Identification of non-substrate-like glycosyltransferase inhibitors from library screening: pitfalls & hits†
Masaki
Ema
a,
Yong
Xu
a,
Sebastian
Gehrke
b and
Gerd K.
Wagner
 *a
*a
aKing's College London, Department of Chemistry, Faculty of Natural & Mathematical Sciences, Britannia House, 7 Trinity Street, London, SE1 1DB, UK. E-mail: gerd.wagner@kcl.ac.uk; Tel: +44 (0)20 7848 1926
bKing's College London, Institute of Pharmaceutical Science, Faculty of Life Sciences & Medicine, UK
First published on 29th November 2017
Abstract
Bacterial glycosyltransferases are potential targets for the development of novel antibiotics and anti-virulence agents. Most existing glycosyltransferase inhibitors are substrate analogues with limited potential for drug development. The identification of alternative inhibitor chemotypes is therefore of great interest for medicinal chemistry, drug discovery and chemical glycobiology. We describe the application of a biochemical glycosyltransferase assay to screen a small compound library containing three distinct chemical scaffolds (nucleosides, steroids and 5-methyl pyrazol-3-ones) against the retaining α-1,4-galactosyltransferase LgtC from Neisseria meningitidis. While no genuine LgtC inhibitory activity was observed in the nucleoside and steroid series, the best hit compounds in the 5-methyl pyrazol-3-one series showed low micromolar activity. We adapted our assay protocol to develop initial structure–activity relationships in this series, and to establish the target selectivity of the most potent inhibitor over two other glycosyltransferases. Our results provide insights into the activity of this class of non-substrate-like glycosyltransferase inhibitors, and highlight important general pitfalls for inhibitor screening against this enzyme family. Key elements of our experimental design, including a validated single-concentration protocol for inhibitor screening, and our process for elimination of false positives, are, in principle, directly transferable to many other sugar-nucleotide-dependent glycosyltransferases.
Introduction
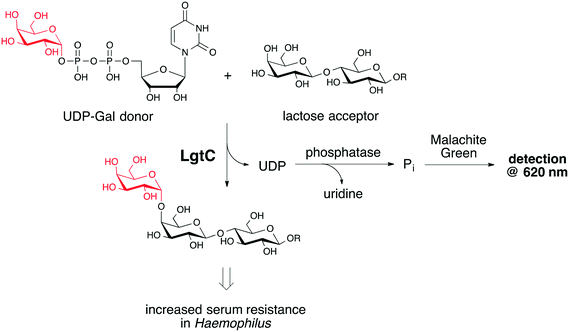
Gram-negative bacterial pathogens such as Haemophilus and Neisseria use certain glycan structures in their cell envelope as virulence factors, e.g. to protect against the immune response of the human host. The biosynthetic machinery required for the biosynthesis of these glycoconjugates therefore offers a rich source of targets for the development of novel antibiotics and anti-virulence agents. An important class of enzymes that fall into this category are glycosyltransferases (GTs), enzymes that catalyse the transfer of a mono- or oligosaccharide from a glycosyl donor to a specific acceptor.1 Most existing inhibitors of bacterial GTs are substrate analogues with limited potential for drug development.2 An important strategy for the identification of alternative GT inhibitor chemotypes is the screening of compound libraries. Despite successful previous examples,3 the screening for GT inhibitors is not trivial, and the identification both of suitable assay conditions, and of suitable screening libraries can be challenging. This is particularly true if a biochemical, rather than a binding, assay is used, due to the complex multi-substrate mechanism of these enzymes.A considerable number of biochemical GT assays have been developed, including radiochemical, immunological, chromatographic, fluorimetric and spectrophotometric formats.4,5 However, many of these assay formats cannot easily be adapted for inhibitor screening because of the requirement for expensive specialized equipment, difficulty to access labelled donor or acceptor, or a general unsuitability of the assay format for high-throughput conditions.4 We have recently described an assay protocol for the