Open Access Article
Open Access ArticleMultiple-unit interlocking enhances the single-stranded tiles assembly of DNA nanostructures†‡
Xiangxiang
Guan
 ab,
Chenyou
Zhu
ab,
Yuanchen
Dong
ab,
Chenyou
Zhu
ab,
Yuanchen
Dong
 *cd,
Dongsheng
Liu
*cd,
Dongsheng
Liu
 *ab and
Chengde
Mao
*ab and
Chengde
Mao
 *e
*e
aKey Laboratory of Bioorganic Phosphorus Chemistry & Chemical Biology (Ministry of Education), Department of Chemistry, Tsinghua University, Beijing 100084, P. R. China. E-mail: liudongsheng@tsinghua.edu.cn
bEngineering Research Center of Advanced Rare Earth Materials, (Ministry of Education), Department of Chemistry, Tsinghua University, Beijing 100084, P. R. China
cCAS Key Laboratory of Colloid Interface and Chemical Thermodynamics, Beijing National Laboratory for Molecular Sciences, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, P. R. China. E-mail: dongyc@iccas.ac.cn
dUniversity of Chinese Academy of Sciences, Beijing 100049, P. R. China
eDepartment of Chemistry, Purdue University, West Lafayette, Indiana 47907, USA. E-mail: mao@purdue.edu
First published on 9th October 2024
Abstract
Single-stranded tiles (DNA brick) assembly has provided a simple and modular tool for constructing nanostructures with the potential for numerous applications. However, in this strategy, the short-strand building blocks are susceptible to environmental fluctuations and bring about rapid dissociation during assembly, resulting in instability and prolonged annealing. Thus, developing new strategies which can enhance the stability and accelerate the assembly process of DNA bricks is important. In this study, we applied the kinetically interlocking multiple-unit (KIMU) strategy to tune the process of DNA brick assembly by adopting long DNA strands as building blocks, ranging from tens of to 1000 nucleotides. We constructed a series of DNA structures with improved stability over DNA bricks. Furthermore, the annealing process could be accelerated by increasing the number of units. Our study demonstrated that DNA assembly based on the KIMU strategy using multiple-unit DNA strands could be a promising method for constructing relatively stable DNA nanostructures.
1. Introduction
During recent decades, DNA technology has been developed to prepare arbitrary 2D and 3D complex structures at the nano-scale. These structures have been successfully employed for various applications, including drug delivery, data storage and lithography.1–3 Since Seeman et al. proposed the concept of DNA nanotechnology and built the first artificial immobile Holliday junction through tile-by-tile assembly, many strategies for DNA assembly have been developed, including DNA origami and single-stranded tiles (DNA Brick) assembly.4–14 In tile-by-tile assembly, DNA strands usually with tens-to-one hundred bases are rationally designed to construct 1D/2D patterns and 3D crystals with extensible dimensions.4,14–18 DNA origami, which is based on the interaction between a long single-strand DNA (long ssDNA) and many short single-stranded DNAs (short ssDNAs), effectively produces a wide range of DNA nanostructures.5,7,19,20 In 2012, Yin et al. devised a single-stranded tiles (DNA brick) strategy, which utilized short ssDNAs as individual bricks with four binding domains to interlock with adjacent strands, to prepare infinite or finite entities.11,12,21–23 Therefore, either longer strands (more than thousands of bases) or short strands (tens of bases) have been typically utilized as building blocks for DNA self-assembly, with relatively little research focusing on medium-length DNA strands.24–29For a short-strand assembly, the DNA Brick method has provided a simple, modular and robust framework for constructing nanostructures with prescribed shapes from short synthetic DNA strands. Strict stoichiometry of DNA components is not required during the assembly and the desired nanostructures can be easily designed. In the brick assembly, each brick is usually composed of tens of nucleotides, which may dissociate quickly under environment fluctuation during the assembly.30 Moreover, potential random interactions may reduce the collision probability between the correct sequences. Therefore, the assembly of DNA bricks requires a prolonged annealing process to form the intended structures, and the resulting assemblies are relatively unstable. The development of new strategies which can facilitate rapid construction and enhance structural stability is important for DNA bricks for improving reliability in future biomedical applications.
