type 3 (CR3);33 scavenger receptor cysteine-rich type 1 (CD163), macrophage scavenger receptor types I and II (MSR1) and scavenger receptor class B (CD36);34 Toll-like receptor 2 (TLR2),35 and low-affinity immunoglobulin gamma Fc region receptor (FCGR).36To confirm that these receptors were involved in the cellular uptake of chiral Au NPs, we next studied the level of the predicted receptors in differentiated human macrophages (THP-1) by RT-qPCR. We selected macrophages due to their protective function, acting as a first line of defense of a living system, i.e., the recognition and clearance of foreign objects from the blood flow.37,38 Further, we incubated human serum-supplied chiral Au NPs with macrophages for 1 and 6 h. We identified two cell receptors potentially involved in uptake: LDL receptor, which showed an enhanced level for L-Au NPs- and decreased level for D-Au NPs-treated cells over time (Fig. 4(a)), and complement receptor type 3, which was enhanced upon treatment with chiral gold nanoparticles (Fig. 4(b)). There was no obvious alteration for other cell receptor expressions (Fig. S7, ESI†). To confirm whether these receptors are involved in the uptake of chiral Au NPs, we next performed in vitro experiments.
Using ICP-MS, we quantified the uptake of Au NPs by macrophages supplied with 10% human serum containing the cell culture medium. We observed that the surface chirality had a significant effect on cellular uptake (Fig. 4(c)), whereupon D-Au NPs were recognized and uptaken by macrophages more than L-Au NPs. Additionally, we observed a higher accumulation of D-Au NPs within macrophages compared to L-Au NPs in TEM (Fig. S8, ESI†). Therefore, we could conclude that chirality has a significant impact on cell recognition. To test our hypothesis of the stereoselective recognition of protein coronas, we determined whether single-protein-incubated chiral gold nanoparticles induced an alteration of cellular uptake. For this purpose, we selected five coronal proteins: complement C3, ApoA1, ApoE, fibronectin, and fibrinogen (Fig. 4(d)–(k)). Based on our previous studies,22,26 we emphasize that the dynamic nature of protein coronas, as well as the altered functionality and adsorbed amount of protein subgroups, could regulate and alter the fate of nanoparticles. Therefore, we used single-protein-supplied serum-free culture media to identify the relevance of certain proteins to chirality-dependent uptake by macrophages. Interestingly, we observed that ApoE (Fig. 4(d)) and ApoA1 (Fig. 4(f)) had a significant impact on the cellular recognition of chiral gold nanoparticles. The presence of these two lipoproteins in the culture media reduced the uptake of L-Au NPs, suggesting that lipoproteins play a major role in the long-life circulation of L-chiral nanomaterials by hindering them from macrophages. Conversely, the presence of these two lipoproteins enhanced the accumulation of D-Au NPs within macrophages. Thus, we decided to determine the influence of LDL receptors on nanoparticle uptake.
We next observed reduced uptakes of ApoE-supplied chiral nanoparticles to 58% and 24% upon blockage of the LDL receptors (Fig. 4(e)). Similarly, 83% and 64% reduced uptakes were observed for chiral nanoparticles supplied with ApoA1 and after blocking of the LDL receptors (Fig. 4(g)). However, complement C3 (Fig. 4(h)) and fibrinogen (Fig. 4(k)) did not show a significant effect on recognition by macrophages. We observed the stereoselective accumulation of chiral gold nanoparticles by macrophages in the presence of IgG (Fig. 4(i)); however, complement C3 alone did not alter the cell recognition. It should be noted that activated complement C3 could tightly bind to both antigens and antibodies, and retains its capacity to react with the C3-cell receptors after being bound to immunoglobulin G (IgG).39 While, fibronectin significantly enhanced the uptake of L-Au NPs (Fig. 4(j)). Taken together, these results show that the chiral Au NPs presented distinct associations with the cell surface. Moreover, coronal proteins are capable of recognizing the surface of mirrored nanoparticles, resulting in different accumulation within the cells. Lipoproteins show a diverse effect, first by reducing the uptake of L-chiral nanoparticles, and secondly by enhancing the accumulation of D-chiral nanoparticles. This finding reveals that lipoproteins are key players in achieving long-life blood circulation or rapid clearance from the blood or liver accumulation40 of administrated Au NPs. Our study also uncovers the importance of competitive interactions on the coronal fingerprints, where the protein–protein interactions due to protein adsorption regulates the behavior of nanomaterials.22 It was reported that the presence of abundant components such as platelet factor 4 might interfere with LDL receptor-mediated cellular uptake,21 corroborating previous findings that platelet factor 4 enhances the uptake of LDLs into cells.41 Similar to our previous study,25 we believe that the collective influence of the stereoselective coronal fingerprints, i.e., lipoproteins, complements, and acute phase proteins on the surface of chiral Au NPs, determine the cell association.
