Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
On-resin Diels–Alder reaction with inverse electron demand: an efficient ligation method for complex peptides with a varying spacer to optimize cell adhesion†
Mareen
Pagel
a,
René
Meier
a,
Klaus
Braun
b,
Manfred
Wiessler
b and
Annette G.
Beck-Sickinger
*a
aInstitute of Biochemistry, Faculty of Biosciences, Pharmacy and Psychology, Brüderstrasse 34, 04130 Leipzig University, Leipzig, Germany. E-mail: abeck-sickinger@uni-leipzig.de
bDeutsches Krebsforschungszentrum, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany
First published on 19th April 2016
Abstract
Solid phase peptide synthesis (SPPS) is the method of choice to produce peptides. Several protecting groups enable specific modifications. However, complex peptide conjugates usually require a rather demanding conjugation strategy, which is mostly performed in solution. Herein, an efficient strategy is described using an on-resin Diels–Alder reaction with inverse electron demand (DARinv). This method is compatible with the standard Fmoc/tBu strategy and is easy to monitor. As a proof of concept a titanium binding peptide was modified with a cyclic cell binding peptide (RGD) by DARinv on a solid support applying different tetrazines and alkenes. The generated bulky DARinv linkers were employed to act as the required spacer for RGD mediated cell adhesion on titanium. In vitro studies demonstrated improved cell spreading on DARinv-conjugated peptides and revealed, in combination with molecular dynamics-simulation, new insights into the design of spacers between the RGD peptide and the surface. Performing the DARinv on resin expands the toolbox of SPPS to produce complex peptide conjugates under mild, catalyst free conditions with reduced purification steps. The resulting conjugate can be effectively exploited to promote cell adhesion on biomaterials.
Introduction
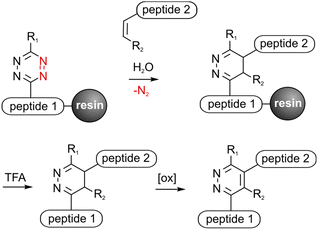
Peptide conjugates are perfect tools to selectively address various targets in medical applications and analytical investigations. Solid phase peptide synthesis (SPPS) provides a variety of orthogonal protecting groups, which facilitate the modification of peptides with fluorophores, drugs, carbohydrates, fatty acids or reporter molecules.1–3 Nevertheless, the efficient synthesis of multifunctional peptides with several complex bioactive moieties such as cyclic peptides is still challenging. Chemoselective click reactions offer the possibility to selectively ligate biomolecules without the use of further protecting groups in benign solvents resulting in high yields.4,5 Therefore click chemistry is a convenient method to upgrade traditional SPPS to obtain multifunctional peptides.6–8 It was shown that copper(I) catalyzed azide–alkyne cycloaddition (CuAAC), oxime ligation and recently the Staudinger–phosphite reaction are applicable for on resin synthesis to obtain branched and functionalized molecules.9–12 Click reactions on a solid support reduce purification steps, enable automation or protect sensitive amino acids like L-3,4-dihydroxylphenylalanine (DOPA) from oxidation.10 Another bioorthogonal cycloaddition is the Diels–Alder reaction with inverse electron demand (DARinv) that can be performed in water without catalysts or hazardous sideproducts and reagents.13 This irreversible reaction between an alkene (dienophile) and a tetrazine (diene) has been successfully used to synthesize peptide conjugates by us and others for various medical applications.14–17 However, solubility problems or instability of tetrazines can lead to difficulties.17,18 An on-resin approach can overcome these limitations and decrease purification and cleavage steps, thus increasing the yield. DARinv-conjugates mostly consist of bulky linker-units between two conjugated molecules with different distances and flexibilities depending on the applied dienes and dienophiles. This could be beneficial for several in vitro investigations where distances are required to enable binding of ligands to proteins or reporter molecules.19–21 RGD-peptides bind to receptors in the cell-membrane and thus can mediate cell-adhesion on surfaces to improve healing and osseointegration of e.g. titanium implants.22,23 It could be shown that a minimum distance between the RGD-peptide and the surface is crucial for successful cell adhesion.24 Hence, DARinv-conjugated RGD-peptides present an efficient approach to promote cell attachment on surfaces such as titanium.Herein, it is demonstrated that the DARinv can be expanded to exploit polymer-bound synthesis compatible with standard Fmoc/tBu-SPPS (Fig. 1). The applicability of two tetrazines and four different dienophiles was investigated by ligating a cell-adhesive- and a titanium-binding-peptide to the solid support. The obtained products with a varying linker length were then applied to improve cell adhesion on coated titanium. Moreover, molecular dynamics (MD)-simulation was used to assess the DARinv-linker conformation and thus discuss the cell response in theory.