identification of non-substrate-like inhibitors of the retaining α-1,4-galactosyltransferase LgtC from Neisseria meningitidis.6 LgtC catalyses the transfer of α-D-galactose from uridine diphosphate α-D-galactose (UDP-Gal) to terminal lactose residues on lipooligosaccharide (LOS) structures of the outer core (Fig. 1),7 resulting in increased serum resistance.8 Inhibitors of LgtC may therefore be useful as anti-virulence agents and tool compounds for anti-virulence drug discovery. Our LgtC assay uses a phosphatase to cleave both phosphates off the secondary reaction product UDP, followed by colorimetric detection of the inorganic phosphate with malachite green (Fig. 1). The assay is inexpensive and operationally simple, and can be carried out in a microplate format.6
In this study, we have adapted this assay format to screen a small compound library against LgtC, in order to identify new inhibitor chemotypes. Most existing LgtC inhibitors are structurally derived from the donor substrate UDP-Gal.7,9 Our library contained three distinct non-substrate-like chemical scaffolds: nucleosides, steroids and pyrazol-3-ones. Nucleosides were selected to investigate if they may be able to act as uncharged mimics of the UDP-Gal donor. Steroids and pyrazol-3-ones were included because of previously reported activities of these structural classes against other GTs. Thus, inhibitory activity against different mammalian and bacterial GTs has been reported for several steroid derivatives, such as finasteride against human UDP-glucuronosyltransferase UGT,10 murgocil against the peptidoglycan GT MurG,11 and lithocholic acid derivatives such as AL10 against α-2,3-sialyltransferase.12,13 Pyrazol-3-ones have been reported as inhibitors of different bacterial GTs, including MurG3 and WaaC.14
We have recently identified a single 5-methyl pyrazol-3-one as an inhibitor of LgtC.15 Herein, we report initial structure–activity relationships (SAR) in this inhibitor series. For these experiments, we have established a single-concentration screening protocol for our assay, and validated it with full IC50 measurements. With this approach, we identified several 5-methyl pyrazol-3-ones with low micromolar activity against LgtC. Following further adaptation of our assay protocol, we also investigated the LgtC target selectivity of the most potent inhibitor over two other glycosyltransferases. No genuine inhibitory activity against LgtC was observed, on the other hand, in either the nucleoside or the steroid series. In the steroid series, we did observe false positives, which prompted us to carefully investigate critical parameters of our screening assay protocol. The results from this study therefore provide not only initial SAR trends in a new class of non-substrate-like GT inhibitors, but also highlight important general pitfalls for inhibitor screening against this enzyme family.
Results
Assay adaptation
For the initial screening experiment, we selected 130 compounds in total, including 20 nucleosides and 90 steroids from a commercial library of known drugs and bioactives, and 20 5-methyl pyrazol-3-ones that had been synthesised in our own laboratory (Table S1, ESI†). Inhibitor candidates were initially tested at a single concentration (50 μM) against LgtC. For this experiment, we adapted our recently reported colorimetric assay protocol.6 LgtC, β-lactose acceptor, inhibitor (in DMSO), calf-intestinal phosphatase (CIP), MnCl2, chicken egg-white lysozyme (CEL) and Triton X-100 were premixed, and the reaction was started by addition of UDP-Gal donor. After incubation for 20 min, the malachite green reagents were added to stop the reaction, and absorbance was measured at 620 nm. With this set-up, 10 inhibitors could be examined, in duplicate, on a single 96-well microplate. To accelerate screening and save on materials, we also attempted to miniaturise the assay, using 384-well microplates. This would have allowed us to reduce the total amount of reagent in each well from 210 μM to 70 μM. Unfortunately, this set-up resulted in poor reproducibility, especially at higher concentrations of inhibitor (Fig. S1, ESI†). We therefore used the standard 96-well plate set-up for all screening experiments.Library screening