Recently, a kinetically interlocking multiple-units (KIMU) theory has been demonstrated to be effective for understanding and tuning the DNA self-assembly.31–34 This theory suggests that increasing the number of noncovalent units could improve the stability of DNA assembly. According to KIMU theory, each brick of length ∼50 nucleotides can also be considered as a single unit. Therefore, it can be anticipated that further conjugation of such units, resulting in a building block of hundreds of nucleotides, could enhance their local concentrations, thus increasing the effective collision between DNA bricks. One study also indicated that introducing a longer seed DNA strand could accelerate the assembly process and extend the temperature region for isothermal annealing,35 which is consistent with the KIMU theory. Furthermore, once the structure has been well assembled with multiple-units strands, the improved stability of the DNA assembly can also be predicted and explained by the kinetically interlocking effect of the increased units. This approach not only enhances the structural integrity of the assembled constructs, but also potentially accelerates the assembly process, paving the way for more efficient and robust applications of DNA nanotechnology. However, while the KIMU theory has already been successfully employed in DNA supramolecular polymerization and DNA hydrogels,31–34,36 systematic investigation of the multiple-units effect on the complex DNA-assembly process and stability is limited.
In this study, we investigated the KIMU effect during DNA self-assembly. We adopted DNA strands of different lengths as building blocks, spanning from tens of nucleotides to about 1000 nucleotides. By regulating the annealing process, we explored the enhanced effect of multiple-unit DNA on fabricating nanostructures, and demonstrated that an increased number of units could enhance the stability of DNA assembly. It should be noted that this is the first systematic study to investigate the effects of a medium-chain length on the stability and folding kinetics of DNA nanostructures. Our study could: (i) provide an alternative tool to tune the assembly process of DNA self-assembly; (ii) give rise to a strategy to construct stable DNA nanostructures in a programmable fashion.
2. Results and discussion
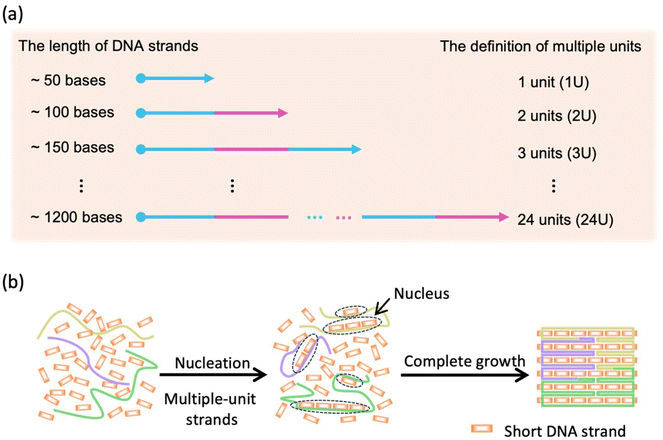
2.1. Construction of a DNA structure based on the KIMU theory
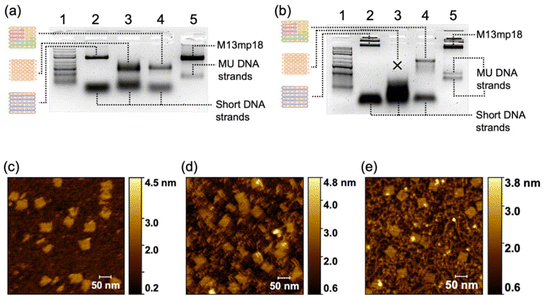
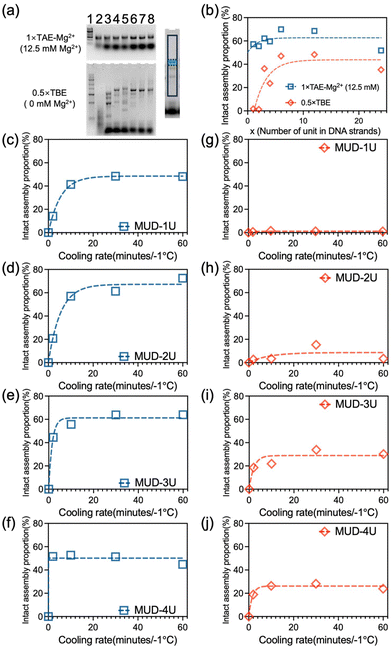
We have chosen a square-shaped structure as a typical example. To reveal the multiple-unit effect, we designed multiple-unit DNA strands (MUD; Scheme 1a and b) to assemble the square structure. It should be noted that in the current study, a DNA segment with length of ∼50 nucleotides (nt) has been defined as a “single unit” according to the traditional brick strategy (Fig. S1a‡). The illustration and the sequences of the DNA nanostructures are depicted in Fig. S2 and detailed in Tables S1–S3.