 Open Access Article
Open Access ArticleAbsolute configuration determination of SMTP-7 via microcrystal electron diffraction (MicroED)†
Bo
Wang
 * and
Yiqing
Lin
* and
Yiqing
Lin
Small Molecule Drug Product Development, Biogen, 115 Broadway, Cambridge, MA 02142, USA. E-mail: bo.wang2@biogen.com
First published on 28th October 2022
Abstract
Direct absolute configuration determination of a drug candidate bearing five chiral centers, SMTP-7, is lacking. With only few-micron-sized powders available, we assigned the absolute configuration of SMTP-7 via MicroED analysis through kinematical refinement with additional chiral information and directly through the recently developed dynamical refinement approach. We showcased the utility of the latest advance in MicroED to unambiguously assign absolute configuration for hard-to-crystallize, complex, and quantity-limited molecules.
Stachybotrys microspora triprenyl phenol (SMTP) is a family of small molecules that are fermented from Stachybotrys microspore by feeding precursor amino acids and amino alcohols.1,2 Among more than 60 SMTP congeners discovered so far, SMTP-7 (BIIB131, TMS-007) exhibits thrombolytic, anti-inflammatory, and antioxidant effects, which demonstrated encouraging clinical Phase II results in Japan, and is currently being developed as the next generation of thrombolytic agent to treat the deadly acute ischemic stroke (AIS).1,3
Prior to the discovery of SMTP, neuritogenic Stachybotrin C was first isolated from Stachybotrys parvispora, and its stereoconfiguration was originally deduced as (8R,9R) via NOESY NMR analysis (Scheme 1).4 After the discovery of SMTP congeners, the stereoconfiguration was first assigned to the structurally simplest one, SMTP-0, to be (8S,9S) using the modified Mosher method via NMR data.5 Later on, the absolute configuration of Stachybotrin C was revised to (8S,9R) via total synthesis analysis,6 and crystal structure determination of a derivative of Stachybotrin C.7 A biosynthesis of the SMTP was proposed, with the chromanlactam containing the isoprene side chain moiety, pre-SMTP, to be structurally fixed.1,8 For example, by feeding L-ornithine or D-ornithine to the fermentation process, SMTP-7 and SMTP-7D can be obtained, respectively. Based on the proposed biosynthesis mechanism and the absolute configuration determination of Stachybotrin C, it was suggested that all the stereoconfiguration of SMTP congeners are (8S,9R).1,7 However, considering the clinical importance of SMTP-7, and the conflicting absolute stereoconfiguration assignment history, a direct and unambiguous absolute configuration assignment of SMTP-7 is much needed. Although vibrational optical activity9 and NMR10 can provide much valuable chiral information, single-crystal X-ray diffraction (scXRD) crystallography is still widely used as the gold standard for absolute structure determination. In this case, the challenge lies in the fact that single crystal samples of SMTP-7 have not been obtained.
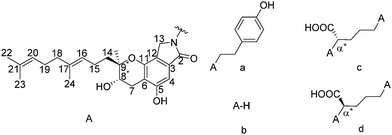 | ||
| Scheme 1 Molecular structures of Stachybotrin C and selected SMTP, (a) Stachybotrin C, (b) SMTP-0, (c) SMTP-7, and (d) SMTP-7D. The stereocenters in investigation are labeled with stars. | ||
Microcrystal electron diffraction (MicroED) provides an alternative solution to crystal structure determination in case scXRD is impossible.11–15 The advantage of MicroED lies in the minimal sample preparation requirements that only sub-micron crystalline powders are sufficient to yield reasonable crystal structures. However, unlike scXRD, kinematically refined ED data does not produce significant anomalous signals to distinguish the enantiomeric pair of solutions.15 Therefore, we previously demonstrated that incorporation of chiral information in material (e.g. chiral salt formation) can be used to derive the absolute configuration.16 Excitingly, computational tools of dynamical refinement on ED data have been recently developed and utilized to directly assign the absolute configuration of model organic molecules,17,18 although a genuinely unknown example is yet to be explored with this approach.19
In this work, we report the crystal structures of SMTP-7 and its diastereomer, SMTP-7D, which were first refined kinematically using micron-sized samples via MicroED. From the structures, we derived the absolute configuration of both compounds with additional chiral information. Furthermore, to obtain the absolute configuration directly without any plausible derivations, and to implement the dynamical refinement on a real-world result-conflicting sample for the first time, we performed dynamical refinement to the MicroED data of SMTP-7/7D, reinforcing the absolute configuration assignments and showcasing the great potential of this approach.