Biodistribution of chiral gold nanoparticles in vivo
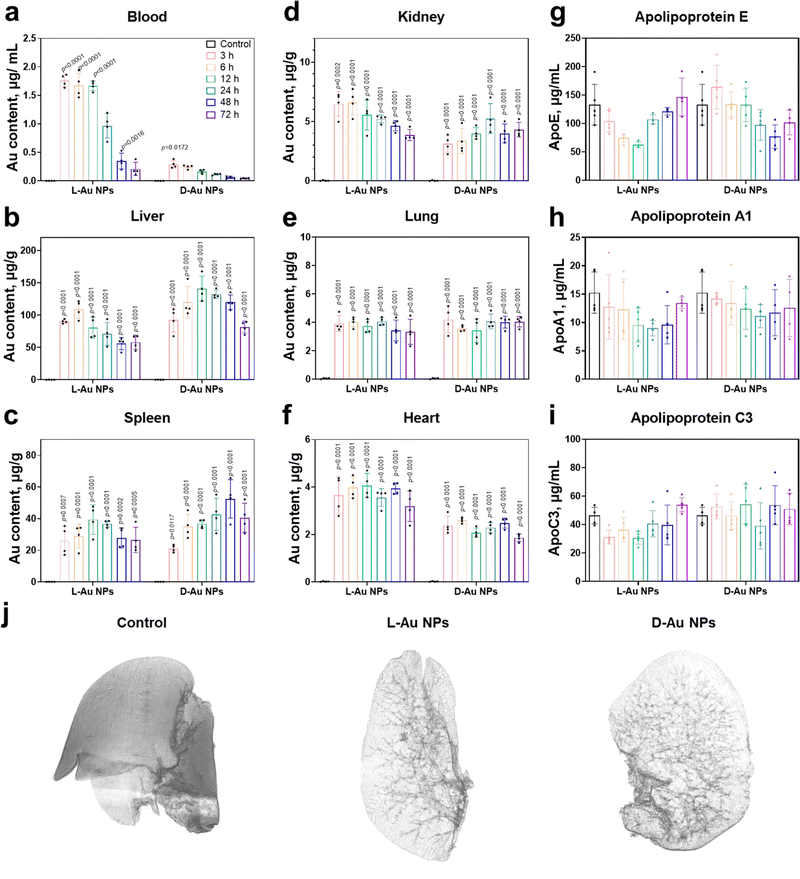
We next attempted to determine whether stereoselective protein coronas could modulate the in vivo biodistribution of chiral Au NPs. Prior to the animal experiments, the safety of the chiral gold nanoparticles was studied (Fig. S9, ESI†). After the intravenous administration of chiral NPs into C57BL/6 mice, the amounts of chiral gold nanoparticles in the blood gradually decreased over time (Fig. 5(a)). Interestingly, L-Au NPs exhibited a longer circulation time than D-Au NPs. We concluded that the surface chirality of nanoparticles affected tissue biodistribution and clearance from the blood. Most of the administrated nanoparticles (30–99%) will end up in the liver.42 Both chiral Au NPs were indeed highly accumulated in the liver (Fig. 5(b)), albeit with different accumulation and clearance rates. Similar to our previous study,22D-Au NPs showed a higher accumulation in the liver compared to L-Au NPs. Moreover, chiral Au NPs showed varied biodistributions in tissues, reaching a maximum accumulation in the spleen (Fig. 5(c)) and kidney (Fig. 5(d)) at different time points. Similar accumulation tendencies of chiral Au NPs were observed in the lung and heart (Fig. 5(e) and (f)). We could conclude that the liver and spleen are the main accumulation organs of the chiral gold nanoparticles upon i.v. injection. Moreover, the nanoparticles trafficked in the blood vessels can also exhibit a small amount of Au accumulated in the lung, heart, and kidneys. Overall, surface chirality essentially affects tissue accumulation and clearance rate in vivo.Next, we asked whether nanoparticle accumulation was correlated with the coronal protein composition. We found that the significant stereoselectivity of lipoproteins bind on the chiral surface of nanoparticles, and further detected changes in the mice serum lipoprotein levels on chiral NPs after treatment with Au NPs at different time points (Fig. 5(g)–(i) and Fig. S10, ESI†). Both ApoE and ApoA1 protein levels were altered within the mouse serum upon treatment, suggesting that these proteins might play significant roles in the recognition of chiral NPs by receptors, and their regulation in the blood circulation and their clearance rate. No obvious difference in the abundance were observed in the coronal fingerprints of chiral gold nanoparticles with the altered biodistribution and clearance rate, the functionality, i.e., dysopsonin and opsonin, of coronal ApoE appeared to be chirality-dependent. It was observed that the ApoE protein may have multiple downstream effects and could alter the identities of coronal proteins.43 ApoE pre-coated gold nanoparticles led to a prolonged blood circulation.44 Moreover, coronal ApoA1 also showed a high relevance to blood circulation lifetime of nanomaterials.45
