Open Access Article
Open Access ArticleElectrochemical recognition and quantification of cytochrome c expression in Bacillus subtilis and aerobe/anaerobe Escherichia coli using N,N,N′,N′-tetramethyl-para-phenylene-diamine (TMPD)†
S.
Kuss
a,
E. E. L.
Tanner
a,
M.
Ordovas-Montanes
b and
R. G.
Compton
 *a
*a
aDepartment of Chemistry, Physical and Theoretical Chemistry Laboratory, University of Oxford, South Parks Road, Oxford, OX1 3QZ, UK. E-mail: richard.compton@chem.ox.ac.uk
bDepartment of Zoology, University of Oxford, Woodstock Road, Oxford, OX2 6GG, UK
First published on 20th September 2017
Abstract
The colorimetric identification of pathogenic and non-pathogenic bacteria in cell culture is commonly performed using the redox mediator N,N,N′,N′-tetramethyl-para-phenylene-diamine (TMPD) in the so-called oxidase test, which indicates the presence of bacterial cytochrome c oxidases. The presented study demonstrates the ability of electrochemistry to employ TMPD to detect bacteria and quantify the activity of bacterial cytochrome c oxidases. Cyclic voltammetry studies and chronoamperometry measurements performed on the model organism Bacillus subtilis result in a turnover number, calculated for single bacteria. Furthermore, trace amounts of cytochrome c oxidases were revealed in aerobically cultured Escherichia coli, which to our knowledge no other technique is currently able to quantify in molecular biology. The reported technique could be applied to a variety of pathogenic bacteria and has the potential to be employed in future biosensing technology.
Introduction
Electrochemistry has become a successful tool for the analysis of biological samples, such as tissue and cancer cells,1 bacteria2 and enzymes.3 As some techniques are able to record currents in the femtoampere range, which relates to only a few thousand electrons transferred per second in a redox process,4 applications of electrochemistry to biological samples not only include the detection of pathogenic organisms,5 but also allow monitoring the release of messenger signals in tissue cells on a molecular level6 and inspire investigations of respiratory chain reactions.7 In such, a sequence of electron transport proteins govern the transport of electrons from substrate molecules onto molecular oxygen in aerobic organisms and cells to generate energy.8 As a redox-driven process, energy conversion in the respiratory chain of organisms is particularly interesting to investigate using electrochemistry, and information about oxygen consumption in living cells,9 as well as influences of compounds on the respiratory chain have been reported.7Quantitative analyses of metabolites released from organisms have been proposed in various ways in literature,10–12 and the expression of a turnover number has become a standard measure in enzymology,13 where it describes the maximum number of substrate molecules converted by an enzyme in a given time.14 The use of turnover numbers can also be applied to bacteria and their ability to enzymatically convert redox active substrate molecules, such as N,N,N′,N′-tetramethyl-para-phenylene-diamine (TMPD), per second at a given concentration during electrochemical measurements. TMPD is particularly interesting, as it is used in cell culture and microbiology to differentiate organisms that exhibit cytochrome c oxidase activity15 and to distinguish between Gram-negative and Gram-positive pathogenic and non-pathogenic bacteria.16,17 It has been well studied by electrochemistry18 and reveals a clean and reversible electrochemical behaviour.19 The concept of TMPD oxidation by microorganisms is widely used and is known as a successful colorimetric indicator for bacterial oxidases, as the radical cation TMPD+˙, formed by oxidation, shows a characteristic deep blue colour. Hence, its transfer to electrochemistry allows quantification of this process and might enable the future detection of various pathogens in vitro.
Herein, we report the electrochemical recognition of the oxidase test, by means of cytochrome c oxidase detection, and its quantitative analysis, expressed as a turnover number, calculated for single bacteria. The oxidation of TMPD by bacterial oxidases, specifically cytochrome c oxidase, a transmembrane protein that plays an important role as an electron acceptor in the respiratory electron transport chain,17 is monitored in Bacillus subtilis (B. subtilis) and Escherichia coli (E. coli). To the best of our knowledge this is the first time cytochrome c oxidase expression can be measured in aerobically grown E. coli, demonstrating the capabilities of electrochemistry applied to biological samples. E. coli are Gram-negative, rod-shaped bacteria, representing an ideal model organism for many studies in molecular biology, because of its high proliferation rate in growth medium in laboratory culture flasks.20 Being part of the normal gut flora in many vertebrates, but also being responsible for about 2 million deaths per year, related to intraintestinal and extraintestinal diseases, E. coli is known for both its commensalism and pathogenicity.21 In contrast to other Gram-negative bacteria, E. coli is a facultative anaerobic organism that, if grown in the presence of oxygen, cytochrome c oxidase expression cannot be detected by the oxidase test.22 The presented study demonstrates the ability of electrochemistry to detect trace amounts of electrochemically active substances and represents an accurate, fast and inexpensive analysis method for pathogenic and non-pathogenic bacteria, based on electroactive species converted by the organisms.