With suitable assay conditions in hand for screening, we tested the three selected compound collections. First, 90 steroid derivatives were selected from a commercial library of known drugs and bioactives. In the single-concentration screen of these 90 steroid derivatives, two derivatives showed significantly higher inhibitory activity than the remaining steroids (79: 38% @ 50 μM; 90: 34% @ 50 μM). Both compounds belong to the class of aminosteroid muscle relaxants (Fig. S2, ESI†). To confirm their inhibitory activity, we determined their full IC50 curves. Encouragingly, in these experiments both compounds showed concentration-dependent activity at concentrations above 100 μM (Fig. S2, ESI†).To investigate the possibility of false positives, e.g. from inhibition of the phosphatase used in the assay and/or interference of the inhibitors with the colorimetric assay read-out, we next carried out control experiments with 79 and 90. First, the assay was repeated with only the phosphatase present, but no LgtC, and the phosphatase substrate UDP instead of UDP-Gal. Disappointingly, in this control experiment both 79 and 90 gave a similar dose–response curve as in the IC50 experiments with LgtC, which indicated that the activity observed in the coupled assay was not due to inhibition of LgtC (Fig. S2, ESI†). To understand if these compounds inhibited the phosphatase or interfered with the malachite green detection reagent, we carried out a second control experiment. Inorganic phosphate (Pi) at different concentrations was reacted with malachite green in the presence or absence of the screening hits. Both 79 and 90 significantly reduced the absorbance under these conditions (Fig. S2, ESI†). Taken together, these results showed that the activity observed for both compounds in the coupled LgtC assay was an assay artefact resulting from interference of 79 and 90 with the colorimetric read-out of our assay. These results highlight not only the need for stringent control experiments with this type of coupled assay, but also the capacity of charged aminosteroids such as 79 and 90 for non-specific aggregation with assay components.
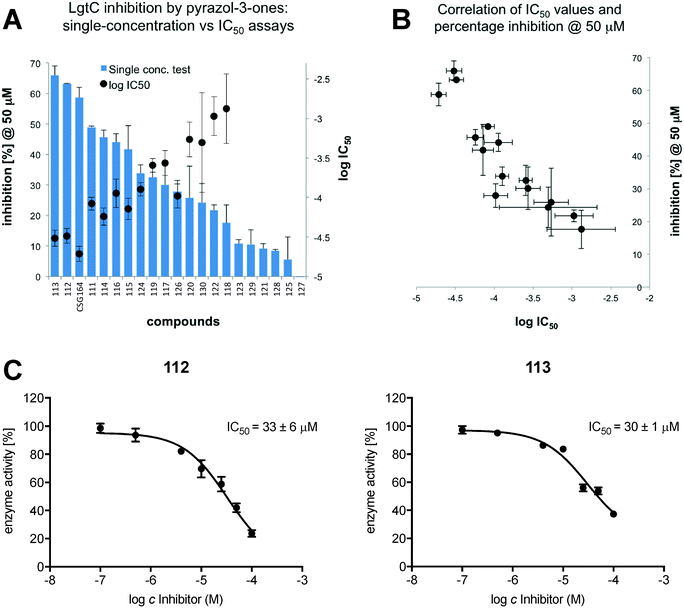
Next, we repeated the screening experiment against LgtC with a collection of 20 nucleoside derivatives (Table S1, ESI†), but did not observe any inhibitory activity. Finally, we repeated the screening experiment against LgtC with 20 5-methyl pyrazol-3-one derivatives (Table S1, ESI†). In this series, diverse levels of activities were observed, ranging from compounds that were practically inactive, such as 127, to compounds such as 113, which were at least equipotent to our prototype inhibitor CSG16415 (Fig. 2A). For compounds that showed inhibition greater than 10% in the single-concentration screen, we determined full IC50 values. Pleasingly, we found a good correlation between inhibitory activity in the single-concentration screen and in full IC50 experiments (Fig. 2B). This result demonstrates that the single-concentration assay format is suitable for discrimination between active and inactive molecules. Importantly, and in contrast to results with aminosteroids 79 and 90, control experiments against phosphatase, with inhibitor at 50 μM, confirmed that for all pyrazol-3-ones 111–130, the activity observed in the coupled assay was due to genuine inhibition of LgtC.
