Computational identification and analysis of signaling subnetworks with distinct functional roles in the regulation of TNF production†‡
Abstract
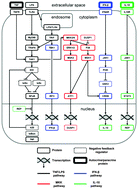
Inflammation is a complex process driven by the coordinated action of a vast number of pro- and anti-inflammatory molecular mediators. While experimental studies have provided an abundance of information about the properties and mechanisms of action of individual mediators, essential system-level regulatory patterns that determine the time-course of inflammation are not sufficiently understood. In particular, it is not known how the contributions from distinct signaling pathways involved in cytokine regulation combine to shape the overall inflammatory response over different time scales. We investigated the kinetics of the intra- and extracellular signaling network controlling the production of the essential pro-inflammatory cytokine, tumor necrosis factor (TNF), and its anti-inflammatory counterpart, interleukin 10 (IL-10), in a macrophage culture. To tackle the intrinsic complexity of the network, we employed a computational modeling approach using the available literature data about specific molecular interactions. Our computational model successfully captured experimentally observed short- and long-term kinetics of key inflammatory mediators. Subsequent model analysis showed that distinct subnetworks regulate IL-10 production by impacting different temporal phases of the cAMP response element-binding protein (CREB) phosphorylation. Moreover, the model revealed that functionally similar inhibitory control circuits regulate the early and late activation phases of nuclear factor κB and CREB. Finally, we identified and investigated distinct signaling subnetworks that independently control the peak height and tail height of the TNF temporal trajectories. The knowledge of such subnetwork-specific regulatory effects may facilitate therapeutic interventions aimed at precise modulation of the inflammatory response.

 Please wait while we load your content...
Please wait while we load your content...