Experimental
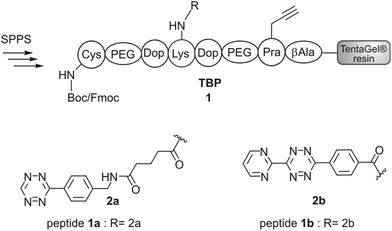
Synthesis of TBP (1) and TBP 2
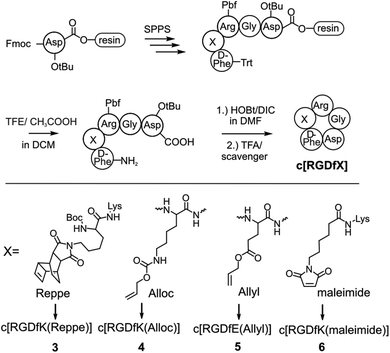
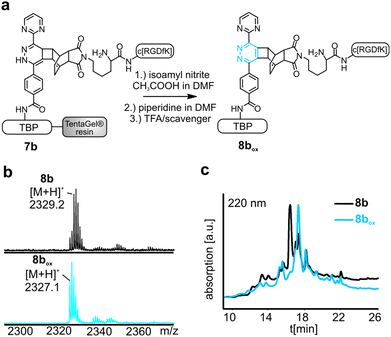
Titanium binding peptides (TBP (1) and TBP 2) were manually synthesized on a TentaGel S RAM (Iris Biotech) resin using the Fmoc/tBu strategy. Deprotection of α-amino groups was performed twice with 30% (v/v) piperidine (Sigma Aldrich) in dimethylformamide (DMF, Biosolve) for 10 min. The peptide was elongated using 2 eq. hydroxybenzotriazole (HOBt, Novabiochem) and diisopropylcarbodiimide (DIC, IrisBiotech) in DMF and 2 eq. of Fmoc-β-Ala-OH, Fmoc-Pra-OH (Iris Biotech), Fmoc-DOPA(acetonide)-OH (Novabiochem), Fmoc-Lys(Mtt)-OH (Bachem) or Fmoc-Lys(Dde)-OH (Iris Biotech), Fmoc-Cys(Trt)-OH or Boc-Cys(Trt)-OH (Iris Biotech, OrPEGen OPC) and Boc-Ser(tBu)-OH (Iris Biotech). For activation of Fmoc-NH-(PEG)2-COOH (13 atoms, Novabiochem) 1.5 eq. 1-[bis-(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU, Novabiochem) and equimolar amounts of N,N-diisopropylethylamine (DIPEA, Roth) were used. As the Lys-side chain protecting group, 4-methyltrityl (Mtt), or N-(1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl) (Dde) was used. Selective cleavage of Mtt was performed with 2% trifluoroacetic acid (TFA, Sigma Aldrich) and 3% triisopropylsilane (TIS, Sigma Aldrich) in dichloromethane (DCM, Biosolve) (v/v/v) for 2 min and was repeated 30 times. Cleaving Dde was realized with 2% hydrazine (Sigma Aldrich) in DMF for 10 min. This step was repeated 12 times. Subsequently, the dienes 2b (4-(6-(pyrimidin-2-yl)-1,2,4,5-tetrazin-3-yl)benzoic acid, synthesized according to Pipkorn et al.) and 2a (5-[4-(1,2,4,5-tetrazin-3-yl) benzyl-amino]-5-oxopentanoic acid (Sigma Aldrich)) were coupled in 2 fold excess with HOBt/DIC for 16 h in DMF.13Synthesis of c[RGDfK(Reppe)] (3), c[RGDfK(Alloc)] (4), c[RGDfE(Allyl)] (5) and c[RGDfK(maleimide)] (6)
The peptide was elongated on an acid labile 2-chlorotrityl-resin (Novabiochem) to obtain Fmoc-D-Phe-Lys(Dde)-Arg(Pbf)-Gly-Asp(tBu)-resin. After Fmoc removal, the N-terminus was protected with triphenylmethyl chloride by DIPEA in DMF. Subsequently, Dde was cleaved from the Lys-side chain, as described, and modified with either the Reppe-dienophile (synthesized according to Pipkorn et al.) or 6-maleimidohexanoic acid by HOBt/DIC activation.13 After cleavage of the protected peptide from the resin with glacial acetic acid and 2,2,2-trifluoroethanol in DCM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, v/v/v), the linear peptide was cyclized with HOBt/DIC in DCM for 16 h. Next, DCM was evaporated and the peptide was fully deprotected with TFA and a scavenger followed by purification. The peptides c[RGDfK(Alloc)] and c[RGDfE(Allyl)] were synthesized by the same protocol (Fmoc-D-Phe-Lys(Alloc)-Arg(Pbf)-Gly-Asp(tBu)-resin, Fmoc-D-Phe-Glu(Allyl)-Arg(Pbf)-Gly-Asp(tBu)-resin), however without side chain modification and trityl-protection.
8, v/v/v), the linear peptide was cyclized with HOBt/DIC in DCM for 16 h. Next, DCM was evaporated and the peptide was fully deprotected with TFA and a scavenger followed by purification. The peptides c[RGDfK(Alloc)] and c[RGDfE(Allyl)] were synthesized by the same protocol (Fmoc-D-Phe-Lys(Alloc)-Arg(Pbf)-Gly-Asp(tBu)-resin, Fmoc-D-Phe-Glu(Allyl)-Arg(Pbf)-Gly-Asp(tBu)-resin), however without side chain modification and trityl-protection.