Profiling of hit compound 113
Next, we further investigated the activity profile of the most potent inhibitor identified from the screen, 5-methyl pyrazol-3-one 113. At concentrations up to 100 μM, 113 showed no activity in control experiments against phosphatase, and did not affect absorbance readings (Fig. S3, ESI†). This provided further confirmation that 113 is a genuine LgtC inhibitor. To assess the target profile of 113 for LgtC, we also tested this inhibitor against two other GTs, the α-(2,3)-sialyltransferase from Pasteurella multocida (SiaT) and the human fucosyltransferase 2 (FUT2). Although both GTs differ from LgtC in their donor substrate specificity (Table 1), our assay can, in principle, also be used for these GTs, as it is based on the generic detection of inorganic phosphate (Pi), irrespective of the nature of the sugar-nucleotide donor from which it is released.6 The application of the assay with SiaT and FUT2 is, however, complicated by the significantly higher Km value of these two enzymes for their respective donor (Table 1). In our standard protocol, LgtC inhibition experiments are carried out with UDP-Gal concentrations slightly above the donor Km. To carry out the inhibition assay with SiaT and FUT2 under comparable conditions, significantly higher donor concentrations were therefore required. This complicated the application of the assay, as its linear range for the correlation between absorbance and analyte concentration is limited to Pi concentrations below 25 μM.6To keep the final Pi concentrations within the linear range of the colorimetric detection step, we therefore adapted our assay protocol. For GTs with a higher donor Km, we introduced a dilution step after the enzymatic reaction, in order to bring the final Pi concentration within the linear range of the assay. To assess if the introduction of such a dilution step might affect inhibition assay results, we first carried out a validation experiment with LgtC and the positive control CSG164. In two separate sets of reaction mixtures, inhibitor, LgtC and β-lactose acceptor were pre-mixed with MnCl2, CEL, CIP and Triton X-100, and the reactions were started by addition of UDP-Gal donor. After 20 minutes incubation, one set of reaction mixtures was analysed as is, while the other was diluted 5-fold with buffer before detection. Upon addition of malachite green reagents and recording of the absorbance at 620 nm, no significant difference in the percentage inhibition of LgtC was observed in the diluted sample, compared to the standard protocol (Fig. S4, ESI†). At 100 μM, CSG164 showed 60% inhibition of LgtC both in the standard protocol and upon 5-fold dilution of the assay solution prior to addition of the malachite green reagents.
With a reliable dilution protocol in hand, 113 was tested against SiaT and FUT2. 113, SiaT or FUT2, acceptor (β-lactose for SiaT, α-lactose for FUT2), metal chloride (MgCl2 for SiaT, MnCl2 for FUT2), CEL, CIP and Triton X-100 were pre-mixed, and the reaction was started by addition of the respective donor. The reaction mixture was incubated for 20 min and diluted as required. Finally, malachite green was added and the absorbance was recorded at 620 nm. Under these conditions, 113 did not reduce the enzymatic activity of SiaT, and had only a modest effect, at the highest concentration, against human FUT2 (Fig. 3A and B). Thus, 113 displays a concentration-dependent inhibitory effect only against LgtC, as the direct comparison of results against all three GTs shows (Fig. 3C).
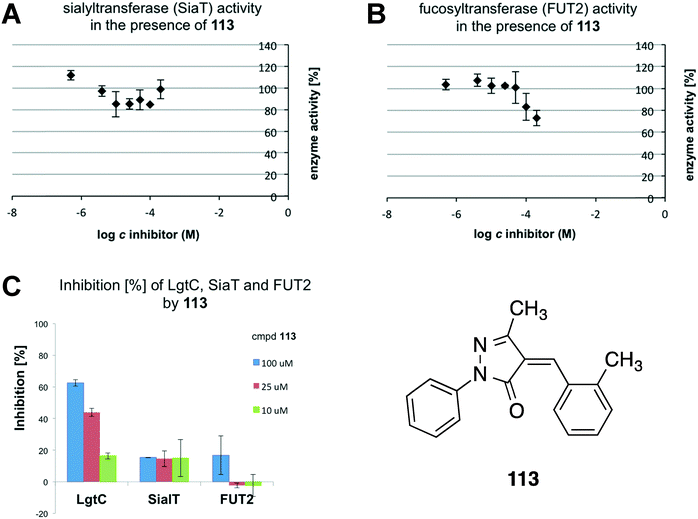 | ||
| Fig. 3 A: Enzyme activity of Pasteurella sialyltransferase SiaT in the presence of pyrazol-3-one 113. B: Enzyme activity of human fucosyltransferase FUT2 in the presence of 113. C: Direct comparison of inhibitory activities of 113 against LgtC, SiaT and FUT2. Percentage inhibition values at 10 μM, 25 μM and 100 μM of 113 are based on residual enzyme activity in the presence of inhibitor, and are extracted from Fig. 2C (for LgtC), 3A (for SiaT) and 3B (for FUT2), respectively. | ||
Conclusion
In this study, we have screened a small collection of non-substrate-like inhibitor candidates against the bacterial glycosyltransferase LgtC, using a biochemical coupled assay. Our results demonstrate that this assay format can be adapted for screening, and highlight several important features for such an application: we show that despite the addition of surfactant to the assay mixture, false positives can arise from interference of inhibitor candidates with the colorimetric malachite green reagent, but also that these assay artefacts can be readily identified through appropriate control experiments. Results in the pyrazol-3-one series demonstrate that a single-concentration set-up is suitable for the identification of potent GT inhibitors, which will facilitate screening of larger libraries.The vast majority of GTs releases a nucleoside diphosphate (NDP) as their secondary reaction product. Our assay is based on the detection of this generic reaction product. Key elements of our experimental design, including the single-concentration screening protocol we have established and validated, and our process for elimination of false positives, are therefore directly transferable to many other GTs, beyond the enzymes in this study. It is useful, in this context, that our assay protocol can be readily adapted for GTs with a significantly higher Km (donor) than LgtC, as is common in this enzyme family. We demonstrate that the limited linear range of the colorimetric detection step can be overcome by introducing a dilution step after the enzymatic reaction. This modification is important because it allows the profiling of inhibitor candidates not only with regard to their potency, but also their GT target selectivity.
5-Methyl pyrazol-3-one 113, which was identified from this screen, is one of the most potent non-substrate-like inhibitors against LgtC reported to date. SAR trends in this series suggest that the ortho position of the benzylidene substituent offers interesting opportunities to modulate inhibitory activity. Non-polar substituents in this position such as 2-CH3 (113) and 2-chloro (112) are advantageous, while a polar group is not (128, 10% inhibition at 50 uM). We have recently shown that the prototype inhibitor CSG164 reacts covalently, through its Michael acceptor system, with a non-catalytic cysteine residue in the LgtC active site.15 Small, non-polar substituents at the ortho position of the benzylidene substituent may contribute to orienting the Michael acceptor system correctly in the LgtC active site, for example by acting as a steric block that restricts rotation about the benzylidene single bond. These results therefore provide an exciting starting point for further optimisation of this new class of non-substrate-like GT inhibitors.
Experimental section
General
Steroids 1–90 and nucleosides 91–108 were selected from the Prestwick Chemical Library (www.prestwickchemical.com). Nucleosides 109–110 were synthesised as previously reported.20 5-Methyl pyrazol-3-ones 111–130 were synthesized by Knoevenagel condensation of a given 5-methyl pyrazol-3-one derivative and the requisite aldhehyde according to our recently reported protocol15 and fully characterised by 1H NMR, 13C NMR and mass spectrometry (ESI). LgtC was expressed and purified as previously reported.9 Recombinant α-(2,3)-sialyltransferase from Pasteurella multocida was purchased from Sigma. Recombinant human fucosyltransferase 2 (FUT2) was purchased from R&D Systems. Assays were carried out on 96-well plates, unless otherwise stated, and analysed at 620 nm on a Polarstar Optima plate reader (BMG Labtech).Inhibition experiments with LgtC
Inhibition experiments with other GTs
Control experiments
Synthesis and characterization of compound 113
Conflicts of interest
The authors declare no competing interests.Acknowledgements
We thank King's College for a King's China Award (to Y. X.), the EPSRC National Mass Spectrometry Facility, Swansea, for the recording of mass spectra, and Mr Hugo Leung and Dr Jingqian Jiang for assistance with the synthetic chemistry. The LgtC plasmid was a gift by Dr Warren Wakarchuk (Toronto). M. E. was supported by a visiting fellowship from the Japanese Patent Office.References
- T. M. Gloster, Advances in understanding glycosyltransferases from a structural perspective, Curr. Opin. Struct. Biol., 2014, 28, 131–141 CrossRef CAS PubMed.
- L. Tedaldi and G. K. Wagner, Beyond substrate analogues: new inhibitor chemotypes for glycosyltransferases, MedChemComm, 2014, 5, 1106–1125 RSC.
- Y. N. Hu, J. S. Helm, L. Chen, C. Ginsberg, B. Gross, B. Kraybill, K. Tiyanont, X. Fang, T. Wu and S. Walker, Identification of Selective Inhibitors for the Glycosyltransferase MurG via High-Throughput Screening, Chem. Biol., 2004, 11, 703–711 CrossRef CAS PubMed.
- G. K. Wagner and T. Pesnot, Glycosyltransferases and their assays, ChemBioChem, 2010, 11, 1939–1949 CrossRef CAS PubMed.
- M. M. Palcic and K. Sujino, Assays for glycosyltransferases, Trends Glycosci. Glycotechnol., 2001, 13, 361–370 CrossRef CAS.
- L. Tedaldi, A. Evitt, N. Göös, J. Jiang and G. K. Wagner, A practical glycosyltransferase assay for the identification of new inhibitor chemotypes, MedChemComm, 2014, 5, 1193–1201 RSC.
- K. Persson, H. D. Ly, M. Dieckelmann, W. W. Wakarchuk, S. G. Withers and N. C. J. Strynadka, Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs, Nat. Struct. Biol., 2001, 8, 166–175 CrossRef CAS PubMed.
- A. L. Erwin, S. Allen, D. K. Ho, P. J. Bonthius, J. Jarisch, K. L. Nelson, D. L. Tsao, W. C. T. Unrath, M. E. Watson, B. W. Gibson, M. A. Apicella and A. L. Smith, Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum, Infect. Immun., 2006, 74, 6226–6235 CrossRef CAS PubMed.
- K. Descroix, T. Pesnot, Y. Yoshimura, S. Gehrke, W. Wakarchuk, M. M. Palcic and G. K. Wagner, Inhibition of galactosyltransferases by a novel class of donor analogues, J. Med. Chem., 2012, 55, 2015–2024 CrossRef CAS PubMed.
- S. J. Lee, J. B. Park, D. Kim, S. H. Bae, Y. W. Chin, E. Oh and S. K. Bae, In vitro selective inhibition of human UDP-glucuronosyltransferase (UGT) 1A4 by finasteride, and prediction of in vivo drug-drug interactions, Toxicol. Lett., 2015, 232, 458–465 CrossRef CAS PubMed.
- P. A. Mann, A. Muller, L. Xiao, P. M. Pereira, C. Yang, S. H. Lee, H. Wang, J. Trzeciak, J. Schneeweis, M. M. Santos, N. Murgolo, X. She, C. Gill, C. J. Balibar, M. Labroli, J. Su, A. Flattery, B. Sherborne, R. Maier, C. M. Tan, T. Black, K. Onder, S. Kargman, F. J. Monsma Jr., M. G. Pinho, T. Schneider and T. Roemer, Murgocil is a Highly Bioactive Staphylococcal-Specific Inhibitor of the Peptidoglycan Glycosyltransferase Enzyme MurG, ACS Chem. Biol., 2013, 8, 2442–2451 CrossRef CAS PubMed.
- K. Chang, L. Lee, J. Chen and W. Li, Lithocholic acid analogues, new and potent α-2,3-sialyltransferase inhibitors, Chem. Commun., 2006, 629–631 RSC.
- C. Chiang, C. Wang, H. Chang, S. V. More, W. Li and W. Hung, A novel sialyltransferase inhibitor AL10 suppresses invasion and metastasis of lung cancer cells by inhibiting integrin-mediated signalling, J. Cell. Physiol., 2010, 223, 492–499 CAS.
- F. Moreau, N. Desroy, J. M. Genevard, V. Vongsouthi, V. Gerusz, G. Le Fralliec, C. Oliveira, S. Floquet, A. Denis, S. Escaich, K. Wolf, M. Busemann and A. Aschenbrenner, Discovery of new Gram-negative antivirulence drugs: Structure and properties of novel E. coli WaaC inhibitors, Bioorg. Med. Chem. Lett., 2008, 18, 4022–4026 CrossRef CAS PubMed.
- Y. Xu, R. Smith, M. Vivoli, M. Ema, N. Goos, S. Gehrke, N. J. Harmer and G. K. Wagner, Covalent inhibitors of LgtC: a blueprint for the discovery of non-substrate-like inhibitors for bacterial glycosyltransferases, Bioorg. Med. Chem., 2017, 25, 3182–3194 CrossRef CAS PubMed.
- L. Ni, H. A. Chokhawala, H. Cao, R. Henning, L. Ng, S. Huang, H. Yu, X. Chen and A. J. Fisher, Crystal Structures of Pasteurella multocida Sialyltransferase Complexes with Acceptor and Donor Analogues Reveal Substrate Binding Sites and Catalytic Mechanism, Biochemistry, 2007, 46, 6288–6298 CrossRef CAS PubMed.
- H. Yu, H. Chokhawala, R. Karpel, H. Yu, B. Wu, J. Zhang, Y. Zhang, Q. Jia and X. Chen, A Multifunctional Pasteurella multocida Sialyltransferase: A Powerful Tool for the Synthesis of Sialoside Libraries, J. Am. Chem. Soc., 2005, 127, 17618–17619 CrossRef CAS PubMed.
- R. J. Kelly, S. Rouquier, D. Giorgi, G. G. Lennon and J. B. Lowe, Sequence and Expression of a Candidate for the Human Secretor Blood Group α(1,2)Fucosyltransferase Gene (FUT2): Homozygosity for an Enzyme-Inactivating Nonsense Mutation Commonly Correlates with the Non-Secretor Phenotype, J. Biol. Chem., 1995, 270, 4640–4649 CrossRef CAS PubMed.
- A. Sarnesto, T. Kohlin, O. Hindsgaul, J. Thurin and M. Blaszezyk-Thurin, Purification of the secretor-type beta-galactoside alpha 1,2-fucosyltransferase from human serum, J. Biol. Chem., 1992, 267, 2737–2744 CAS.
- J. Jiang, V. Kanabar, B. Padilla, F. Man, S. C. Pitchford, C. P. Page and G. K. Wagner, Uncharged nucleoside inhibitors of β-1,4-galactosyltransferase with activity in cells, Chem. Commun., 2016, 52, 3955–3958 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00550d |
| This journal is © The Royal Society of Chemistry 2018 |