To determine the stereoselective accumulation of chiral Au NPs, the liver was further imaged using synchrotron radiation micro-CT. This advanced imaging technique enables high-resolution, non-destructive imaging of biological and material samples. By analyzing the distribution of chiral Au NPs in the liver, valuable information pertaining to their pharmacokinetics could be obtained. Our findings demonstrated that both chiral NPs exhibited a distribution pattern concentrated within or nearby liver blood vessels (see Fig. 5(j) and supplementary Movies 1–3, ESI†), with D-Au NPs showing a higher accumulation according to the stronger X-ray adsorption intensity. Interestingly, we observed an enhanced accumulation of D-Au NPs in the liver, which might partially be explained by the deeper penetration of these nanoparticles in liver lobes, followed by their transportation via blood capillaries. The micro-CT results thus offered evidence about the distinct blood circulation and tissue accumulation mediated by the surface chirality of nanomaterials.
Our findings further support the idea that lipoproteins including ApoE and ApoA1, play different roles on the surface of chiral nanoparticles, acting as dysopsonins and opsonins. In comparison, complements and fibronectin showed chirality-driven enhanced cell associations of specific nanoparticles. The way how serum proteins interact with nanoparticles possibly affects the function of proteins and influences their cellular uptake and tissue distribution. These findings highlight the key role of a complex interplay between the chiral surface of nanoparticles and the stereoselective corona fingerprints, including the amount, function, and orientation of proteins obtained at the nano-bio interface and their fate in vivo.
The concept of stereoselectivity, which describes a biomolecule's preference for a particular surface chirality, in chiral nanomedicine remains elusive. The biological activity and interaction of biomolecules, including proteins and enzymes, depend on their stereoselectivity. Therefore, understanding how chiral surfaces might impact the orientation, binding affinity, and conformation of biomolecules, leading to an altered behavior in vivo yet needs to be attained. Although chiral nanoparticles have the potential for determining the adsorption of different proteins due to their chiral surface, but further research on how chiral nanomaterials impact the function of biomolecules is needed. On the other hand, the transfer of chirality from molecules to nanomaterials is attracting more attention, leading to an enhanced understanding of chirality phenomena and the fabrication of new nanomaterials.46 We encourage researchers to study how a single property such as chirality leads to a distinct fate of chiral nanoparticles in vivo. With respect to the chirality phenomenon of nanoparticles, open questions still remain, including whether biomolecules can selectively bind to chiral surfaces, and whether the interactions with nanoparticles can induce chirality-specific change in structure and functions in proteins.