After unsuccessful single crystal growth screenings of SMTP-7, we redirected our structural efforts to MicroED analysis since the micron-sized powder materials of both SMTP-7 and SMTP-7D were tested to be crystalline (Fig. S1 and S2, ESI†). Using the powders directly, the crystal structures of SMTP-7 and SMTP-7D were determined from the MicroED datasets using a pipeline method previously described.20 Implementing this methodology, the SMTP-7/7D structures, as shown in Fig. 1, were refined kinematically and finalized with scXRD processing tools.
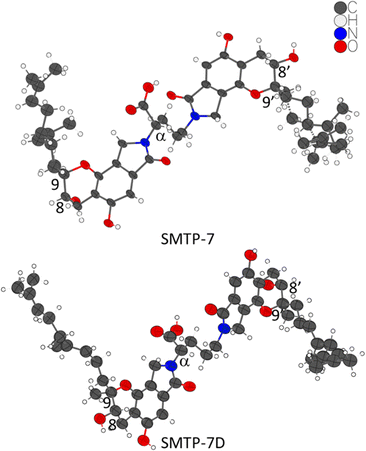 | ||
| Fig. 1 One asymmetric unit of the crystal structures of SMTP-7 and SMTP-7D. The thermal ellipsoids are at 30% probability level. The chiral centers in question are labeled. | ||
We obtained a pair of enantiomeric solutions of (8S,9R,αS,8′S,9′R) and (8R,9S,αR,8′R,9′S) for SMTP-7. Considering L-ornithine (S-configuration) was fed into the fermentation process to produce SMTP-7, if there is no epimerization that inverts the carboxylic acid group during the fermentation process, the stereocenter at the α position preserves as S. Therefore, the absolute configuration of SMTP-7 must be (8S,9R,αS,8′S,9′R). On the other hand, although unlikely and unreported in SMTP discovered so far,1 if there is an enzyme-catalyzed epimerization process that favors the inversion involving the carboxylic group from the S-configuration to the R-configuration, which cannot be completely ruled out since chiral centers adjacent to carbonyls are known to be the substrate of racemases and epimerases,21 the SMTP-7 should, then, be assigned to (8R,9S,αR,8′R,9′S). Since the drug substance is chirally pure, we can first rule out the racemization case. If the αR-favored epimerization process is true, SMTP-7D, which was produced from feeding the D-ornithine (R-configuration), would retain the R-configuration at the α-position. From the kinematically refined MicroED structure of SMTP-7D, the two possible solutions are (8S,9R,αR,8′S,9′R) and (8R,9S,αS,8′R,9′S). In the hypothesized epimerization case, the absolute configuration of SMTP-7D should be (8S,9R,αR,8′S,9′R). However, in the proposed biosynthesis of the SMTP congeners, the pre-SMTP moiety is considered to be structurally fixed. But the pre-SMTP assignment of (8R,9S) in the “epimerized” SMTP-7 is against the assignment of (8S,9R) in SMTP-7D. Therefore, the conflicting result suggested that such epimerization is unlikely. Based on the arguments, SMTP-7 was assigned to (8S,9R,αS,8′S,9′R) and SMTP-7D is assigned to (8S,9R,αR,8′S,9′R). To validate the assignment experimentally, vibrational circular dichroism (VCD) spectrum was collected for SMTP-7. By comparing the calculated VCD spectrum to the experimental data, the analysis substantiated the assignment of (8S,9R,αS,8′S,9′R) to SMTP-7 (see ESI†).