Diels–Alder reaction with inverse electron demand (DARinv) on resin to yield peptides 8a–c
TentaGel resin loaded with 1a and respectively 1b was swollen in H2O. Then the resin was incubated with 1.5 eq. of a 30 mM aqueous solution of peptides 3–6 at room temperature on a shaker in an open reaction vessel (N2 release). When the reaction was complete, the resin was washed with water, DMF and DCM. If the N-terminus was protected by Fmoc, a deprotection step with piperidine, as stated above, followed. The peptide was cleaved from the resin and isolated as described.Monitoring of the Diels–Alder reaction with inverse electron demand (DARinv) on resin by photometry
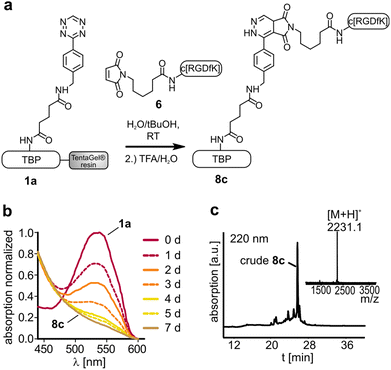
Diene modified resin (approximately 1 μmol) was transferred into a 96-well plate and swollen in water. Subsequently, the water was removed and the dissolved dienophile (30 mM; 100 μL) was added. An absorption scan from 430–600 nm of the diene modified resin was carried out with a plate reader (Tecan, Infinite® 200 PRO series). If the absorption maximum at 540 nm was diminished, the reaction was complete (8a: 5 h, 8b: 2 h, 8c: 7 d).Thiol-maleimide Michael addition to yield peptide 8d
Peptide 8d was synthesized by ligation of TBP 2 (sequence: Ser-PEG-DOPA-Cys-DOPA-PEG-Pra-β-Ala) and c[RGDfK(maleimide)] (6) in H2O/tBuOH for 20 h. The peptide was purified as described.Cleavage of peptides from the resin and purification
Final and sample cleavage of the peptides was performed with TFA/scavenger (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) by shaking for 2 h at RT. Scavenger mixtures were used as follows: TIS/H2O (1
1 v/v) by shaking for 2 h at RT. Scavenger mixtures were used as follows: TIS/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) for peptide 6 and TBP 2, thioanisole/thiocresol (1
1 v/v) for peptide 6 and TBP 2, thioanisole/thiocresol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) and thioanisole/1,2-ethanedithiol (7
1 v/v) and thioanisole/1,2-ethanedithiol (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v) for 3–5, conjugates 8a,b and H2O for peptides 1a,b and 8c. Peptides were precipitated and washed with diethyl ether. The dissolved peptide was analyzed and lyophilized. Isolation of the obtained peptides was carried out by RP-HPLC on a Phenomenex Jupiter Proteo column (90 Å/4 μm, 22 mm × 250 mm) using linear gradients of eluent B in eluent A (A: 0.1% TFA in H2O, B: 0.08% TFA in ACN). The identity and purity of the isolated products was verified by MALDI-ToF (Bruker Daltonics) and ESI-HCT (high-capacity ion trap, Bruker Daltonics) mass spectrometry and analytical RP-HPLC (Varian VariTide RPC (200 Å, 6 μm), Phenomenex Jupiter Proteo (90 Å/4 μm and 300 Å, 5 μm) and Grace Vydac (300 Å, 5 μm)) with linear gradients of eluents A and B. The detected m/z signals were in agreement with the calculated molecular weights and the peptide products were obtained with a purity of ≥90% (Table 1).
3 v/v) for 3–5, conjugates 8a,b and H2O for peptides 1a,b and 8c. Peptides were precipitated and washed with diethyl ether. The dissolved peptide was analyzed and lyophilized. Isolation of the obtained peptides was carried out by RP-HPLC on a Phenomenex Jupiter Proteo column (90 Å/4 μm, 22 mm × 250 mm) using linear gradients of eluent B in eluent A (A: 0.1% TFA in H2O, B: 0.08% TFA in ACN). The identity and purity of the isolated products was verified by MALDI-ToF (Bruker Daltonics) and ESI-HCT (high-capacity ion trap, Bruker Daltonics) mass spectrometry and analytical RP-HPLC (Varian VariTide RPC (200 Å, 6 μm), Phenomenex Jupiter Proteo (90 Å/4 μm and 300 Å, 5 μm) and Grace Vydac (300 Å, 5 μm)) with linear gradients of eluents A and B. The detected m/z signals were in agreement with the calculated molecular weights and the peptide products were obtained with a purity of ≥90% (Table 1).