Conclusions
Increasing interest in the interaction of engineered nanomaterials with biological systems has led to studies of the correlation of protein coronas with nanoparticles’ fate in vivo. This study focused on the effect of surface chirality on protein corona formation and the resulting impact on cell recognition, tissue accumulation, and the clearance rates of chiral NPs. We observed that lipoproteins, such as ApoE and ApoA1, could display altered functionality when they interact with the chiral surface of gold nanoparticles, which can be recognized and bound to LDL receptors, resulting in a stereoselective fate within living organisms. Our findings demonstrate that the surface chirality of nanomaterials determines the protein corona composition, including lipoproteins, complements, and acute phase proteins that play crucial roles in stereoselective biodistribution in vivo. These results highlight the importance of understanding how protein corona regulates the delivery of nanocarriers to targeted tissues. Such knowledge will significantly contribute to the design and development of biomedical nanomaterials and helps realize better safety during the applications.Author contributions
This project was designed by D. B., L. W., Y. Zhou and was supervised by C. C. and Y. Zhao. Main experiments were performed by D.B. and L. W. with great assistance from K. L. Synthesis of chiral gold nanoparticles and their characterizations were performed and analyzed by D. B., M. P., and R. C. ICP-MS experiments were performed and analyzed by D. B., K. L., H. Y. and R. C. SR micro-CT experiments were performed and analyzed by D. B., L. W., K. L., and R. C. with the guide from W. H. and Q. Y. Manuscript was prepared by D. B. and L. W. with input from other authors. All authors have approved it for publication in the present form.Conflicts of interest
All authors declare no competing interests.Acknowledgements
The authors are grateful to Dr B. Li (IHEP) and R. Qiao (NCNST) for their kind help with ICP-MS experiment; X. Zhang (PKU) for help with LC-MS/MS experiment. We appreciate funding from the National Key Research and Development Program of China (2020YFA0710702, 2021YFA1200900, 2021YFE0112600), the National Natural Science Foundation of China (31971322, 21773172), Start Funding of Wenzhou Institute of UCAS (WIUCASQD2019001), Major instrument project of National Natural Science Foundation of China (22027810), Beijing Municipal Health Commission (2021-1G-1191), the Research Center of Clinical Functional Materials and Diagnosis & Treatment Devices of Zhejiang Province (WIBEK181006), Major Science and Technology Project of Wenzhou Science and Technology (ZG2022017). The authors also acknowledge the support from the Directional Institutionalized Scientific Research Platform of Beijing Synchrotron Radiation Facility of CAS.References
- J. E. Hein and D. G. Blackmond, Acc. Chem. Res., 2012, 45, 2045–2054 CrossRef CAS.
- D. Trauner, Angew. Chem., Int. Ed., 2009, 48, 6589 CrossRef CAS.
- E. Hendry, T. Carpy, J. Johnston, M. Popland, R. V. Mikhaylovskiy, A. J. Lapthorn, S. M. Kelly, L. D. Barron, N. Gadegaard and M. Kadodwala, Nat. Nanotechnol., 2010, 5, 783–787 CrossRef CAS PubMed.
- H.-E. Lee, H.-Y. Ahn, J. Mun, Y. Y. Lee, M. Kim, N. H. Cho, K. Chang, W. S. Kim, J. Rho and K. T. Nam, Nature, 2018, 556, 360–365 CrossRef CAS PubMed.
- S. Ostovar pour, L. Rocks, K. Faulds, D. Graham, V. Parchaňský, P. Bouř and E. W. Blanch, Nat. Chem., 2015, 7, 591–596 CrossRef CAS PubMed.
- L. Durán Pachón, I. Yosef, T. Z. Markus, R. Naaman, D. Avnir and G. Rothenberg, Nat. Chem., 2009, 1, 160–164 CrossRef PubMed.
- W. Feng, J.-Y. Kim, X. Wang, H. A. Calcaterra, Z. Qu, L. Meshi and N. A. Kotov, Sci. Adv., 2017, 3, e1601159 CrossRef PubMed.
- J. X. Xu, M. S. Alom, R. Yadav and N. C. Fitzkee, Nat. Commun., 2022, 13, 7313 CrossRef CAS PubMed.