‡ Basically, the square is designed with 12 helices (H) by 107 bases (B), which is constructed by folding three multiple-unit DNA strands (360 nt, 410 nt, 614 nt, representing 7, 8, and 12 units, respectively) in conjunction with 42 short DNA strands. A similar square structure based on the brick assembly was also designed with dimensions of 12 H × 105 B, composed of 71 short DNA strands. We also designed CDO-square structures (Fig. S1b‡) with an identical seam and sequence arrangement through an M13mp18 scaffold as an extreme example of MUD.To elucidate the assembly behavior of the MUD, after an annealing process, we characterized the assemblies with agarose-gel electrophoresis and atomic force microscopy (AFM). As depicted in Fig. 1a, under TAE–Mg2+, aside from the band corresponding to the starting DNA strands, a new band was observed (lane 4), indicative of successful assembly. The high proportion of intact assemblies confirmed the efficient assembly elicited using the MUD strategy, which is comparable with strategies using DNA bricks and DNA origami. Subsequently, the morphology of the MUD samples was also carefully examined by AFM. As shown in Fig. 1c, the primary nanostructures exhibited the designed square shape, approximately 35 nm × 36 nm, suggesting that the MUD sequences had become effectively folded into the intended nanostructures. For comparison, the DNA bricks-square and CDO-square presented a similar morphology. Notably, a conventional DNA-origami strategy utilized the M13mp18 with a length of 7249 bases as the scaffold. The AFM image (Fig. 1e) showed many tangled threads, which was the excess part of the scaffold, and resulted in a relatively slower migration rate in the gel image (Fig. 1a). Therefore, the MUD strategy clearly led to the formation of DNA nanostructures, showing comparable quality to those constructed via the methods of DNA bricks and DNA origami.
Then, we demonstrated the improved stability of the DNA structures constructed by MUD. Herein, we employed agarose-gel electrophoresis with a magnesium-free buffer to estimate the structural stability of the DNA square. As shown in Fig. 1b, the products band of the MUD-square was smeared slightly downwards, but the main expected band remained predominant. In contrast, the product of the DNA bricks-square was nearly undetectable, indicating the disassembly of the structure under this condition. Furthermore, the MUD-square appeared comparable to the CDO-square, which served as a positive control. On the other hand, the expected product of the strategy using all-short ssDNA bricks was unstable and almost disappeared under agarose-gel electrophoresis. Moreover, we further examined the ion stability of DNA assemblies by using AFM after incubating the DNA assemblies in 0.5× TBE buffer for 3.5 h (Fig. S3‡). Intact square-shaped structures of predetermined size were clearly observed in both the CDO-square and MUD-square, whereas the DNA bricks-square showed evidence of broken holes and smaller fragments. Therefore, the MUD-square exhibited significantly improved stability than the DNA bricks-square, and was comparable with the CDO-square. Herein, according to the KIMU theory, the multiple-unit DNA strands increases the local concentration of units, enabling well-assembled DNA structures to recover more readily through kinetic interlocking after local dissolution under conditions of cation dilution. This interlocking, facilitated by the interactions among multiple units, enhances the stability of the previous assembly composed entirely of short DNA strands.