| Entry | Sequence | M [Da] | [M + H]+ | Elutiona (%ACN) | Purity |
|---|---|---|---|---|---|
| Peptides were analyzed by MALDI-ToF MS (Bruker, Daltonics) and RP-HPLC. The peptide purity was evaluated by two different HPLC systems using different columns and gradients of eluent B (0.08 % TFA in ACN) in eluent A (0.1 % TFA in H2O) on a Phenomenex Jupiter Proteo (90 Å, 5 μm)a, Phenomenex Jupiter Proteo (300 Å, 4 μm)b and VariTide (200 Å, 6μm)c; doxidized product isolated, PEG (polyethylene glycol), Y* (L-3,4-dihydroxyphenylalanine), G* (propargyglycine), βA (beta alanine). n.i. = not isolated. | |||||
| 3 | c[RGDfK(Reppe)] | 915.5 | 916.5 | 44b | >95 |
| 4 | c[RGDfK(Alloc)] | 687.3 | 688.0 | 40 | >95 |
| 5 | c[RGDfE(Allyl)] | 644.3 | 645.0 | 40 | >95 |
| 6 | c[RGDfK(maleimide)] | 796.4 | 797.5 | 37 | >95 |
| 8a | C-PEG-Y*-K(2a-c[RGDfK(Reppe)])-Y*-PEG-G*-βA-NH2 | 2349.1 | 2350.1 | 36 | >90 |
| 8b d | C-PEG-Y*-K(2b-c[RGDfK(Reppe)])-Y*-PEG-G*-βA-NH2 | 2326.1 | 2327.0 | 41c | >90 |
| 8c | C-PEG-Y*-K(2a-c[RGDfK(maleimide])-Y*-PEG-G*-βA-NH2 | 2230.0 | 2231.0 | n.i. | n.i. |
| 8d | S-PEG-Y*-C(c[RGDfK(maleimide)])-Y*-PEG-G*-βA-NH2 | 1933.9 | 1935.0 | 33 | >95 |
Cellular assays
SaOS-2 cells (Sarcoma osteogenic, kindly provided by Prof. Scharnweber, Dresden, Germany) were cultured under a humidified atmosphere at 37 °C and 5% CO2 in McCoy's 5A containing 15% heat inactivated fetal calf serum (FCS), 1% (v/v) glutamine and 1% (v/v) penicillin/streptomycin (by PAA, Lonza and Biochrome).Titanium foil (Sigma Aldrich, thickness: 0.127 mm) was cut into round pieces and etched in H2SO4 (30%)/H2O2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for 7 min. The slides were then washed with water and PBS buffer (Dulbecco's phosphate) and sterilized in 70% ethanol/water (v/v) under ultrasonic irradiation for 15 min. After washing twice with PBS, the Ti-plates were incubated overnight at room temperature with a 1 μM solution of peptides dissolved in PBS as well as PBS (untreated Ti) and fibronectin (25 μg mL−1 in PBS) as controls.
1, v/v) for 7 min. The slides were then washed with water and PBS buffer (Dulbecco's phosphate) and sterilized in 70% ethanol/water (v/v) under ultrasonic irradiation for 15 min. After washing twice with PBS, the Ti-plates were incubated overnight at room temperature with a 1 μM solution of peptides dissolved in PBS as well as PBS (untreated Ti) and fibronectin (25 μg mL−1 in PBS) as controls.
To investigate the initial cell behavior to the synthesized peptide coatings, cells were resuspended in media without FCS (to prevent adsorption of serum proteins) and seeded on coated and washed Ti-plates. After adhesion for 6 h, the plates were washed twice with PBS and subsequently the cells were fixed with 4% paraformaldehyde in PBS for 30 min. The plates were washed with PBS and the cells were permeabilized with 1% triton X-100 in PBS for 1 min. Next, the slides were washed and cells were stained with phalloidin-tetramethylrhodamine-isothiocyanate (phalloidin-TRITC, Sigma Aldrich) and HOECHST 33342 (Sigma Aldrich). Fluorescence microscopy (Axio Observer microscope, Zeiss) was performed with mounted Ti-plates on glass slides. For each plate, three representing pictures were taken at 20 fold magnification. The average cell area was determined by manually outlining each cell on one representing picture of each triplicate (using the Software Axio Vision 4.8, Zeiss). Data were analyzed from at least 3 independent experiments and presented as mean ± SEM.
Molecular dynamics of the linker
Molecular dynamics simulations were carried out using the Gromacs-4.6 package.25 The simulation system consisted of the respective linker with a C-terminal amidated and N-terminal acetylated lysine on the RGD-peptide side and a C-terminal amidated and N-terminal acetylated lysine/cysteine on the titanium binding site. The linker was parameterized with antechamber26 and acpype27 for the GAFF force field. The charges were derived with the antechamber sqm method. The simulations were carried out under periodic boundary conditions in TIP3P water with 150 mM NaCl at 300 K for 50 ns after equilibration with NVT and NPT ensemble for 50 ps each. The trajectories were analyzed with the g_dist tool by measuring the Cα–Cα distance and plotting the values in a histogram with a bin size of 0.5 Å.Results and discussion
Peptide modification with tetrazines
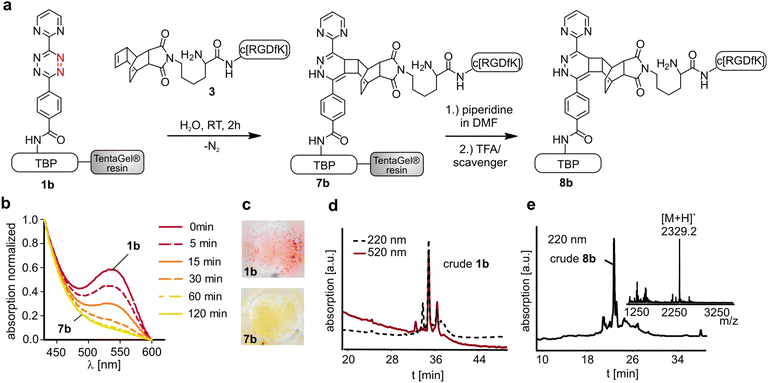
The titanium binding peptide (TBP, 1) was synthesized on a TentaGel resin to enable the DARinv on the resin in aqueous solution. The sequence was built up of spacer units such as polyethylene glycol (PEG) and β-Ala. DOPA (L-3,4-dihydroxyphenylalanine), which shows high affinity and stability to titanium and other surfaces, was used to anchor the peptide to the biomaterial.28 Functional groups were introduced by using Lys, Pra (L-propargylglycine) and Cys to allow the multifunctional modifications of TBP.29 Two dienes (2a,b) were coupled to TBP after selective side chain deprotection of Lys (Fig. 2). The result of the reaction was studied by RP-HPLC and MALDI-ToF-MS analysis (Fig. 4d). The tetrazine stability in 1a and 1b was tested in solution and on resin over 7 d in an aqueous environment and yielded low fragmentation (see the ESI†). As a side reaction, the undesired DARinv between the alkyne functionality of Pra and the applied tetrazines was observed, mainly for peptide 1a in solution. On resin, this reaction occurred only to a small extent. Cleavage of 1b from the resin resulted in fragmentation induced by water, which was slightly increased after incubation of the resin in water for 7 d.30 Nevertheless, possible degradation of tetrazines by the cleavage of the peptide from the resin is circumvented by the DARinv on resin.Peptide modification with dienophiles