At present, most 3D ED structures were processed using kinematic refinement.15 Due to the stronger interaction of electrons with matter, multiple scattering occurs in the 3D ED experiment (e.g. MicroED). To reduce the impact of this effect on experimental diffraction intensities, technique such as precession electron diffraction, rotation electron diffraction, and continuous rotation method have been developed and applied to successful structure determination with ED data.22 However, the opportunity to obtain the absolute structure is missed when the multiple scattering events were neglected. On the other hand, the dynamical refinement was recently made possible to account for the multiple scattering effect on various materials such as pharmaceutical cocrystals,18 metal–organic framework,23 zeolites24via software development of PETS 2 and Jana2006.25–27 It has been shown with model chiral compounds that dynamical refinement can be used to determine the absolute structure and improve the model accuracy of chiral crystals.17,18 In this work, we aim to apply this approach to SMTP-7 and SMTP-7D, two relatively large small molecule drug candidates, not only to substantiate the absolute configuration assignments but also to validate the dynamical refinement methodology.
After converting and processing the raw MicroED data with PETS 2, we implemented dynamical refinement on the two kinematically refined structures of SMTP-7/7D using Jana2020.25,26 By comparing the SMTP-7 enantiomeric models, it was shown that the (8S,9R,αS,8′S,9′R) model has lower R-factors than the enantiomer in all 21 datasets being processed (selected ten results are shown in Fig. 2). And the (8S,9R,αR,8′S,9′R) SMTP-7D model has lower R-factors in all four datasets. In all cases, the structural models were refined with all structural parameters being fixed to facilitate the absolute structure assignment, due to the large molecule size and number of parameters. This restraint refinement process was shown here to be efficient in differentiating the enantiomeric models using the dynamically refined wRall as the indicator. Note that the analysis was performed based on individual datasets on different powder particles from the same chirally pure batch. For samples with low chiral purity, one may consider sampling multiple datasets on distinct powders to ensure the absolute structure determination is not result from a minor chiral component. After all, our analysis strongly substantiates the absolute configuration assignment of SMTP-7/7D. Mutually, the result validates the robustness and readiness of implementing dynamical refinement on MicroED datasets to obtain absolute structures of crystalline powders.
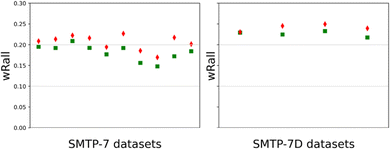 | ||
| Fig. 2 Post-dynamical refinement R-factor (wRall) comparison between the correct (green square) and inverted (red diamond) models of SMTP-7 and SMTP-7D. | ||
Besides providing valuable stereoconfiguration information, the MicroED crystal structures of SMTP-7/7D also exhibit interesting packing behaviors of both compounds. SMTP-7 adopts the P21212 space group, with Z′ = 1 and Z = 4. Interestingly, the crystal packs in such a way that the chromanlactam moieties are close intermolecularly so that it favors the pi–pi interaction from the aromatic rings. Intriguingly, seven out of the ten oxygen atoms in one molecule participate in the hydrogen bonding networks, except both in the carboxylic group which contains the best hydrogen bond donor. Instead, the carboxylic group is oriented in the layers of the isoprene side chains. When using a water probe with a radius of 1.4 Å, 0.4% unit cell volume voids were calculated, which are all located in the hydrophobic layers with the isoprene groups. The void space indicates that the space was not tightly packed by the long alkyl groups. Therefore, the disordering, which was modeled for one side of SMTP-7 (ratio of site occupancy of the two disordered alkyl groups is 51%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 49%), can be understood that the disorder favors entropy gain to contribute to the stability of this free acid form. SMTP-7D adopts the P212121 space group, with Z′ = 1 and Z = 4, interestingly, this diastereomer has the same hydrogen-bonding participating pattern with the carboxylic group pointing into the layers composed of the isoprene side chains. When using a water probe with a radius of 1.4 Å, no voids were calculated, which is in alignment with that no disorder was observed for the isoprene side chains.