- M. Qiu, Y. Tang, J. Chen, R. Muriph, Z. Ye, C. Huang, J. Evans, E. P. Henske and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2116271119 CrossRef CAS.
- W. Ngo, S. Ahmed, C. Blackadar, B. Bussin, Q. Ji, S. M. Mladjenovic, Z. Sepahi and W. C. W. Chan, Adv. Drug Delivery Rev., 2022, 185, 114238 CrossRef CAS PubMed.
- B. Du, M. Yu and J. Zheng, Nat. Rev. Mater., 2018, 3, 358–374 CrossRef.
- D. Baimanov, R. Cai and C. Chen, Bioconjugate Chem., 2019, 30, 1923–1937 CrossRef CAS PubMed.
- J. Ren, R. Cai, J. Wang, M. Daniyal, D. Baimanov, Y. Liu, D. Yin, Y. Liu, Q. Miao, Y. Zhao and C. Chen, Nano Lett., 2019, 19, 4692–4701 CrossRef CAS PubMed.
- J. Lazarovits, S. Sindhwani, A. J. Tavares, Y. Zhang, F. Song, J. Audet, J. R. Krieger, A. M. Syed, B. Stordy and W. C. W. Chan, ACS Nano, 2019, 13, 8023–8034 CrossRef CAS PubMed.
- R. Cai, J. Ren, M. Guo, T. Wei, Y. Liu, C. Xie, P. Zhang, Z. Guo, J. Chetwynd Andrew, C. Ke Pu, I. Lynch and C. Chen, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2200363119 CrossRef CAS PubMed.
- K. Ezzat, M. Pernemalm, S. Pålsson, T. C. Roberts, P. Järver, A. Dondalska, B. Bestas, M. J. Sobkowiak, B. Levänen, M. Sköld, E. A. Thompson, O. Saher, O. K. Kari, T. Lajunen, E. Sverremark Ekström, C. Nilsson, Y. Ishchenko, T. Malm, M. J. A. Wood, U. F. Power, S. Masich, A. Lindén, J. K. Sandberg, J. Lehtiö, A.-L. Spetz and S. El Andaloussi, Nat. Commun., 2019, 10, 2331 CrossRef.
- X. Liu, W. Wei, Z. Liu, E. Song, J. Lou, L. Feng, R. Huang, C. Chen, P. C. Ke and Y. Song, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2108131118 CrossRef CAS PubMed.
- Y. Cong, D. Baimanov, Y. Zhou, C. Chen and L. Wang, Adv. Drug Delivery Rev., 2022, 191, 114615 CrossRef CAS.
- M. Cao, R. Cai, L. Zhao, M. Guo, L. Wang, Y. Wang, L. Zhang, X. Wang, H. Yao, C. Xie, Y. Cong, Y. Guan, X. Tao, Y. Wang, S. Xu, Y. Liu, Y. Zhao and C. Chen, Nat. Nanotechnol., 2021, 16, 708–716 CrossRef CAS.
- M. Mahmoudi, Nat. Commun., 2022, 13, 49 CrossRef CAS PubMed.
- W. Ngo, J. L. Y. Wu, Z. P. Lin, Y. Zhang, B. Bussin, A. Granda Farias, A. M. Syed, K. Chan, A. Habsid, J. Moffat and W. C. W. Chan, Nat. Chem. Biol., 2022, 18, 1023–1031 CrossRef CAS.
- D. Baimanov, J. Wang, J. Zhang, K. Liu, Y. Cong, X. Shi, X. Zhang, Y. Li, X. Li, R. Qiao, Y. Zhao, Y. Zhou, L. Wang and C. Chen, Nat. Commun., 2022, 13, 5389 CrossRef CAS PubMed.
- Z. Guo, P. Zhang, S. Chakraborty, A. J. Chetwynd, F. A. Monikh, C. Stark, H. Ali-Boucetta, S. Wilson, I. Lynch and E. Valsami-Jones, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2105245118 CrossRef CAS.