Different dienophiles were introduced to the cyclic cell binding peptide c[RGDfK]. The peptide was synthesized on an acid labile chlorotrityl resin (Fig. 3). The modification with different dienophiles was either realized by the incorporation of alkene containing amino acids (4, 5) or by Lys-side chain modification (3, 6). The dienophile functionalized peptides were cleaved under mild acidic conditions to retain side chain protecting groups. Subsequently, the selective head to tail cyclization was performed in solution. Peptides were finally purified by RP-HPLC after complete deprotection by TFA. The Reppe-anhydride, which has already been used in previous studies, was coupled at the Lys-side chain of the c[RGDfK]-peptide to yield compound 3.31–33 To facilitate the introduction of dienophiles during SPPS, we used terminal alkenes, which are present in common protecting groups like Alloc and Ally. A 6-maleimidohexanoic acid modified peptide (6) was synthesized to explore the feasibility of this easily accessible dienophile for the DARinv on resin (Fig. 3).Ligation of peptides by DARinv on resin
To perform the DARinv on the solid phase, the dienophile 3 was dissolved in H2O (c = 30 mM) and added to the resin bound peptides 1a and b. A disadvantage of reactions on the solid phase is the often complicated monitoring of the reaction. However, herein we demonstrate a direct photometric measurement of the reaction process on resin. The DARinv was monitored by measuring the specific absorbance of the tetrazines at 540 nm directly on the resin by photometry (Fig. 4b and c). The cycloaddition with peptide 1b was complete after 2 h. Tetrazine 2a resulted in a slower reaction with full conversion after 5 h (see the ESI†), which can be explained by the presence of stronger electron withdrawing groups in 2b compared to 2a. Higher reactivity of tetrazines often results in lower stability.34 During the DARinv on resin with peptide 3 we could not detect the differences in diene stability. However, for reactions with longer incubation times, as for peptide 6, lower stability of tetrazine 2b compared to 2a was observed (see the ESI†). Furthermore, combining the DARinv with additional reactions as the CuAAC lead to stability problems for compound 8b originated from tetrazine 2b as well (see the ESI†). Contrarily, 8a was stable under the CuAAC on resin and was recently used to synthesize a multifunctional cell adhesive titanium coating.29 This suggests that the choice of the tetrazine can strongly influence the stability of the resulting DARinv-product as shown also for metabolic degradation.14 After completion of the reaction, the dienophile containing peptide 3, which was used in excess, was fully recovered by lyophilization. Cleavage from the resin and subsequent analysis showed the generation of the desired dihydropyridazine 8b (Fig. 4e). Notably, the DARinv-product was stable under the conditions of the Fmoc-cleavage with piperidine in DMF, which underlines the compatibility with Fmoc/tBu-based-SPPS.Conjugate 8b was partially oxidized to the corresponding pyridazine (Fig. 1). Complete oxidation can be achieved by incubation with isoamyl nitrite and acetic acid, which can be performed on resin and in solution (Fig. 5).35
Terminal alkenes were successfully applied to react with tetrazines by a DARinv.36,37 Incubation of the tetrazine containing peptide 1a and dienophile 4 or 5 respectively could show the formation of the desired DARinv product, which was however cleaved after cycloaddition (for the hypothesized mechanism see the ESI†). The reaction of these terminal alkene bearing peptides with tetrazine 1b resulted only in low conversion of around 10% as a result of low reactivity and stability of the tetrazine (see the ESI†). Alternative tetrazines could be applied to overcome this problem to establish an easy introduction of dienophiles in SPPS. Moreover, the suggested mechanism could be probably applied as a decaging method as described recently for proteins or drugs, since the active chemotherapeutic RGD-peptide is released upon fragmentation.38,39 Maleimides are classical dienophiles in normal Diels–Alder reactions.40 However, the DARinv between peptide 1a and the maleimide modified RGD (6) resulted in the stable product 8c. The reaction completed rather slowly after 7 d (Fig. 6). This reactivity towards tetrazines should be considered when using maleimide modified tetrazines or a combination of DARinv with a thiol-maleimide addition.41–43 Moreover, this finding gives rise to the potential use of maleimides as easily accessible dienophiles in the DARinv. However, alternative tetrazines should be tested to increase the reaction rate. The outcome of all the studied reactions between the different tetrazines and dienophiles is summarized in Table 2.
The DARinv on a solid support displays the following advantages. Tetrazines are often hardly soluble in aqueous solution. Therefore, the reaction has to be carried out in organic solvents or the synthesis strategy has to be changed by the introduction of solubility increasing molecules such as short PEG units.18 Performing the DARinv with a tetrazine containing peptide bound to a solid phase overcomes this problem. Besides the advantages of water as a solvent for environmental and biochemical issues it is also favorable for kinetic reasons.44 It is known that water can increase the proximity of hydrophobic molecules like the used diene and dienophiles.31 Hence the reaction can be accelerated.45 Additionally, the water-swellable TentaGel resin facilitates the synthesis of rather long or branched peptides compared to the synthesis carried out on non-PEG-based resins. Furthermore, cleavage and purification steps are reduced since the crude resin bound peptides can be directly used for the ligation. This is especially beneficial for peptides that consist of rather cost intensive amino acids such as DOPA. DARinv-reagents as the dienophile carrying RGD peptide are easily recovered by lyophilization and can be thus used in excess without loss of yield owing to purification steps.
DARinv-linkers as spacers to improve cell adhesion
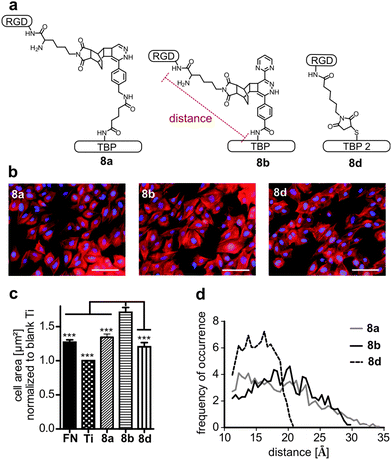
RGD-mediated cell adhesion is strongly dependent on the distance between the integrin ligand and the corresponding surface. Various spacers as polyglycine, polyproline, aminohexanoic acid or PEG have been used so far.46,47 Using bulky dienes and dienophiles circumvents the introduction of additional spacer moieties. Hence we applied the DARinv with rather large dienes and dienophiles to efficiently introduce a distinct spacer. To test the influence of the varying lengths of tetrazines 2a and 2b, cell adhesion studies with the peptide conjugates 8a and 8b were performed. Furthermore, peptide 8d, synthesized by maleimide–thiol Michael addition, was tested to compare the tested DARinv-linker with a shorter spacer (Fig. 7a). Negative controls, containing RAD were synthesized to proof the RGD-mediated cell adhesion (see the ESI†). The increased cell size is an indicator of successful cell attachment and is dependent on the RGD-surface distance.47 Therefore, cell spreading was determined by measuring the average cell area on coated Ti. Interestingly, conjugate 8b showed the significantly highest average cell size compared to all the tested coatings, although its spacer is shorter than that of peptide 8a (Fig. 7b). By MD-simulation the maximum length (from Cα of Lys in RGD to Cα of Lys in TBP and respectively to Cα of Cys in TBP 2) as well as the most likely present spacer distance were calculated. These studies revealed that the longest spacer is in fact present in 8a with 34 Å. However, this linker is more prone to collapse and interacts intramolecularly, thus it only reaches a distance of around 12 Å for the most stable conformations (Table 3, see also the ESI†). Peptide 8b consists of a spacer with a maximum length of 30 Å. The mostly occurring distance is 20 Å, thus cell spreading is more advanced on 8b than on 8a. The linker of 8d was, as anticipated, the shortest with a maximum length of 21 Å and the most stable conformations showed a distance of 16 Å. Although the most likely distance of 8d is higher than that of 8a, cell spreading is more pronounced on 8a. This can be explained by the higher average length of 19 Å for 8a compared to 16 Å for 8d (Fig. 7d and Table 3).| Maximum distance [Å] | Most frequent distance [Å] | Average distance [Å] | |
|---|---|---|---|
| 8a | 34 | 12 | 19 |
| 8b | 30 | 20 | 19 |
| 8d | 21 | 16 | 15 |
Previous studies already have suggested that a longer, more rigid and extended spacer (e.g. polyproline) favors RGD-mediated cell adhesion, however until now this has not been assessed by MD-simulation.46 In addition, the herein described MD-simulations support the assumption that too long spacers impair cell adhesion because of their pronounced flexibility.46,48 Nevertheless, a long and flexible linker should be favored over a too short spacer as their average length may be higher. These results highlight that estimating the distance of a RGD peptide to its surface only by the extended conformation of a linker molecule is not sufficient. It is rather suggested that the three-dimensional length is evaluated. We suggest that a suitable linker consists of two short flexible ends, connected by a long rigid structure (see the ESI†). The flexible parts in the spacer may be important to freely orientate the RGD-peptide on the surface towards the cell membrane.
Conclusion
The Diels–Alder reaction with inverse electron demand (DARinv) could be successfully employed to ligate peptides on a solid support. This method describes an efficient strategy to complex peptide conjugates without using catalysts or additive reagents and it is easily monitored. Low solubility as well as yield loss and potential fragmentation through cleaving tetrazine modified peptides from the resin are thereby circumvented. Furthermore, maleimide was found to be an easily accessible dienophile alternative for DARinv ligations on resin. Large DARinv-reagents could be applied to generate a spacer that might be useful for several biochemical applications. MD-simulations gave insights into the theoretical conformations of different DARinv-linkers, thereby providing crucial information for the choice of a distinct diene–dienophile pair. Thereby, tetrazine 2b in the DARinv product 8b resulted in the most enhanced cell adhesion because of its less flexible structure. With the here presented DARinv on resin the toolbox of orthogonal reactions for SPPS is broadened.Acknowledgements
We are thankful for the financial support from the DFG (TRR67, A4), the European Union and the Free State of Saxony as well as of the Graduate School BuildMoNa. For technical support in cell culture Kristin Löbner is acknowledged. The authors would like to thank Regina Reppich-Sacher for excellent assistance in mass spectrometry analysis. Ronny Müller and Christina Dammann are acknowledged for help in peptide synthesis.References
- A. Isidro-Llobet, M. Alvarez and F. Albericio, Chem. Rev., 2009, 109, 2455–2504 CrossRef CAS PubMed.
- D. Böhme and A. G. Beck-Sickinger, J. Pept. Sci., 2015, 21, 186–200 CrossRef PubMed.
- V. M. Ahrens, R. Frank, S. Boehnke, C. L. Schutz, G. Hampel, D. S. Iffland, N. H. Bings, E. Hey-Hawkins and A. G. Beck-Sickinger, ChemMedChem, 2015, 10, 164–172 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS.
- D. Schumacher and C. P. R. Hackenberger, Curr. Opin. Chem. Biol., 2014, 22, 62–69 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- S. Hofmann, S. Maschauer, T. Kuwert, A. G. Beck-Sickinger and O. Prante, Mol. Pharmaceutics, 2015, 12, 1121–1130 CrossRef CAS PubMed.
- W. Tang and M. L. Becker, Chem. Soc. Rev., 2014, 43, 7013–7039 RSC.
- C. W. Tornoe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef CAS PubMed.
- H. O. Ham, S. H. Park, J. W. Kurutz, I. G. Szleifer and P. B. Messersmith, J. Am. Chem. Soc., 2013, 135, 13015–13022 CrossRef CAS PubMed.
- I. E. Decostaire, D. Lelievre, V. Aucagne and A. F. Delmas, Org. Biomol. Chem., 2014, 12, 5536–5543 CAS.
- J. Bertran-Vicente, M. Schumann, P. Schmieder, E. Krause and C. P. Hackenberger, Org. Biomol. Chem., 2015, 13, 6839–6843 CAS.
- R. Pipkorn, W. Waldeck, B. Didinger, M. Koch, G. Mueller, M. Wiessler and K. Braun, J. Pept. Sci., 2009, 15, 235–241 CrossRef CAS PubMed.
- R. Selvaraj, B. Giglio, S. Liu, H. Wang, M. Wang, H. Yuan, S. R. Chintala, L. P. Yap, P. S. Conti, J. M. Fox and Z. Li, Bioconjugate Chem., 2015, 26, 435–442 CrossRef CAS PubMed.
- R. Hassert, M. Pagel, Z. Ming, T. Haupl, B. Abel, K. Braun, M. Wiessler and A. G. Beck-Sickinger, Bioconjugate Chem., 2012, 23, 2129–2137 CrossRef CAS PubMed.
- K. Braun, M. Wiessler, R. Pipkorn, V. Ehemann, T. Bauerle, H. Fleischhacker, G. Muller, P. Lorenz and W. Waldeck, Int. J. Med. Sci., 2010, 7, 326–339 CrossRef CAS PubMed.
- B. M. Zeglis, F. Emmetiere, N. Pillarsetty, R. Weissleder, J. S. Lewis and T. Reiner, ChemistryOpen, 2014, 3, 48–53 CrossRef CAS PubMed.
- A. Niederwieser, A. K. Spate, L. D. Nguyen, C. Jungst, W. Reutter and V. Wittmann, Angew. Chem., Int. Ed., 2013, 52, 4265–4268 CrossRef CAS PubMed.
- M. Kantlehner, P. Schaffner, D. Finsinger, J. Meyer, A. Jonczyk, B. Diefenbach, B. Nies, G. Holzemann, S. L. Goodman and H. Kessler, ChemBioChem, 2000, 1, 107–114 CrossRef CAS PubMed.
- D. M. Lewallen, D. Siler and S. S. Iyer, ChemBioChem, 2009, 10, 1486–1489 CrossRef CAS PubMed.
- A. S. Ham, A. L. Klibanov and M. B. Lawrence, Langmuir, 2009, 25, 10038–10044 CrossRef CAS PubMed.
- G. Vidal, T. Blanchi, A. J. Mieszawska, R. Calabrese, C. Rossi, P. Vigneron, J. L. Duval, D. L. Kaplan and C. Egles, Acta Biomater., 2013, 9, 4935–4943 CrossRef CAS PubMed.
- J. Auernheimer, D. Zukowski, C. Dahmen, M. Kantlehner, A. Enderle, S. L. Goodman and H. Kessler, ChemBioChem, 2005, 6, 2034–2040 CrossRef CAS PubMed.
- J. H. Beer, K. T. Springer and B. S. Coller, Blood, 1992, 79, 117–128 CAS.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- J. M. Wang, W. Wang, P. A. Kollman and D. A. Case, J. Mol. Graphics Modell., 2006, 25, 247–260 CrossRef CAS PubMed.
- A. Sousa da Silva and W. Vranken, BMC Res. Notes, 2012, 5, 1–8 Search PubMed.
- H. Lee, N. F. Scherer and P. B. Messersmith, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12999–13003 CrossRef CAS PubMed.
- M. Pagel, R. Hassert, T. John, K. Braun, M. Wiessler, B. Abel and A. G. Beck-Sickinger, Angew. Chem., Int. Ed., 2016, 55, 4826–4830 CrossRef CAS PubMed.
- R. Hoogenboom, B. C. Moore and U. S. Schubert, J. Org. Chem., 2006, 71, 4903–4909 CrossRef CAS PubMed.
- S. Horner, C. Uth, O. Avrutina, H. Frauendorf, M. Wiessler and H. Kolmar, Chem. Commun., 2015, 51, 11130–11133 RSC.
- K. Braun, M. Wiessler, V. Ehemann, R. Pipkorn, H. Spring, J. Debus, B. Didinger, M. Koch, G. Muller and W. Waldeck, Drug Des., Dev. Ther., 2009, 2, 289–301 CrossRef.
- M. Wiessler, W. Waldeck, C. Kliem, R. Pipkorn and K. Braun, Int. J. Med. Sci., 2010, 7, 19–28 CrossRef CAS.
- M. R. Karver, R. Weissleder and S. A. Hilderbrand, Bioconjugate Chem., 2011, 22, 2263–2270 CrossRef CAS PubMed.
- H. S. Beckmann, A. Niederwieser, M. Wiessler and V. Wittmann, Chemistry, 2012, 18, 6548–6554 CrossRef CAS PubMed.
- A. K. Späte, V. F. Schart, S. Schöllkopf, A. Niederwieser and V. Wittmann, Chemistry, 2014, 20, 16502–16508 CrossRef PubMed.
- Y. J. Lee, Y. Kurra, Y. Yang, J. Torres-Kolbus, A. Deiters and W. R. Liu, Chem. Commun., 2014, 50, 13085–13088 RSC.
- R. M. Versteegen, R. Rossin, W. ten Hoeve, H. M. Janssen and M. S. Robillard, Angew. Chem., Int. Ed., 2013, 52, 14112–14116 CrossRef CAS PubMed.
- J. Li, S. Jia and P. R. Chen, Nat. Chem. Biol., 2014, 10, 1003–1005 CrossRef CAS PubMed.
- A. D. de Araujo, J. M. Palomo, J. Cramer, O. Seitz, K. Alexandrov and H. Waldmann, Chemistry, 2006, 12, 6095–6109 CrossRef PubMed.
- Z. Wu, S. Liu, M. Hassink, I. Nair, R. Park, L. Li, I. Todorov, J. M. Fox, Z. Li, J. E. Shively, P. S. Conti and F. Kandeel, J. Nucl. Med., 2013, 54, 244–251 CrossRef CAS PubMed.
- S. Liu, M. Hassink, R. Selvaraj, L. P. Yap, R. Park, H. Wang, X. Chen, J. M. Fox, Z. Li and P. S. Conti, Mol. Imaging, 2013, 12, 121–128 CAS.
- J. D. Thomas, H. Cui, P. J. North, T. Hofer, C. Rader and T. R. Burke Jr., Bioconjugate Chem., 2012, 23, 2007–2013 CrossRef CAS PubMed.
- G. K. van Der Wel, J. W. Wijnen and J. B. Engberts, J. Org. Chem., 1996, 61, 9001–9005 CrossRef CAS PubMed.
- G. Graziano, J. Phys. Org. Chem., 2004, 17, 100–101 CrossRef CAS.
- D. Pallarola, A. Bochen, H. Boehm, F. Rechenmacher, T. R. Sobahi, J. P. Spatz and H. Kessler, Adv. Funct. Mater., 2014, 24, 943–956 CrossRef CAS PubMed.
- J. W. Lee, Y. J. Park, S. J. Lee, S. K. Lee and K. Y. Lee, Biomaterials, 2010, 31, 5545–5551 CrossRef CAS PubMed.
- U. Hersel, C. Dahmen and H. Kessler, Biomaterials, 2003, 24, 4385–4415 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ob00314a |
| This journal is © The Royal Society of Chemistry 2016 |