49%), can be understood that the disorder favors entropy gain to contribute to the stability of this free acid form. SMTP-7D adopts the P212121 space group, with Z′ = 1 and Z = 4, interestingly, this diastereomer has the same hydrogen-bonding participating pattern with the carboxylic group pointing into the layers composed of the isoprene side chains. When using a water probe with a radius of 1.4 Å, no voids were calculated, which is in alignment with that no disorder was observed for the isoprene side chains.
Absolute configuration determination of chiral molecules is a key step in pharmaceutical development and chiral chemistry. Due to the structural determination challenges associated with the clinically important drug candidate SMTP-7, we conducted the extensive and novel absolute structure determination analysis that led to the assignment of (8S,9R,αS,8′S,9′R) to SMTP-7 and (8S,9R,αR,8′S,9′R) to SMTP-7D. Using the two complex drug candidates as realistic examples, we demonstrated that, with only micron-sized powder available, it is totally practical to determine the absolute configuration with high confidence. One can either implement kinematical refinement with additional chiral information or perform dynamical refinement directly. Since 3D ED (e.g. MicroED) is relatively new to pharmaceutical and structural scientists, we want to use these two realistic examples to raise awareness of the recent advance in the ED field that can tremendously benefit drug development and structural chemistry research.
BW conceptualized and wrote the manuscript. YL secured financial support and oversaw the project. All authors contributed to the article and approved the submitted version.
We thank Dr Jessica Bruhn and her group at NanoImaging Services for MicroED data collection and processing. We thank Dr Paul Klar's python script and advice on ED data converting and processing. We thank Prof. Lukáš Palatinus for allowing us to use PETS 2 for this research. We thank Jordan Nafie and Dr Rina Dukor at BioTools for VCD data collection and analysis. We thank Prof. Lukáš Palatinus, Prof. Michal Dušek, Prof. Václav Petříček, Dr Paul Klar, and Dr Petr Brázda at FZU for hosting the 2021 ED workshop.
Conflicts of interest
There are no conflicts to declare.Notes and references
- K. Hasumi and E. Suzuki, Int. J. Mol. Sci., 2021, 22, 954 CrossRef CAS PubMed.
- W. Hu, R. Narasaki, S. Ohyama and K. Hasumi, J. Antibiot., 2001, 54, 962–966 CrossRef CAS PubMed.
- H. Sawada, N. Nishimura, E. Suzuki, J. Zhuang, K. Hasegawa, H. Takamatsu, K. Honda and K. Hasumi, J. Cereb. Blood Flow Metab., 2014, 34, 235–241 CrossRef CAS PubMed.
- Y. Nozawa, M. Ito, K. Sugawara, K. Hanada and K. Mizoue, J. Antibiot., 1997, 50, 641–645 CrossRef CAS PubMed.
- K. Hasumi, K. Hasegawa and Y. Kitano, J. Antibiot., 2007, 60, 463–468 CrossRef CAS PubMed.
- M. Jacolot, M. Jean, N. Tumma, A. Bondon, S. Chandrasekhar and P. van de Weghe, J. Org. Chem., 2013, 78, 7169–7175 CrossRef CAS PubMed.
- Y. Kuroda, K. Hasegawa, K. Noguchi, K. Chiba, K. Hasumi and Y. Kitano, J. Antibiot., 2018, 71, 584–591 CrossRef CAS PubMed.
- Y. Nishimura, E. Suzuki, K. Hasegawa, N. Nishimura, Y. Kitano and K. Hasumi, J. Antibiot., 2012, 65, 483–485 CrossRef CAS PubMed.