2.2. Generalizability of the MUD strategy
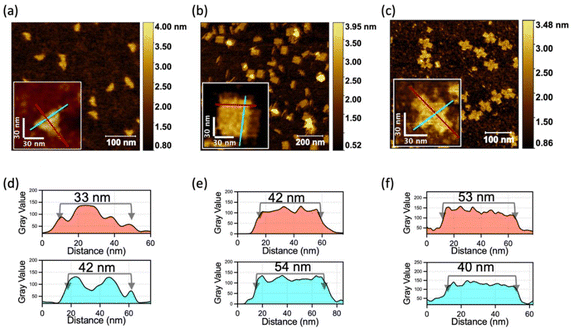
Next, we demonstrated the generalizability of the multiple-unit DNA-assembly strategy in the structural design. We designed three nanostructures: triangle-shaped MUD, rectangle-shaped MUD and cross-shaped MUD. The same as the MUD-square, they were constructed by several 300–400 bases-long multiple-unit DNA strands with short DNA strands (as designed in Fig. S4–S6‡). Through agarose-gel electrophoresis, it was verified that all these assemblies had been successfully formed into intact nanostructures with considerable proportions (Fig. S6a‡). Subsequently, through AFM (Fig. 2), it was confirmed that the three structures were well assembled and matched the predesigned dimensions. Furthermore, these assemblies also maintained a high proportion of an intact assembly structure under a magnesium-free condition (Fig. S7b‡). This result confirmed that the nanostructures constructed by the multiple-unit DNA-assembly strategy possessed universally satisfactory ion stability and broad applicability in various structural designs.2.3. Effect of the number of units
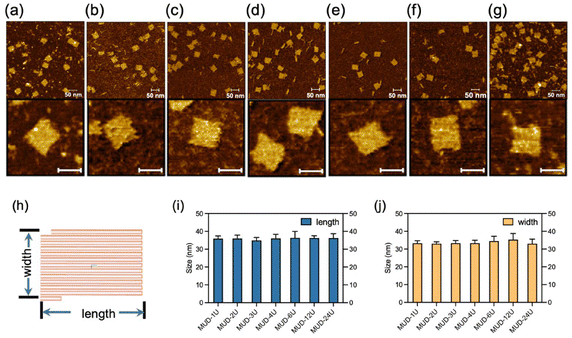
After a significant yield, good stability and structural integrity had been demonstrated in the MUD strategy, next we explored the effect of the length of MUD strands. We systematically designed a series of DNA nanostructures that were assembled with different MUD strands termed “MUD-xU”, where x denotes the number of units (Fig. S8‡). A single-stranded DNA segment of length ∼50 nt can be defined as a single unit. Accordingly, DNA strands with about 100 nt, 150 nt and longer length were designed to represent 2-unit, 3-unit DNA strands and so on, respectively (as illustrated in Scheme 1a). It should be noted that the single-stranded DNA in the DNA bricks strategy and the staples in the DNA origami strategy can be also regarded as a single-unit DNA. The nanostructures of MUD-xU were designed to be of size 36 nm × 32 nm according to distinct strategies. In our study, MUD-1U utilized ssDNA bricks, while the others incorporated multiple-unit strands, specifically 12 MUD-2U, eight MUD-3U, six MUD-4U, four MUD-6U, two MUD-12U, and one MUD-24U, respectively. It is worth noting that the MUD-24U sample was constructed by assembling one long 24-fold-unit DNA strand with several short DNA strands, and exemplified the traditional approach to DNA origami.Then, we performed AFM characterization on these assemblies. As shown in Fig. 3(a–g), the fully assembled rectangle-shaped nanostructures constituted the majority of all the assemblies across all MUD samples. Furthermore, the magnified AFM images of MUD samples (Fig. 3h–n) revealed identical morphologies. AFM images demonstrated the same size in both dimensions in accordance with the predicted designs, and thus confirmed the structural integrity of the entire series of MUD-xU samples. These results demonstrated the success of the multiple-unit DNA-assembly strategy across a wide range of nucleotide lengths, from tens to 1000.