- K. Hou, J. Zhao, H. Wang, B. Li, K. Li, X. Shi, K. Wan, J. Ai, J. Lv, D. Wang, Q. Huang, H. Wang, Q. Cao, S. Liu and Z. Tang, Nat. Commun., 2020, 11, 4790 CrossRef CAS PubMed.
- R. Cai, J. Ren, Y. Ji, Y. Wang, Y. Liu, Z. Chen, Z. Farhadi Sabet, X. Wu, I. Lynch and C. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 1997–2008 CrossRef CAS PubMed.
- D. Baimanov, J. Wu, R. Chu, R. Cai, B. Wang, M. Cao, Y. Tao, J. Liu, M. Guo, J. Wang, X. Yuan, C. Ji, Y. Zhao, W. Feng, L. Wang and C. Chen, ACS Nano, 2020, 14, 5529–5542 CrossRef CAS PubMed.
- X. Wang, X. Wang, M. Wang, D. Zhang, Q. Yang, T. Liu, R. Lei, S. Zhu, Y. Zhao and C. Chen, Small, 2018, 14, 1703982 CrossRef PubMed.
- X. Wang, M. Wang, R. Lei, S. F. Zhu, Y. Zhao and C. Chen, ACS Nano, 2017, 11, 4606–4616 CrossRef CAS PubMed.
- F. Hao, F. Geng, X. Zhao, R. Liu, Q. S. Liu, Q. Zhou and G. Jiang, NanoImpact, 2021, 22, 100321 CrossRef CAS.
- J. Deng, H. Zheng and C. Gao, Mater. Chem. Front., 2017, 1, 542–549 RSC.
- D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen and C. V. Mering, Nucleic Acids Res., 2019, 47, D607–D613 CrossRef CAS PubMed.
- J. M. Castellano, R. Deane, A. J. Gottesdiener, P. B. Verghese, F. R. Stewart, T. West, A. C. Paoletti, T. R. Kasper, R. B. DeMattos, B. V. Zlokovic and D. M. Holtzman, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 15502–15507 CrossRef CAS PubMed.
- G. D. Ross,Encyclopedia of Immunology, Elsevier, Oxford, 2nd edn, 1998, pp.29–634 Search PubMed.
- A. Alquraini and J. El Khoury, Curr. Biol., 2020, 30, R790–R795 CrossRef CAS PubMed.
- K. A. Fitzgerald and J. C. Kagan, Cell, 2020, 180, 1044–1066 CrossRef CAS PubMed.
- F. Junker, J. Gordon and O. Qureshi, Front. Immunol., 2020, 11, 1393 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. W. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- R. Nezlin, The Immunoglobulins, Academic Press, New York, 1998, 219–254 Search PubMed.
- S. A. Dilliard, Q. Cheng and D. J. Siegwart, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2109256118 CrossRef CAS PubMed.
- T. Nassar, B. S. Sachais, S. E. Akkawi, M. A. Kowalska, K. Bdeir, E. Leitersdorf, E. Hiss, L. Ziporen, M. Aviram, D. Cines, M. Poncz and A. A.-R. Higazi, J. Biol. Chem., 2003, 278, 6187–6193 CrossRef CAS PubMed.
- Y.-N. Zhang, W. Poon, A. J. Tavares, I. D. McGilvray and W. C. W. Chan, J. Controlled Release, 2016, 240, 332–348 CrossRef CAS PubMed.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. M. Lima, E. A. Appel, F. Dormont, J.-M. Lim, R. Karnik, R. Langer and O. C. Farokhzad, Nat. Commun., 2017, 8, 777 CrossRef PubMed.
- X. Lu, P. Xu, H.-M. Ding, Y.-S. Yu, D. Huo and Y.-Q. Ma, Nat. Commun., 2019, 10, 4520 CrossRef PubMed.
- M. Li, X. Jin, T. Liu, F. Fan, F. Gao, S. Chai and L. Yang, Nat. Commun., 2022, 13, 4137 CrossRef.
- B. L. Li, J. J. Luo, H. L. Zou, Q.-M. Zhang, L.-B. Zhao, H. Qian, H. Q. Luo, D. T. Leong and N. B. Li, Nat. Commun., 2022, 13, 7289 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nh00124e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |