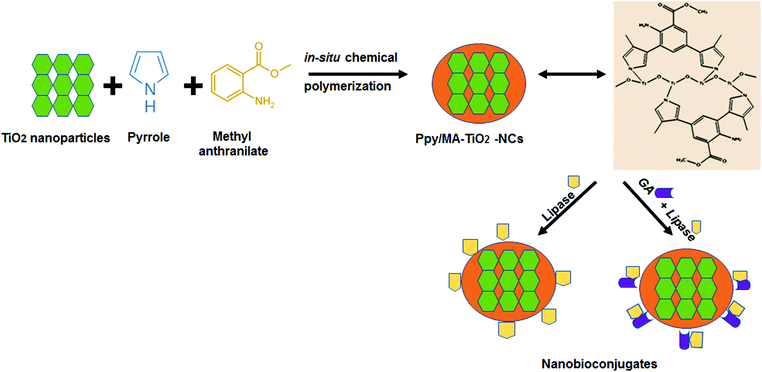
A polypyrrole–methyl anthranilate functionalized worm-like titanium dioxide nanocomposite as an innovative tool for immobilization of lipase: preparation, activity, stability and molecular docking investigations†
Shamoon
Asmat
a,
Qayyum
Husain
 *a and
Mohd Shoeb
Khan
b
*a and
Mohd Shoeb
Khan
b
aDepartment of Biochemistry, Faculty of Life Sciences, Aligarh Muslim University, Aligarh-202002, India. E-mail: qayyumbiochem@gmail.com; Tel: +91-9897701792
bDepartment of Chemistry, Aligarh Muslim University, Aligarh-202002, India
First published on 13th November 2017
Abstract
Enzymatic biocatalysis has a vast reputation on an industrial scale. The major hindrance faced by enzymes is their denaturation under various conditions. In this study, we report that the activity and stability of enzymes can be enhanced by their immobilization on a novel synthesized polypyrrole–methyl anthranilate-titanium oxide nanocomposite containing amine groups. We demonstrated this by successfully immobilizing lipase from Rhizopus oryzae on a Ppy–MA/TiO2 NC by physical adsorption and a glutaraldehyde-activated covalent coupling procedure. The catalytic efficiencies of the free and immobilized preparations were determined for the hydrolysis of p-nitrophenyl palmitate. The covalently immobilized lipase displayed a significantly higher activity yield (effectiveness factor of 0.97) in comparison with the adsorbed counterpart (effectiveness factor of 0.86). The binding of lipase to the Ppy–MA/TiO2 NC was confirmed by transmission electron microscopy, Fourier transform infrared spectroscopy and scanning electron microscopy. TGA and DTA were performed to investigate the thermal stability of the synthesised biocatalysts. The storage stability, solvent tolerance, and reusability of the resulting nanobiocatalyst and the effect of pH and temperature on its catalytic activities were also investigated. The prepared nanobiocatalysts displayed remarkably improved activity in terms of solvent tolerance (activity recovery of 150% and 125% in acetone and isopropanol, respectively) in comparison with its free counterpart. We envisage that the covalent binding method played a profound role in enhancing the properties of the enzyme immobilized on the Ppy–MA/TiO2 NC. In view of the abovementioned results, a hydrolytic enzyme immobilized on this functionalized TiO2 nanocarrier can be used as a stable and effective nanobiocatalyst with vital implications for industrial biotechnology.
1. Introduction
Enzymes are potent biomolecules that are extensively used in numerous fields such as biotechnology, biomedical sciences, biochemical engineering, bioinformatics, biochemistry and microbiology owing to their excellent catalytic efficiency.1 In particular, lipase (E.C. 3.1.1.3), which is a protein of the hydrolase class, has attracted substantial attention in the dairy, flavour, food, pharmaceuticals and leather industries as a versatile biological catalyst for organic synthesis in mild reaction conditions in various media.2–7 For these applications of lipase, loss of enzymatic activity during its use is frequently observed, which occasionally hinders its industrial applications. In addition, its activity is influenced by temperature and the chemical environment such as the pH, metal ions and organic solvents. Moreover, potential biocatalytic applications of the soluble form of lipase have been greatly hindered by subtle proteomic features, which lead to poor activity, solubility and stability in several organic media.Immobilization is frequently used as an effective means of improving the catalytic properties and stability of enzymes.8–10 As promising and versatile ligands, NCs with a narrow pore size distribution, large surface area, and high mechanical and thermal stability have gained increasing importance in industrial applications for enzyme immobilization, which is largely due to their reproducible and simple preparation protocol.11,12 Moreover, the binding of biomolecules to nanosized supports results in the formation of interactions that may improve the rigidity and stabilize the tertiary and quaternary structures of enzymes.13 NCs are now used in conjugation with biological materials, such as enzymes, proteins, and nucleic acids, as they result in the effortless recovery of a broad range of these nanomaterials from reaction systems, which thereby reduces the operational costs of the processes. However, to meet the needs of practical applications, a novel, versatile, low-cost and especially environmentally friendly approach for obtaining nanobiocatalysts is still desired.
Lately, inorganic NCs based on organic polymers have gained importance because of their much broader range of applications, in particular in sensor devices, biomimics and biomedical substances. Among various polymers, polypyrrole (PPy) has been extensively employed for enzyme immobilization owing to its biocompatibility, high electrical conductivity, and facile and cost-effective synthesis in comparison with other polymers.14,15 However, after polymerization it is quite tedious to process as it forms an infusible and insoluble mass, which thus restricts its applicability. Hence, it is desirable to copolymerize pyrrole with other polymers in order to improve its processability without impairing its stability, conductivity and mechanical strength. Recently, the exploitation of titanium-based NPs has become a versatile technique for creating fascinating supports for enzyme stabilization owing to their small size and large surface area.16–18 Titanium-based nanosized materials explicitly influence the thermal and mechanical properties of substances such as their flexibility and firmness and create a biocompatible environment for high-yield enzyme immobilization.19 Nevertheless, to the best of our knowledge, titania modified with a copolymer of pyrrole and methyl anthranilate has never been utilised as a matrix for enzyme immobilization. Continuing efforts are essential for research on the immobilization of enzymes on this new support.
In this study, novel copolymer of pyrrole and methyl anthranilate was synthesised in collaboration with stable TiO2 NPs for its possible application in Rhizopus oryzae lipase (ROL) immobilization. Owing to the beneficial properties of titania nanomaterials and polypyrrole–methyl anthranilate, NCs comprising titanium-based nanomaterials with this copolymer are expected to further enhance the stability and catalytic activity of immobilized lipase. Synergistic interactions between the polymers and inorganic substances endow the polymer NCs with enhanced strength and stiffness and exceptional conductivity. The resulting novel hybrid NC was characterized by X-ray diffraction (XRD) and Fourier transform infrared spectroscopy (FT-IR) in order to determine the binding between the support and the enzyme. The surface morphology and size distribution of the facilely synthesised Ppy–MA/TiO2 NC and the immobilized formulation were characterized by scanning electron microscopy (SEM) and transmission electron microscopy (TEM). The thermal behaviour of the prepared nanoconjugate was examined by differential thermal analysis (DTA) and thermogravimetric analysis (TGA), which demonstrated enhanced thermal stability. The immobilized enzyme exhibited significantly enhanced activity in the presence of organic solvents in comparison with its non-immobilized counterpart and maintained 90% of its initial activity even after 10 repeated uses. Furthermore, the biocatalysts exhibited enhanced stability in the pH range from 8 to 10 and in the temperature range from 30 to 65 °C, which are vital features that open routes for industrial uses of this biocatalytic system. Therefore, the immobilization of lipase is necessary not only for increasing the stability and activity of enzymes but also for the easy recovery and repeated use of enzymes.
2. Materials and methods
2.1 Chemicals and reagents
Pyrrole, methyl anthranilate, glutaraldehyde 25% (v/v) and p-nitrophenyl palmitate (p-NPP) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ppy–MA was stored under argon in the dark and was purified by vacuum distillation. H2SO4, iron sulfate heptahydrate, and H2O2 (30% v/v) were procured from Merck (USA). Crystalline TiO2 was acquired from MK Nano, Canada, and was used as received. Lipase (Rhizopus oryzae EC 3.1.1.3) was obtained from Sisco Research Laboratories Chemicals (Mumbai, India). All the reagents and chemicals used in this study were of analytical grade.2.2 Synthesis of Ppy–MA/TiO2 copolymer NC via chemical oxidative polymerization
In an exemplary experimental procedure for the synthesis of the NC, a chemical oxidative polymerization method was utilized for the copolymerization of Ppy–MA. The procedure involved the copolymerization of pyrrole and methyl anthranilate in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and was carried out as follows. Methyl anthranilate (6.5 mL, 25 mmol) was added to a dilute solution of H2SO4 (5.2 mL, 50 mmol), followed by the addition of pyrrole (3.25 mL, 25 mmol) with continuous stirring. Then, iron sulfate heptahydrate (278 mg, 0.5 mmol) was added to the reaction mixture with vigorous stirring. At the same time, 10% (w/w) TiO2 NPs and 30% H2O2 (6.95 mL, 68 mmol) were slowly added to the reaction mixture over a period of 5 h. The solution was continuously stirred for a further 8 h under air at room temperature to cause the precipitation of a black solid. The reaction mixture was quenched with crushed ice, and the solid product was collected on a glass filter, washed with ethanol, and dried in a dynamic vacuum at room temperature to give a black powdery NC of the copolymer.
1 and was carried out as follows. Methyl anthranilate (6.5 mL, 25 mmol) was added to a dilute solution of H2SO4 (5.2 mL, 50 mmol), followed by the addition of pyrrole (3.25 mL, 25 mmol) with continuous stirring. Then, iron sulfate heptahydrate (278 mg, 0.5 mmol) was added to the reaction mixture with vigorous stirring. At the same time, 10% (w/w) TiO2 NPs and 30% H2O2 (6.95 mL, 68 mmol) were slowly added to the reaction mixture over a period of 5 h. The solution was continuously stirred for a further 8 h under air at room temperature to cause the precipitation of a black solid. The reaction mixture was quenched with crushed ice, and the solid product was collected on a glass filter, washed with ethanol, and dried in a dynamic vacuum at room temperature to give a black powdery NC of the copolymer.
2.3 Immobilization procedure of lipase onto Ppy–MA/TiO2 NC
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 30 min. The mixture was subjected to magnetic stirring at 4 °C overnight to obtain the maximum amount of precipitate. The obtained precipitate was dissolved in 50 mM Tris–HCl buffer with a pH of 8 and was dialyzed against an assay buffer. The enzyme solution thus obtained was used for immobilization studies. A 5 mL sample of a 2 mg mL−1 lipase solution was prepared, and the resulting solution was added to 5 mL of 10 mg mL−1 activated Ppy–MA/TiO2 NC in 0.1 M Tris–HCl buffer with a pH of 8. After the solution was continuously stirred at room temperature for 24 h, an immobilized nanoconjugate was isolated. The solution was washed three times with 0.05 M Tris–HCl buffer with a pH of 8. The NC was obtained via centrifugation at 3000 rpm for 6 min and resuspended in 0.05 M phosphate buffer with a pH of 7.2.
000 rpm at 4 °C for 30 min. The mixture was subjected to magnetic stirring at 4 °C overnight to obtain the maximum amount of precipitate. The obtained precipitate was dissolved in 50 mM Tris–HCl buffer with a pH of 8 and was dialyzed against an assay buffer. The enzyme solution thus obtained was used for immobilization studies. A 5 mL sample of a 2 mg mL−1 lipase solution was prepared, and the resulting solution was added to 5 mL of 10 mg mL−1 activated Ppy–MA/TiO2 NC in 0.1 M Tris–HCl buffer with a pH of 8. After the solution was continuously stirred at room temperature for 24 h, an immobilized nanoconjugate was isolated. The solution was washed three times with 0.05 M Tris–HCl buffer with a pH of 8. The NC was obtained via centrifugation at 3000 rpm for 6 min and resuspended in 0.05 M phosphate buffer with a pH of 7.2.
2.4 Assay of enzymatic activity
A JASCO UV/vis spectrophotometer was used to determine the hydrolytic activity of the lipase by measuring the absorbance at 410 nm produced by the release of p-nitrophenol. The substrate chosen for the determination of lipase activity was p-NPP. The substrate solution was prepared with 300 mg Triton X-100, p-NPP (150 mM in isopropanol) and 135 mL 25 mM Tris–HCl buffer with a pH of 7.0. The reaction mixture contained 0.15 mL lipase solution and 3 mL substrate solution. The reaction was carried out by incubating the reaction mixture at 37 °C for 15 min in a water bath under continuous stirring, and thereafter 0.25 mL 0.1 M Na2CO3 was introduced to terminate the reaction. The activity yield (%) was the ratio between the activity of immobilized lipase and the activity of the same amount of free lipase in solution as that immobilized onto the Ppy–MA/TiO2 NC. One unit of lipase activity (U) is defined as the amount of enzyme that causes the hydrolysis of 1 μmol of p-nitrophenol from p-NPP per min under optimal experimental conditions.The immobilization efficiency was estimated in terms of the enzyme activity yield expressed as a percentage as follows (Table S1, ESI†):
| Activity yield (%) = B/A × 100% |
| Loading efficiency (%) = (CoVo − CiVi)/CoVo × 100% |
B is the enzymatic activity recorded in the nanoconjugate, and A is the difference between the enzymatic activity of the loaded enzyme and that observed in the wash solution. The effectiveness factor (η) was estimated using the ratio of theoretically bound ROL to actually bound ROL.20
2.5 Characterization of synthesised nanobiocatalyst
2.6 Effect of temperature on the activity of free and immobilized lipase preparations
The optimum temperature for the free and immobilized lipase preparations was investigated in the range of 20–70 °C in 20 mM phosphate buffer (pH 7) by means of an assay of hydrolytic activity. The activity at 40 °C was used as a control (100%) for the determination of the remaining (%) activity of free and immobilized enzyme formulations.2.7 Effect of pH on the activity of free and immobilized lipase preparations
The effect of pH on the activity of the free enzyme and immobilized lipase preparations was investigated in buffers with a pH in the range from 4 to 10 via an assay of hydrolytic activity. The enzymatic activity at a pH of 8 and 9 was used as a reference (100%) for the calculation of the remaining (%) activity of free and immobilized enzyme preparations, respectively. The concentration of the p-NPP solution in each case was kept constant under the assay conditions.2.8 Determination of kinetic parameters
Changes in the kinetic parameters of the hydrolysis of p-NPP involving free and immobilized lipase preparations were calculated. For the determination of the kinetic parameters for the hydrolytic activity of the enzyme preparations, a fixed amount of the enzyme was reacted with varying concentrations of p-NPP (2 to 15 mM). The values of the Michaelis–Menten parameters Km and Vmax were determined from a Lineweaver–Burk plot for the corresponding pH and temperature.2.9 Assay of thermal stability
Studies of thermal stability were performed by the pre-incubation of free and immobilized lipase preparations in phosphate buffer (50 mM, pH 7.0) at 60 °C for 2 h. Aliquots of each preparation were taken at indicated times and chilled quickly in ice for 5 min, followed by the measurement of the residual enzymatic activity at 30 °C as the samples were brought to room temperature. The activity achieved without incubation at 60 °C was used as a control (100%) for the estimation of the residual percentage activity.2.10 Storage stability
Free and immobilized lipase formulations were incubated in sodium phosphate buffer (0.05 M, pH 7.0) at 30 °C for a period of 45 days. Aliquots from each preparation (2 mL) were taken in triplicate at intervals of 5 days and were then analysed for residual enzymatic activity in the hydrolysis of p-NPP. The activity determined on the first day was used as a control (100%) for the calculation of the remaining percentage activity.2.11 Analysis of reusability
To investigate the reusability of adsorbed and covalently immobilized lipase, experiments were performed in triplicate under standard assay conditions on immobilized enzyme preparations. Each reaction was terminated after 15 min. At the end of each cycle, the immobilized lipase was separated, washed with phosphate buffer with a pH of 7.0, stored at 4 °C and used again for 10 successive days. The nanobiocatalysts were recovered by centrifugation at 5000 rpm for 15 min and were resuspended in the assay buffer. The activity of immobilized lipase on the first day was used as a control (100%) to calculate the remaining percentage activity after each use.2.12 Solvent tolerance
The nanobiocatalysts were filtered, and the pellets were washed with double-distilled water three times and subsequently with ethanol. They were then air-dried by incubation at 45 °C for 8 h. Free lipase and the immobilized preparation were incubated in 0.1 mL of various solvents, namely, acetonitrile, acetone, isopropanol, butanol, nitroaniline, toluene and chlorobenzene, at 30 °C for 1 h. The residual enzymatic activity after 1 h with respect to the lipase activity at time zero, which was considered to be 100%, was estimated. The reference model for the effect of organic solvents comprised the bioconjugate incubated in 50 mM sodium phosphate buffer.2.13 In silico screening
Molecular docking studies of the Ppy–MA/TiO2 nanocomposite were performed using MMV software. The retrieved protein associated with Rhizopus lipase was imported, arranged and improved by this software. This software was also utilized to determine its flexibility for achieving a durable potential binding site with a receptor. An investigation of the repeating units in the Ppy–MA/TiO2 nanocomposite was performed using ChemDraw Ultra3D software. The structure of the nanocomposite was actively minimized with the aid of an MM2 force field with an RMS gradient of 0.0001. The energy profiles of ligand–receptor interactions were assessed using GEMDOCK, and the programs mentioned above were utilized to achieve molecular docking and visualization of the docked poses.2.14 Protein estimation
The concentration of proteins was estimated according to the standard method described by Lowry et al.212.15 Statistical analysis
The data obtained from various studies were plotted using Origin 8 and expressed with the standard deviation of error. Each value represents the mean value for experiments performed in triplicate, and statistical analysis was performed for comparison between free and immobilized lipase. Values of p of ≤0.05 were considered to be statistically significant.3. Results and discussion
3.1 Lipase immobilization on Ppy–MA/TiO2 NC
The Ppy–MA/TiO2 NC was effectively synthesized by the copolymerization of pyrrole and methyl anthranilate via a facile in situ chemical oxidation procedure in the presence of TiO2 (Scheme 1).The product, namely, Ppy–MA/TiO2 NC, that was synthesised in this work was utilized as a support for the adsorption and covalent immobilization of lipase. The plentiful surface amino groups present on the Ppy–MA/TiO2 composite were used as a base for its surface modification. The immobilization of lipase was achieved via the use of glutaraldehyde, which is a versatile crosslinking agent that has been shown to be efficient for the multipoint covalent attachment of enzymes [Scheme 1]. The loading efficiencies of physically adsorbed and covalently modified lipase bound to the nanosupport were found to be 79.5% and 87.2%, respectively, as estimated from the equation given in Section 2.4. Multipoint attachment is of great significance for obtaining the complete benefit of immobilization and enhancing the stability of the enzyme.22 The physically adsorbed enzyme preparation exhibited an activity yield of 86%, whereas the covalently modified enzyme formulation exhibited a remarkably high activity yield of 97%.
3.2 Characterization of the synthesised nanobiocatalysts
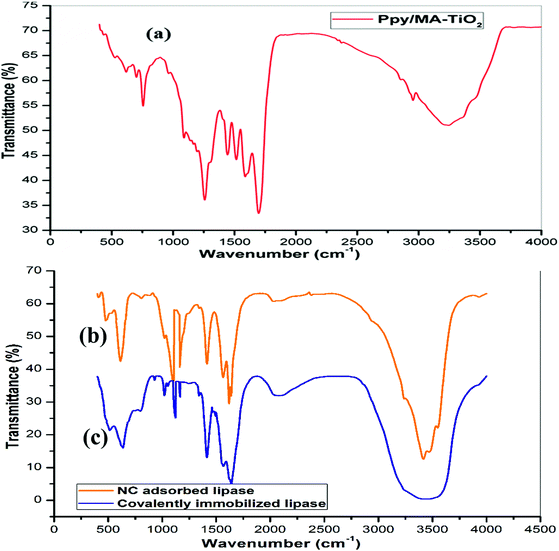
To determine the presence of characteristic functional groups and reveal the chemical structure of the compound, FT-IR analysis was performed. The FT-IR spectra of the Ppy–MA–TiO2 NC, NC-adsorbed lipase and covalent immobilized enzyme preparation are shown in Fig. 1. The characteristic bands at 526 and 1441 cm−1 relate to the stretching vibrations of pyrrole rings. The characteristic bands at 1564 and 1473 cm−1 are due to the antisymmetric and symmetric ring stretching modes of Ppy, respectively.23 The characteristic bands at 792, 1281, 1193, and 1087 cm−1 in the spectrum of Ppy–MA were assigned to in-plane and out-of-plane vibrations of![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H groups and confirm a noticeable increase in charge delocalization on the polymeric backbone as an aftereffect of interfacial interactions with the Ppy–MA matrix and TiO2 nanoparticles (Fig. 1a). The FT-IR spectrum of NC-adsorbed lipase (Fig. 1b) displays characteristic peaks at wavenumbers of 1084 cm−1 (C–O bond stretching vibrations), 1650 cm−1 (N–H bending vibrations), and 2922 cm−1 (C–H stretching vibrations) and in the range of 3422–3148 cm−1 (–OH stretching vibrations and –NH stretching vibrations). Additional peaks observed in the range of 500–1500 cm−1 are characteristic of ring stretching and in-plane deformation of C–H groups. The FT-IR spectra of both immobilized lipase preparations confirmed the presence of a number of functional groups in the structure of the enzyme, which were predominantly involved in the binding of the enzyme to the Ppy–MA/TiO2 NC.24 The stretching of carbonyl groups in lipase was detected for both types of immobilized formulation by a broadening of peaks in the range from 3345 cm−1 to 3650 cm−1. There was a considerable increase in the intensity of peaks at 3483, 2922, 1652, and 650 cm−1, which confirms the effective immobilization of lipase.25 However, the peak intensity declined in the case of covalently immobilized lipase (Fig. 1c), which confirmed that the interaction between the enzyme and nanosupport was stable.
C–H groups and confirm a noticeable increase in charge delocalization on the polymeric backbone as an aftereffect of interfacial interactions with the Ppy–MA matrix and TiO2 nanoparticles (Fig. 1a). The FT-IR spectrum of NC-adsorbed lipase (Fig. 1b) displays characteristic peaks at wavenumbers of 1084 cm−1 (C–O bond stretching vibrations), 1650 cm−1 (N–H bending vibrations), and 2922 cm−1 (C–H stretching vibrations) and in the range of 3422–3148 cm−1 (–OH stretching vibrations and –NH stretching vibrations). Additional peaks observed in the range of 500–1500 cm−1 are characteristic of ring stretching and in-plane deformation of C–H groups. The FT-IR spectra of both immobilized lipase preparations confirmed the presence of a number of functional groups in the structure of the enzyme, which were predominantly involved in the binding of the enzyme to the Ppy–MA/TiO2 NC.24 The stretching of carbonyl groups in lipase was detected for both types of immobilized formulation by a broadening of peaks in the range from 3345 cm−1 to 3650 cm−1. There was a considerable increase in the intensity of peaks at 3483, 2922, 1652, and 650 cm−1, which confirms the effective immobilization of lipase.25 However, the peak intensity declined in the case of covalently immobilized lipase (Fig. 1c), which confirmed that the interaction between the enzyme and nanosupport was stable.
To confirm the successful amalgamation of the organic polymer on the TiO2 nanoparticles, comparative X-ray diffraction patterns of Ppy–MA and the Ppy–MA/TiO2 NC were recorded. The X-ray diffractograms of Ppy–MA and the Ppy–MA/TiO2 NC noticeably display the main distinctive peaks of Ppy in the diffractogram of the Ppy–MA/TiO2 NC, which indicates the effective enclosure of TiO2 nanoparticles by the in situ polymerization of Ppy–MA (Fig. 2). The characteristic peaks of Ppy–MA that were observed at 2θ values of 25.692°, 43.930°, and 64.280° are in accordance with previously reported X-ray diffractograms of polypyrrole.26,27 The X-ray diffractograms of Ppy–MA and the Ppy–MA/TiO2 NC visibly show the main characteristic peaks of Ppy in the diffractogram of the Ppy–MA/TiO2 NC, which indicates the successful enclosure of TiO2 NPs via the in situ polymerization of Ppy–MA. To investigate the elemental distribution and structure of the NC, EDX elemental mapping was employed.28Fig. 2c illustrates the occurrence of each element in the Ppy/MA/TiO2 NC as determined by EDAX. As an illustrative example, the EDX spectrum and elemental mappings of C, O, N, and Ti were recorded for a sample of the Ppy/MA/TiO2 NC (Fig. 2c). The bar graph in the inset displays the relative weight percentages of the constituents of the nanocomposite. It can clearly be seen that titanium, as a support material, was covered by regions of a phase rich in pyrrole and methyl anthranilate owing to the effective coating with pyrrole and methyl anthranilate, which was also well supported by the results of TEM analysis.
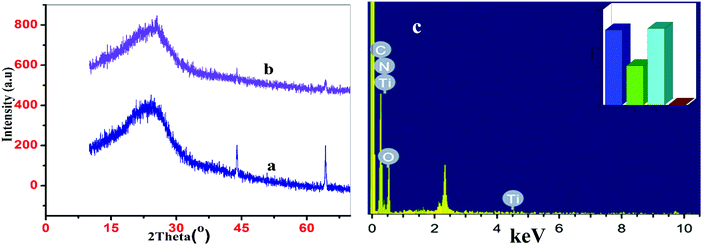 | ||
| Fig. 2 XRD diffractograms of (a) Ppy/MA copolymer and (b) the synthesised Ppy/MA/TiO2 NC. (c) EDX analysis of the nanocomposite. | ||
The thermal behaviour of the adsorbed and covalently modified Ppy–MA/TiO2 nanobioconjugates was compared with that of the free lipase by TGA and DTA analysis under a nitrogen atmosphere (Fig. 3). In comparison with the immobilized enzyme formulations, free lipase decomposed much more rapidly, which could possibly be linked to the denaturation of the protein conformation of lipase. The nanobiocatalysts exhibited two major stages of weight loss. The initial weight loss, which occurred at about 100–220 °C in the TGA analysis with a corresponding endothermic peak in DTA, can be ascribed to the loss of water molecules and unreacted constituents.29 The next weight loss in the range of 430–590 °C, which was supported by a second DTA endothermic peak, was assigned to the decomposition of the Ppy–MA polymer and smaller polymeric chains. The TGA curves in Fig. 3 indicated that the covalently immobilized bioconjugate remained most stable up to 800 °C in comparison with the adsorbed preparation and free enzyme, which was also supported by the DTA analysis. The high thermal stability of the immobilized nanobiocatalysts may be ascribed to strong interactions between the enzyme and NC,30 which has already been confirmed by the FT-IR studies mentioned above.
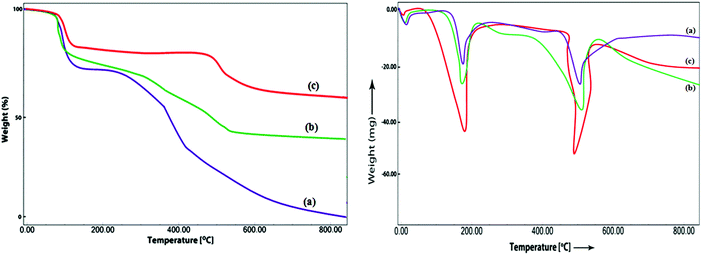 | ||
| Fig. 3 TGA and DTA thermograms of (a) free lipase, (b) physically adsorbed lipase and (c) covalently immobilized lipase. | ||
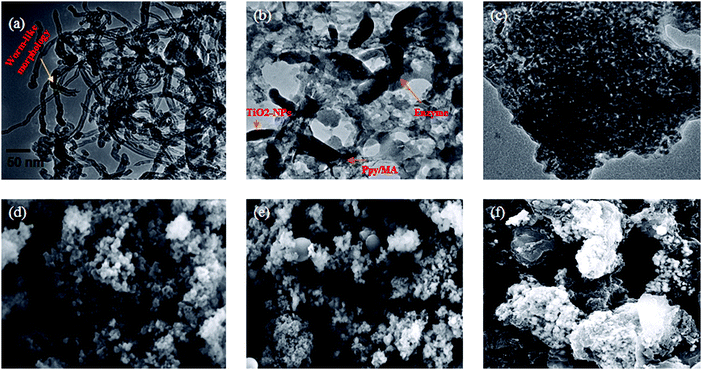
The morphologies of NCs with and without conjugation to lipase are displayed in Fig. 4. The TEM micrograph of Ppy–MA–TiO2 (Fig. 4a) shows a worm-like structure in the NC, which indicates that TiO2 nanoparticles (blackish shades) were homogeneously distributed in a matrix of polypyrrole–methyl anthranilate copolymer with an average particle size in the range of ∼38–50 nm. The TiO2 nanoparticles were not simply mixed with the Ppy–MA copolymer but were rather bound by the Ppy–MA matrix and displayed strong conjugation between Ppy–MA and TiO2 NPs. This could be ascribed to the formation of hydrogen bonds between N–H groups and oxygen atoms on the TiO2 surface. However, the TEM micrographs of physically adsorbed and covalently immobilized Ppy–MA/TiO2 NCs show (Fig. 4) dense granular agglomerates with average particle sizes of ∼45–65 nm and ∼62–90 nm, respectively. It can be determined by physical evaluation of the micrographs that after the binding of lipase to the NC there appeared to be no alteration in the morphology of the NC when its size increased, which indicates that the reaction only took place on the surface of the nanoparticles.
SEM images of the Ppy–MA/TiO2 NC both alone and with immobilized lipase are shown in Fig. 4(d, e and f), respectively. After the synthesis of the NC, TiO2 nanoparticles were found to be trapped in a matrix of Ppy–MA copolymer, which may possibly be due to the strong interaction between these components. SEM micrographs of the Ppy–MA/TiO2 NC show rough and bumpy structures, which suggest that the NC assisted in the deposition of greater amounts of lipase. Furthermore, comparative analyses of the SEM micrographs of the nanobiocatalyst and Ppy–MA/TiO2 NC confirmed that a thick sheet of lipase was deposited throughout the Ppy–MA/TiO2 NC. In conclusion of the results, it is quite clear that immobilization of the enzyme was successfully performed on the NC, and we propose that Ppy–MA/TiO2 NCs have superior affinity for the deposition of lipase.
3.3 Studies of optimum pH and temperature
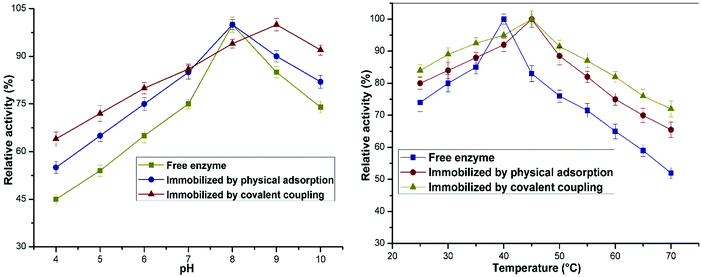
The pH was found to have a notable effect on the catalytic activity of the soluble as well as the immobilized lipase preparation (Fig. 5a). The optimum pH for achieving the highest enzymatic activity of the free and adsorbed enzyme preparations was 8. The optimum pH for the covalently immobilized enzyme was 9 (in comparison with 8 for the free enzyme). This is consistent with previously reported results of enzyme immobilization.31 This may be due to the shielding effect exerted by the substantial immobilization of the enzyme on the organic–inorganic carrier and the resulting space constraints; these factors may reduce the denaturation of the enzyme upon a sudden change in pH. It should be noted that the adsorbed and immobilized nanoconjugates displayed higher sensitivity in the alkaline region in comparison with the free enzyme. These results confirm that lipase immobilized on the Ppy–MA/TiO2 NC displayed greater tolerance of pH, which may have a vital effect on the applications of enzymes in various industries.Fig. 5b shows that the soluble lipase attained a maximum catalytic activity at an optimum temperature of 40 °C, and a gradual decrease in residual activity was observed at temperatures beyond this value. The immobilized lipase preparations exhibited a maximum activity at 45 °C, and a shift was thus observed between the optimum temperatures before and after the immobilization of lipase. The immobilized nanobiocatalyst exhibited elevated residual activity at high temperatures in comparison with the solution of the free enzyme. This shift after the immobilization of the enzyme on the nanosupport might be due to the increased stability of the enzyme, which facilitated the formation of the enzyme–substrate complex without affecting the accessibility of substrates to the active site.32 It was observed that there was a change in optimum temperatures for both enzyme preparations, which was also described in an earlier work.33 The free enzyme displayed acceptable thermal stability at an optimum temperature of about 35–40 °C; however, for the immobilized formulation the optimum reaction temperature was between 40 °C and 65 °C. In particular, when the temperature was increased to 70 °C, the covalently immobilized lipase exhibited a comparatively high hydrolytic activity of 72%, whereas its soluble counterpart retained only 50% of its activity. These results showed that lipase had outstanding thermal resistance after immobilization onto the Ppy–MA/TiO2 NC. The retention of stability and activity displayed by the immobilized enzyme preparation over a wide range of temperatures can be explained by the phenomenon of multipoint attachment, which endows the enzyme with rigidity and thus avoids conformational changes in the enzyme.
3.4 Kinetic parameters
The kinetic parameters for free and immobilized lipase preparations were calculated by monitoring the hydrolysis of a p-NPP substrate at varying concentrations (2.00 to 15.00 mM) in phosphate buffer. The Km value is related to the affinity between the enzyme and substrate. A lower value of Km implies greater affinity between the enzyme and its substrate, whereas a higher value of Km implies that the affinity is less. For both immobilized enzyme formulations a decline in the apparent value of Km was detected, which indicated enhanced affinity in the formation of the enzyme–substrate complex (Table 1). Similar results were also reported in the literature, which suggests that this may be due to the more effective conformation of immobilized lipase with a more suitable orientation, which leads to improved affinity for the substrate and more accessible active sites.34| Enzyme preparation | K m (mM) | V max (mM min−1) | V max/Km (min−1) |
|---|---|---|---|
| ROL | 6.6 | 7.9 | 1.19 |
| PAL | 5.4 | 5.1 | 0.94 |
| CIL | 3.5 | 3.9 | 1.14 |
The Vmax value designates the maximum reaction rate when all the enzyme sites are saturated with the substrate. It is well known that the greater is the Vmax value, the higher is the activity of the enzyme. The Vmax value decreased for the hydrolysis of p-NPP in case of both physically adsorbed and covalent immobilized enzymatic preparation (Table 1).
Similar results that showed lower Vmax values after enzyme immobilization were also reported in some previous studies.35 This decrease may be associated with conformational changes that occurred in the structure of the enzyme after immobilization. The catalytic efficiency (Vmax/Km) of the enzyme immobilized by physical adsorption was somewhat less than that of the free enzyme. This finding is in accordance with earlier studies,36 in which a decline was reported in the catalytic efficiencies of enzymes when they were immobilized by physical adsorption on single-walled carbon nanotubes. However, the preparation that was covalently immobilized using glutaraldehyde displayed increased catalytic efficiency, which indicated that the functional changes induced by immobilization were minimal.
3.5 Thermal stability
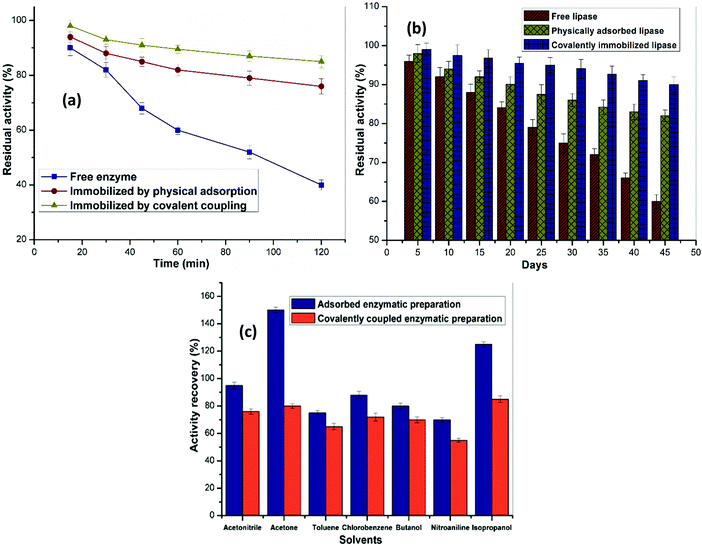
The stability of an immobilized enzyme is crucial for its reusability and practical applications. To examine the thermal stability of immobilized lipase, its residual activity was measured by incubation at 60 °C for a defined time interval, and the results are depicted in Fig. 6. Free lipase retained 41% of its catalytic activity after 2 h at 65 °C, whereas the adsorbed and covalently modified immobilized formulations displayed remarkable activity retention of 76% and 86%, respectively, under identical incubation conditions. These results could presumably be associated with stabilization of the formation of multipoint bonds between the nanosupport and enzyme molecules by adsorption, chelation, or covalent bonding.373.6 Storage stability
The storage stability was studied after free and immobilized lipase preparations were stored at 30 °C for a period of 45 days. Fig. 6 shows the comparative residual enzymatic activities. The results confirmed that the immobilized lipase exhibited higher retention of its initial hydrolytic activity. After storage for 30 days, the covalently attached lipase on the Ppy/MA–TiO2 NC retained superior activity of 94% after 45 days, whereas the adsorbed enzyme retained about 90% of its initial activity. In contrast, free lipase only retained nearly 60% of its initial activity. The relatively greater stability was associated with the increase in rigidity and the stronger interaction of lipase with the support, which increased the rigidity of the enzyme. The results in Fig. 6 illustrate that ROL immobilized on the PPy/MA–TiO2 NC displayed high durability and also that loss of the protein was minimal in comparison with the physically adsorbed enzyme on the NC.3.7 Solvent tolerance
The effect of several organic solvents on the stability of both the immobilized lipase preparations was assessed. The hydrolytic activity of lipase both adsorbed and covalently modified by glutaraldehyde on the Ppy/MA–TiO2 NC in various organic solvents (acetone, toluene, etc.) after incubation was compared with that of a control with no solvent (100%). The enzyme formulation that was immobilized non-covalently exhibited improved tolerance of the various solvents in comparison with covalently immobilized lipase with respect to its free form. Remarkably, the relative activity retention of lipase that was covalently immobilized on the synthesised Ppy/MA–TiO2 NC was less (80% and 85%) after incubation in acetone and isopropanol, respectively. On the other hand, there was a remarkable increase in the residual activity of the non-covalently immobilized lipase (150% and 125%) after incubation in the abovementioned solvents. It is interesting to note that lipase when physically adsorbed on the Ppy/MA–TiO2 NC exhibited greater solvent tolerance in comparison with the enzyme that was covalently immobilized on the nanosupport, despite the fact that it displayed lower catalytic activity (Fig. 6). This may possibly have been due to stronger hydrophobic interactions of the enzyme with the surface of the Ppy/MA–TiO2 NC, which could prevent the loss of enzyme. These outcomes demonstrate that it was not possible to devise a general rule for the behaviour of the immobilized enzyme formulations in terms of their stability and activity in organic solvents. This behaviour may possibly be assigned to the fact that the organic solvents maintained the inner rigidity of lipase and did not easily displace the water layer from the surface of the enzyme. To date, there have been several reports of the stability of lipases in organic solvents that support our findings.38–40 As it is well known that the catalytic performance of enzymes relies on their tolerance of different solvent systems, the fact that our formulation displayed exceptionally increased stability in acetone (150 ± 2.2%) and isopropanol (125 ± 2.0%) in comparison with its free form suggests its potential as a nanobiocatalyst for transesterification reactions and biodiesel production.3.8 Reusability of immobilized enzyme preparations
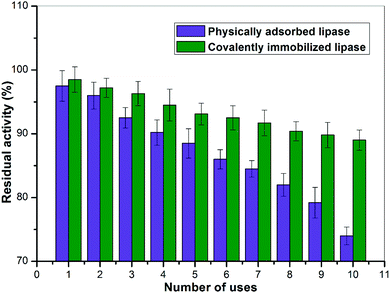
The feasibility of an industrial application based on immobilized enzyme preparations depends on whether the enzyme can be regenerated and consequently on whether the nanobiocatalyst can be repeatedly utilized. The operational stability of ROL both non-covalently and covalently immobilized on the Ppy/MA–TiO2 NC was investigated. As seen in Fig. 7, both immobilized enzyme formulations retained more than 70% of their activity after 10 repeated uses. The covalent attachment process had a substantial stabilizing effect on the ROL in the case of the NC that was used. Covalently immobilized lipase on the Ppy/MA–TiO2 NC retained about 90% of its initial activity after 10 repeated uses, whereas the non-covalently immobilized enzyme lost more than 25% of its activity under identical conditions of repeated use. The stabilizing effect of the Ppy/MA–TiO2 nanosupport as an immobilization matrix that was seen here was in agreement with those previously observed for various enzymes covalently immobilized on nanomaterials.41,42 Thus, the excellent ability of lipase to be repeatedly used represents a profitable advantage for its utilization in an economically feasible enzyme-catalyzed process for industrial applications.3.9 In silico screening
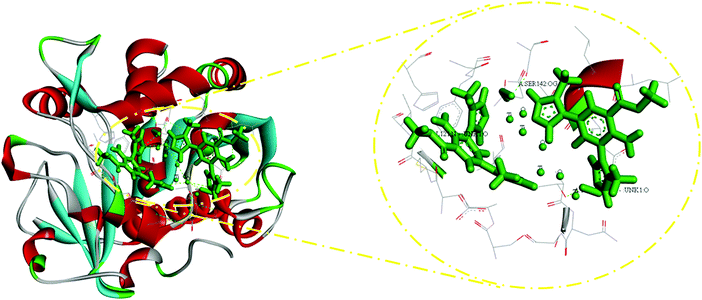
Molecular docking studies were executed in order to justify the approaches used to explain the results observed for lipase. Hence, the data for the Ppy–MA/TiO2 nanocomposite were examined on a structural basis by both molecular modelling and a docking study of Rhizopus lipase (PDB: 4L3W) using MVD and Discovery Studio software with the aim of predicting its affinity, orientation, and surrounding surfaces. The different bonds, i.e., van der Waals forces, hydrogen bonds and hydrophobic behaviour with amino acids, were in favourable agreement with the binding affinities predicted by molecular docking studies, which supports previous reports.43Via molecular docking investigations, it was found that the A: THR110:OG1–:UNK1:O A:VAL121:N–:UNK1:O A:SER142:OG–:UNK1:O:UNK1:O–A:SER142:OG0:UNK1:O–A:THR110:OG1 residues (Fig. 8) interacted with high frequency and in an appropriate orientation with the NC. The binding affinity for docked Ppy–MA/TiO2 with the retrieved protein, as predicted by Hex software, was estimated to be −181.01 kcal mol−1. The lower binding energy suggests that the combination of the NC with lipase was more stable and that Ppy–MA/TiO2 was suitable for the immobilization of the enzyme. Hence, as a novel nanosupport the Ppy–MA/TiO2 NC efficiently enhances the stability of the enzyme and enables the repeated use of the enzyme, and thus has improved usability in industrial applications.4. Conclusions
In the present work, we have demonstrated the development of an innovative nanobiocatalytic system via immobilization of Rhizopus oryzae lipase on a novel Ppy–MA/TiO2 NC synthesized by chemical oxidative polymerization. We have established that lipase both adsorbed and immobilized via coupling with glutaraldehyde on Ppy/MA–TiO2 retained its activity at high temperatures (76% and 84% of its initial activity at 65 °C, respectively) when compared with that of the free enzyme (activity recovery of ∼40%). The pH and thermal stabilities of both forms of immobilized lipase formulation were also enhanced. The nanobiocatalyst retained about 85% of its activity after incubation in a series of organic solvents, and only minor loss of activity was observed for immobilized lipase incubated in toluene. In addition, for some solvents such as isopropanol and acetone the activity recovery was as high as 125% and 150%, respectively. The improved catalytic behaviour observed for lipase immobilized on Ppy/MA–TiO2 indicates that the functionalized nanosupport is suitable for the development of an efficient nanobiocatalytic system that can efficiently work in an aqueous as well as in an organic medium.Abbreviations
| Ppy–MA/TiO2 | Polypyrrole–methyl-anthranilate-functionalized titanium dioxide NCs |
| NCs | Nanocomposites |
| NPs | Nanoparticles |
| SEM | Scanning electron microscopy |
| EDAX | Energy-dispersive X-ray spectroscopy |
| TEM | Transmission electron microscopy |
| TGA | Thermogravimetric analysis |
| DTA | Differential thermal analysis |
| FT-IR | Fourier transform infrared spectroscopy. |
Conflicts of interest
The authors declare that there is no conflict of interest among authors and with any funding agency.Acknowledgements
Shamoon Asmat is grateful to the USIF, AMU for providing the facilities of TEM and SEM for this study.References
- U. T. Bornscheuer, G. W. Huisman, R. J. Kazlauskas, S. Lutz, J. C. Moore and K. Robins, Nature, 2012, 485, 185–194 CrossRef CAS PubMed.
- M. Zheng, X. Xiang, S. Wang, J. Shi, Q. Deng, F. Huang and R. Cong, Process Biochem., 2017, 53, 102–108 CrossRef CAS.
- J. H. Kim, S. Park, H. Kim, H. J. Kim, Y.-H. Yang, Y. H. Kim, S.-K. Jung, E. Kan and S. H. Lee, Carbohydr. Polym., 2017, 157, 137–145 CrossRef CAS PubMed.
- J. Gao, Y. Wang, Y. Du, L. Zhou, Y. He, L. Ma, L. Yin, W. Kong and Y. Jiang, Chem. Eng. J., 2017, 317, 175–186 CrossRef CAS.
- J. Gao, W. Kong, L. Zhou, Y. He, L. Ma, Y. Wang, L. Yin and Y. Jiang, Chem. Eng. J., 2017, 309, 70–79 CrossRef CAS.
- Y. Jiang, X. Liu, Y. Chen, L. Zhou, Y. He, L. Ma and J. Gao, Bioresour. Technol., 2014, 153, 278–283 CrossRef CAS PubMed.
- M. Widersten, Curr. Opin. Chem. Biol., 2014, 21, 42–47 CrossRef CAS PubMed.
- P.-C. Chen, X.-J. Huang and Z.-K. Xu, Phys. Chem. Chem. Phys., 2015, 17, 13457–13465 RSC.
- S. A. Ansari and Q. Husain, Biotechnol. Adv., 2012, 30, 512–523 CrossRef CAS PubMed.
- R. Ahmad and M. Sardar, Biochem. Anal. Biochem., 2015, 4, 1 Search PubMed.
- C. Hou, Z. Qi and H. Zhu, Colloids Surf., B, 2015, 128, 544–551 CrossRef CAS PubMed.
- Q. Husain, Biointerface Res. Appl. Chem., 2016, 6, 1585–1606 Search PubMed.
- M. Ali, Q. Husain, N. Alam and M. Ahmad, Water, Air, Soil Pollut., 2017, 228, 22 CrossRef.
- M. Dervisevic, E. Dervisevic, H. Azak, E. Çevik, M. Åženel and H. B. Yildiz, Sens. Actuators, B, 2016, 225, 181–187 CrossRef CAS.
- Z. Yang, C. Zhang, J. Zhang and W. Bai, Biosens. Bioelectron., 2014, 51, 268–273 CrossRef CAS PubMed.
- J. Bai and B. Zhou, Chem. Rev., 2014, 114, 10131–10176 CrossRef CAS PubMed.
- Y. Jiang, W. Tang, J. Gao, L. Zhou and Y. He, Enzyme Microb. Technol., 2014, 55, 1–6 CrossRef CAS PubMed.
- Q. Husain, Biocatalysis, 2017, 3, 37–53 Search PubMed.
- P. Zucca and E. Sanjust, Molecules, 2014, 19, 14139–14194 CrossRef.
- U. Jan, Q. Husain and M. Saleemuddin, Biotechnol. Appl. Biochem., 2001, 34, 13–17 CrossRef CAS PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CAS.
- G. Zhao, Y. Li, J. Wang and H. Zhu, Appl. Microbiol. Biotechnol., 2011, 91, 591–601 CrossRef CAS PubMed.
- G. Cho, B. M. Fung, D. T. Glatzhofer, J.-S. Lee and Y.-G. Shul, Langmuir, 2001, 17, 456–461 CrossRef CAS.
- F. Su, G. Li, Y. Fan and Y. Yan, Sci. Rep., 2016, 6 Search PubMed.
- R. Yadav, N. Labhsetwar, S. Kotwal and S. Rayalu, J. Nanopart. Res., 2011, 13, 263–271 CrossRef CAS.
- S. A. Waghuley, S. M. Yenorkar, S. S. Yawale and S. P. Yawale, Sens. Actuators, B, 2008, 128, 366–373 CrossRef CAS.
- H. Yuvaraj, M. H. Woo, E. J. Park, Y. T. Jeong and K. T. Lim, Eur. Polym. J., 2008, 44, 637–644 CrossRef CAS.
- W. Li, L. Kuai, Q. Qin and B. Geng, J. Mater. Chem. A, 2013, 1, 7111–7117 CAS.
- S. A. Ansari and Q. Husain, J. Mol. Catal. B: Enzym., 2011, 70, 119–126 CrossRef CAS.
- J. Sun, R. Yendluri, K. Liu, Y. Guo, Y. Lvov and X. Yan, Phys. Chem. Chem. Phys., 2017, 19, 562–567 RSC.
- G. Di Nardo, C. Roggero, S. Campolongo, F. Valetti, F. Trotta and G. Gilardi, Dalton Trans., 2009, 6507–6512 RSC.
- N. Ortega, M. Perez-Mateos, M. A. C. Pilar and M. A. D. Busto, J. Agric. Food Chem., 2008, 57, 109–115 CrossRef PubMed.
- S. Asmat, Q. Husain and A. Azam, RSC Adv., 2017, 7, 5019–5029 RSC.
- M.-Y. Zhuang, Q.-L. Zhou, X.-Y. Wang, J.-X. Zhang, L. Xue, R. Wang, J.-X. Zhang and Y.-W. Zhang, Nanosci. Nanotechnol. Lett., 2016, 8, 251–254 CrossRef.
- M.-H. Liao and D.-H. Chen, Biotechnol. Lett., 2001, 23, 1723–1727 CrossRef CAS.
- J. T. Cang-Rong and G. Pastorin, Nanotechnology, 2009, 20, 255102 CrossRef PubMed.
- J. C. Wu, S. S. Lee, M. M. B. Mahmood, Y. Chow, M. M. R. Talukder and W. J. Choi, J. Mol. Catal. B: Enzym., 2007, 45, 108–112 CrossRef CAS.
- L. Fernandez-Lopez, N. Rueda, R. Bartolome-Cabrero, M. D. Rodriguez, T. L. Albuquerque, J. C. S. dos Santos, O. Barbosa and R. Fernandez-Lafuente, Process Biochem., 2016, 51, 48–52 CrossRef CAS.
- N. Rueda, T. L. Albuquerque, R. Bartolome-Cabrero, L. Fernandez-Lopez, R. Torres, C. Ortiz, J. dos Santos, O. Barbosa and R. Fernandez-Lafuente, Molecules, 2016, 21, 646 CrossRef PubMed.
- V. G. Tacias-Pascacio, S. Peirce, B. Torrestiana-Sanchez, M. Yates, A. Rosales-Quintero, J. J. Virgen-OrtÃz and R. Fernandez-Lafuente, RSC Adv., 2016, 6, 100281–100294 RSC.
- J. Cui, L. Cui, S. Jia, Z. Su and S. Zhang, J. Agric. Food Chem., 2016, 64, 7179–7187 CrossRef CAS PubMed.
- K. Banjanac, M. Mihailović, N. Prlainović, M. Stojanović, M. Carević, A. Marinković and D. Bezbradica, J. Chem. Technol. Biotechnol., 2016, 91, 439–448 CrossRef CAS.
- S. K. Chaturvedi, N. Zaidi, P. Alam, J. M. Khan, A. Qadeer, I. A. Siddique, S. Asmat, Y. Zaidi and R. H. Khan, PLoS One, 2015, 10, e0136528 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7nj02951a |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2018 |