To investigate the assembly further quantitatively, agarose-gel electrophoresis was used to characterize the efficiency of the assembly (upper-left panel in Fig. 4a). The MUD-1U was based on the DNA-bricks strategy, which uses equimolar-ratio short oligonucleotides, while the other structures use the excess 5-fold short ssDNA, compared with medium-long ssDNA. Therefore, the band of short ssDNA in MUD-1U assembly was lighter than those of others. We analyzed band density and calculated the assembly proportion by determining the percentage ratio between the intensity of the target band, indicated by the green area and that of the selected lane area within the black-boxed region shown in the right panel of Fig. 4a. All the MUD samples displayed similar behavior in the proportions of complete assemblies (box symbols in Fig. 4b). Meanwhile, to investigate the ion stability of MUD-xU samples, we utilized agarose-gel electrophoresis with 0.5 × TBE (Mg2+ = 0 mM) buffer (lower-left panel in Fig. 4a). The results (diamond symbols in Fig. 4b) demonstrated that MUD-1U and MUD-2U were nearly zero with respect to the proportion of intact assemblies. Specifically, MUD-1U, assembled using the DNA-bricks strategy, showed a pattern of downward-blurred bands that extended to a relatively lower position, signifying a more random and severe degradation process. In contrast, MUD-2U, when subjected to magnesium-free conditions in agarose-gel electrophoresis, dissolved into fragments indicative of DNA assemblies with specific numbers of double-units, as evidenced by discontinuous bands. For DNA strands with length of more than triple units, the assemblies can maintain a relatively high proportion of complete assemblies. Moreover, as the unit numbers in the DNA strands continued to increase, the proportions of intact assemblies of MUD-xU samples remained unchanged. These results suggested when the number of units exceed three, the nanostructures constructed by our multiple-unit strategy become increasingly ion-stable. To further understand the effect of magnesium ions during electrophoresis, we also examined agarose gel with intermediate concentrations of Mg2+ (2 mM and 7 mM, Fig. S9‡), which showed that the MUD structures suffered gradual destruction under electrophoresis with a decreasing concentration of Mg2+.
We also investigated the annealing speed for these MUD-xU (x = 1–4) structures and conducted the assembly process with different cooling rates (Fig. S10–S14‡). As Fig. 4(c–f) shows, for all MUD-xU samples, the slower the cooling rate, the higher was the proportion of full nanostructures. Meanwhile, as the number of units in the MUD-xU assemblies increased, the rate also increased. In general, when the cooling process slows down, the inner parts of the DNA strands get more opportunity to fully interact with one another, resulting in an obvious increase in the assembly yield. With further extension of the length of DNA strands, the number of covalent crosslinking units increases, which has a dramatically enhancing effect on the collision probability in terms of the KIMU theory. Hence, quick annealing is enough for a DNA-assembly system with long DNA strands. For the MUD-4U structure, the MUD strand contains enough units to increase the local concentration of the building block to collide in a short time, and reaches the optimal yield after a rapid annealing process. We also observed that the extended annealing time may increase the possibility of non-specific interactions between different strands, which results in a slight decrease in the proportion of the target assembly (Fig. 4f). Moreover, as shown in Fig. 4(g–j), the proportion of intact MUD-xU under agarose-gel electrophoresis without magnesium indicated that the resistance to degradation gradually increased with an increase in the number of units, aligning with the overall results presented in Fig. 4b. These data indicated that the MUD strategy had a significant acceleration effect on the assembly process of MUD-xU by increasing the number of covalent crosslinking units.
In addition, we tested the folding of MUC-xU structures in different magnesium concentrations. As illustrated in Fig. S11–S15,‡ the MUD-xU nanostructures were successfully assembled under magnesium-ion concentrations exceeding 10 mM. Their yields nearly peaked when the magnesium-ion concentration in the assembly buffer reached 20 mM. Consequently, we selected 20 mM as the magnesium-ion concentration for the aforementioned study. It is notable that multiple-unit DNA strands were investigated by denatured polyacrylamide-gel electrophoresis (Fig. S16‡). A single band corresponding to the expected length, as verified by marker positions, was presented in each lane, and we considered this to indicate that the quality of MUD strands was sufficient for assembly.