- Y. He, B. Wang, R. K. Dukor and L. A. Nafie, Appl. Spectrosc., 2011, 65, 699–723 CrossRef CAS PubMed.
- J. M. Seco, E. Quiñoá and R. Riguera, Chem. Rev., 2004, 104, 17–118 CrossRef CAS.
- D. Shi, B. L. Nannenga, M. G. Iadanza and T. Gonen, eLife, 2013, 2, e01345 CrossRef PubMed.
- B. L. Nannenga, D. Shi, A. G. W. Leslie and T. Gonen, Nat. Methods, 2014, 11, 927–930 CrossRef CAS PubMed.
- B. L. Nannenga and T. Gonen, Nat. Methods, 2019, 16, 369–379 CrossRef CAS PubMed.
- C. G. Jones, M. W. Martynowycz, J. Hattne, T. J. Fulton, B. M. Stoltz, J. A. Rodriguez, H. M. Nelson and T. Gonen, ACS Cent. Sci., 2018, 4, 1587–1592 CrossRef CAS PubMed.
- T. Gruene, J. J. Holstein, G. H. Clever and B. Keppler, Nat. Rev. Chem., 2021, 1–9 Search PubMed.
- B. Wang, J. F. Bruhn, A. Weldeab, T. S. Wilson, P. T. McGilvray, M. Mashore, Q. Song, G. Scapin and Y. Lin, Chem. Commun., 2022, 58, 4711–4714 RSC.
- P. Klar, Y. Krysiak, H. Xu, G. Steciuk, J. Cho, X. Zou and L. Palatinus, ChemRxiv, 2021 DOI:10.26434/chemrxiv-2021-4jh14.
- P. Brázda, L. Palatinus and M. Babor, Science, 2019, 364, 667–669 CrossRef PubMed.
- A. Saha, S. S. Nia and J. A. Rodríguez, Chem. Rev., 2022, 122, 13883–13914 CrossRef CAS PubMed.
- J. F. Bruhn, G. Scapin, A. Cheng, B. Q. Mercado, D. G. Waterman, T. Ganesh, S. Dallakyan, B. N. Read, T. Nieusma, K. W. Lucier, M. L. Mayer, N. J. Chiang, N. Poweleit, P. T. McGilvray, T. S. Wilson, M. Mashore, C. Hennessy, S. Thomson, B. Wang, C. S. Potter and B. Carragher, Front. Mol. Biosci., 2021, 8, 354 Search PubMed.
- M. E. Tanner, Acc. Chem. Res., 2002, 35, 237–246 CrossRef CAS PubMed.
- B. L. Nannenga, G. Bu and D. Shi, Front. Mol. Biosci., 2018, 5, 114 CrossRef CAS PubMed.
- J. Hynek, P. Brázda, J. Rohlíček, M. G. S. Londesborough and J. Demel, Angew. Chem., Int. Ed., 2018, 57, 5016–5019 CrossRef CAS PubMed.
- M. Debost, P. B. Klar, N. Barrier, E. B. Clatworthy, J. Grand, F. Laine, P. Brázda, L. Palatinus, N. Nesterenko, P. Boullay and S. Mintova, Angew. Chem., Int. Ed., 2020, 59, 23491 CrossRef CAS PubMed.
- V. Petříček, M. Dušek and L. Palatinus, Z. Kristallogr. – Cryst. Mater., 2014, 229, 345–352 CrossRef.
- L. Palatinus, V. Petříček and C. A. Corrêa, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 235–244 CrossRef CAS PubMed.
- L. Palatinus, P. Brázda, M. Jelínek, J. Hrdá, G. Steciuk and M. Klementová, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2019, 75, 512–522 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Material and characterizations, details of MicroED, VCD, and dynamical refinement analysis. CCDC 2208444 and 2208429. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2cc05218k |
| This journal is © The Royal Society of Chemistry 2022 |