3. Conclusions
Using ssDNA of different lengths, we constructed a series of DNA assemblies based on a MUD strategy and investigated the process of DNA assembly. A typical square-shaped DNA nanostructure based on the MUD strategy was prepared with intact morphology and high efficiency of assembly, comparable with that obtained using DNA bricks or conventional DNA origami. In addition, multiple nanostructures based on the MUD strategy were designed and constructed, which verified the generalizability of the multiple-unit DNA-assembly strategy. Furthermore, MUD with a varying number of units was also fabricated to systematically examine the impact on the stability and assembly process of DNA nanostructures. We also demonstrated that multiple-unit DNA can accelerate the assembly process by increasing the unit number. Our study reveals the enhancement effect of multiple units for the stability of the DNA assemblies composed of DNA strands with lengths ranging from tens of bases to 1000 bases, and offers an alternative strategy to effectively fabricate a stable DNA structure. The current cost may be slightly higher than the brick assembly, but a reduction can be anticipated in the near future with developments in DNA synthesis.Data availability
All data are provided in the manuscript and the ESI.‡Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the National Basic Research Plan of China (2023YFA0915201), Beijing Municipal Science & Technology Commission and Zhongguancun Science Park Administrative Committee (Z231100007223003), Beijing Natural Science Foundation (JQ24007), National Natural Science Foundation of China (21821001), and the National Science Foundation (CCF2107393) for financial support.References
- L. F. Song, F. Geng, Z. Y. Gong, X. Chen, J. J. Tang, C. Y. Gong, L. B. Zhou, R. Xia, M. Z. Han, J. Y. Xu, B. Z. Li and Y. J. Yuan, Nat. Commun., 2022, 13, 5361 CrossRef CAS PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- W. Sun, J. Shen, Z. Zhao, N. Arellano, C. Rettner, J. S. Tang, T. Y. Cao, Z. Y. Zhou, T. Ta, J. K. Streit, J. A. Fagan, T. Schaus, M. Zheng, S. J. Han, W. M. Shih, H. T. Maune and P. Yin, Science, 2020, 368, 874–877 CrossRef CAS PubMed.
- E. Winfree, F. R. Liu, L. A. Wenzler and N. C. Seeman, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- S. M. Douglas, H. Dietz, T. Liedl, B. Hoegberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 CrossRef CAS PubMed.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, Science, 2011, 332, 342–346 CrossRef CAS PubMed.
- Y. He, T. Ye, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Nature, 2008, 452, 198–U141 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- Q. Jiang, C. Song, J. Nangreave, X. Liu, L. Lin, D. Qiu, Z.-G. Wang, G. Zou, X. Liang, H. Yan and B. Ding, J. Am. Chem. Soc., 2012, 134, 13396–13403 CrossRef CAS PubMed.
- Y. Ke, L. L. Ong, W. M. Shih and P. Yin, Science, 2012, 338, 1177–1183 CrossRef CAS PubMed.
- B. Wei, M. Dai and P. Yin, Nature, 2012, 485, 623–626 CrossRef CAS PubMed.
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS PubMed.
- H. Yan, S. H. Park, G. Finkelstein, J. H. Reif and T. H. LaBean, Science, 2003, 301, 1882–1884 CrossRef CAS PubMed.
- T. J. Fu and N. C. Seeman, Biochemistry, 1993, 32, 3211–3220 CrossRef CAS PubMed.
- X. P. Yang, L. A. Wenzler, J. Qi, X. J. Li and N. C. Seeman, J. Am. Chem. Soc., 1998, 120, 9779–9786 CrossRef CAS.
- J. P. Zheng, J. J. Birktoft, Y. Chen, T. Wang, R. J. Sha, P. E. Constantinou, S. L. Ginell, C. D. Mao and N. C. Seeman, Nature, 2009, 461, 74–77 CrossRef CAS PubMed.
- Y. G. Ke, Y. Liu, J. P. Zhang and H. Yan, J. Am. Chem. Soc., 2006, 128, 4414–4421 CrossRef CAS PubMed.
- S. Ramakrishnan, G. Krainer, G. Grundmeier, M. Schlierf and A. Keller, Small, 2017, 13, 1702100 CrossRef PubMed.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS PubMed.
- Y. G. Ke, L. L. Ong, W. M. Shih and P. Yin, Science, 2012, 338, 1177–1183 CrossRef CAS PubMed.
- L. L. Ong, N. Hanikel, O. K. Yaghi, C. Grun, M. T. Strauss, P. Bron, J. Lai-Kee-Him, F. Schueder, B. Wang, P. F. Wang, J. Y. Kishi, C. Myhrvold, A. Zhu, R. Jungmann, G. Bellot, Y. G. Ke and P. Yin, Nature, 2017, 552, 72–77 CrossRef CAS PubMed.
- P. Yin, R. F. Hariadi, S. Sahu, H. M. T. Choi, S. H. Park, T. H. LaBean and J. H. Reif, Science, 2008, 321, 824–826 CrossRef CAS PubMed.
- X. Y. G. Posnjak, P. Butler, O. Bienek, M. Dass, S. Lee, I. D. Sharp and T. Liedl, Science, 2024, 384, 781–785 CrossRef PubMed.
- F. A. S. Engelhardt, F. Praetorius, C. H. Wachauf, G. Bruggenthies, F. Kohler, B. Kick, K. L. Kadletz, P. N. Pham, K. L. Behler, T. Gerling and H. Dietz, ACS Nano, 2019, 13, 5015–5027 CrossRef CAS PubMed.
- Z. Zhao, M. Zhang, J. M. Hogle, W. M. Shih, G. Wagner and M. L. Nasr, J. Am. Chem. Soc., 2018, 140, 10639–10643 CrossRef CAS PubMed.
- J. Liu and H. Gu, STAR Protoc., 2021, 2, 100531 CrossRef CAS PubMed.
- J. Liu, L. Chen, T. Zhai, W. Li, Y. Liu and H. Gu, J. Am. Chem. Soc., 2022, 144, 16598–16603 CrossRef CAS PubMed.
- P. M. Nafisi, T. Aksel and S. M. Douglas, Synth. Biol., 2018, 3, ysy015 CrossRef CAS PubMed.
- Y. Kim and P. Yin, Angew. Chem., Int. Ed., 2020, 59, 700–703 CrossRef CAS PubMed.
- R. Chen, Y. Li, Y. Jin, Y. Sun, Z. Zhao, Y. Xu, J.-F. Xu, Y. Dong and D. Liu, Carbohydr. Polym., 2023, 310, 120703 CrossRef CAS PubMed.
- J. Shi, H. Jia, H. Chen, X. Wang, J.-F. Xu, W. Ren, J. Zhao, X. Zhou, Y. Dong and D. Liu, CCS Chem., 2019, 1, 296–303 CrossRef CAS.
- J. Shi, C. Zhu, Q. Li, Y. Li, L. Chen, B. Yang, J.-F. Xu, Y. Dong, C. Mao and D. Liu, Macromol. Rapid Commun., 2021, 42, 2100182 CrossRef CAS PubMed.
- Y. Li, Y. Ding, B. Yang, T. Cao, J. Xu, Y. Dong, Q. Chen, L. Xu and D. Liu, CCS Chem., 2023, 5, 434–444 CrossRef CAS.
- Y. Zhang, A. Reinhardt, P. Wang, J. Song and Y. Ke, Angew. Chem., Int. Ed., 2020, 59, 8594–8600 CrossRef CAS PubMed.
- X. Li, X. Dai, Y. Pan, Y. Sun, B. Yang, K. Chen, Y. Wang, J.-F. Xu, Y. Dong, Y. R. Yang, L.-T. Yan and D. Liu, J. Am. Chem. Soc., 2022, 144, 21267–21277 CrossRef CAS PubMed.
Footnotes |
| † We would like to dedicate this article in honour of Professor George Whitesides to celebrate his 85th birthday. |
| ‡ Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03288h |
| This journal is © The Royal Society of Chemistry 2024 |