Results and discussion
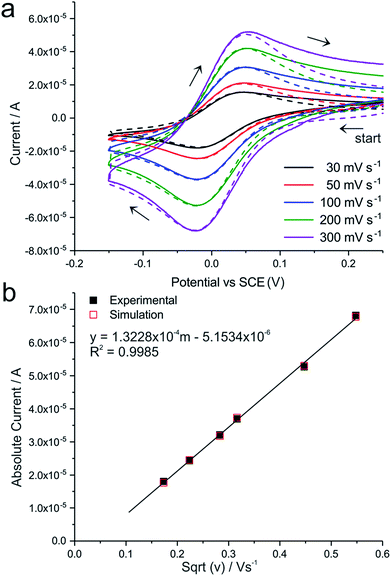
In this section, we first analyse and identify TMPD-BF4 as a suitable redox mediator for the electrochemical analysis of bacteria. As a proof of concept, we demonstrate the electrochemical recognition of the oxidase test in the species B. subtilis, resulting in a turnover number, calculated for single bacteria. Finally, we present the detection of cytochrome c oxidase in aerobically and anaerobically grown E. coli bacteria, emphasizing the sensitivity of this method.The compound N,N,N′,N′-tetramethyl-para-phenylene-diamine (TMPD) is rapidly oxidized by atmospheric oxygen, therefore the radical cation salt N,N,N′,N′-tetramethyl-para-phenylene-diamine tetrafluoroborate (TMPD-BF4) was synthesized, to assure solution stability and higher accuracy during bioelectrochemical measurements. TMPD-BF4 was synthesized following the procedure of Yamauchi et al.23 A product composition of ∼75% TMPD-BF4 and 25% TMPD in solution was determined (Fig. S1†), and its electrochemical behaviour was analysed using cyclic voltammetry. Fig. 1a shows cyclic voltammograms recorded using a 3 mm gold macroelectrode in 2.1 mM TMPD-BF4 in PBS buffer solution (0.17 M, pH 7.4a), which functions as supporting electrolyte. Various scan rates, ranging from 30 to 300 mV s−1, were applied. As the singly oxidized form of TMPD, the TMPD+˙ radical cation can be reduced to TMPD and oxidized to TMPD2+:
| TMPD+˙ + e− ⇄ TMPD | (1) |
| TMPD2+ + e− ← TMPD+˙ | (2) |
To account for minor solution changes related to the oxidation of TMPD by oxygen, the initial concentration ratio of TMPD to TMPD+˙ was slightly adjusted for each scan without changing the overall shape or position of the simulated curves. This allowed an offset correction for the presented curves and a capacitive contribution between 10 to 30 μF was attributed. From the simulations, the standard electrochemical rate constant ks was determined to be equal or greater than 0.025 cm s−1, and a standard electrode potential E0 of 0.013 (±0.001) V vs. SCE was determined. The resulting theoretical voltammograms (Fig. 1a, dotted lines) are in good agreement with the experimental measurements (full lines). The electrochemical characterization of TMPD-BF4, its solubility and stability make it a suitable redox mediator for the analysis of pathogenic and non-pathogenic bacterial oxidases.
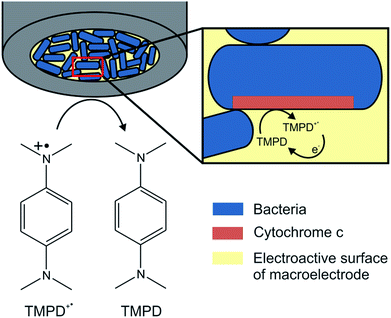
For the localized electrochemical recognition of cytochrome c oxidases in bacteria, target cultures were immobilized onto a gold macroelectrode by dropcasting and immersed into a solution of PBS, containing 1.7 mM TMPD+˙ (Fig. 2). The radical cation can be reduced electrochemically at the electrode to TMPD, providing the substrate for the target bacteria to oxidise and convert TMPD back to TMPD+˙.
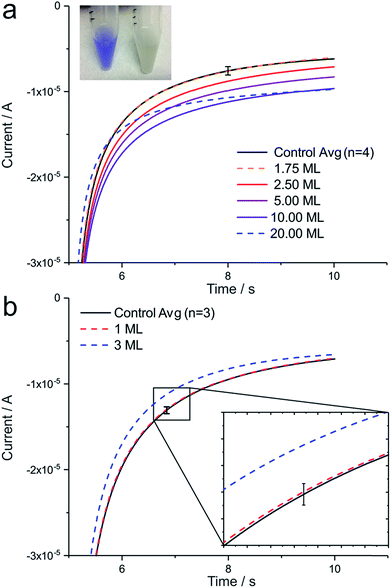
The chemical oxidation of TMPD to TMPD+˙ can be observed in a test tube as a colour change, as shown in Fig. 3a (inset), and is known as “oxidase test” in enzymology and microbiology. To transfer this process to an electrochemical set up, B. subtilis bacteria (cultured on agar) at different concentrations, ranging from 3.5 × 106 cells to 4 × 107 cells (1.75 to 20 monolayers (ML)), were dropcasted onto a working electrode. By applying an oxidative potential far exceeding the standard potential of TMPD during chronoamperometry, any remaining TMPD in the diffusion layer of the electrode is converted to TMPD+˙. An immediately following step potential to the reductive regime generates TMPD as an artificial electron donor for B. subtilis' cytochrome c oxidases. Fig. 3a shows an increase in electrochemical current in the presence of B. subtilis bacteria at the electrode, compared to the control, representing an unmodified electrode surface. A level of detection (LOD) of 5 × 106 bacteria at the electrode was determined, corresponding to a surface concentration of about 2.5 monolayers (ML), whereas electrode blockage was observed at a concentration of 20 ML. When cultured in broth instead of agar plates, B. subtilis results in a negative oxidase test (Fig. 3a, inset, test tube right) and can be employed as a negative control, as shown in Fig. 3b. Here, a concentration of 1 ML does not result in a measurable current increase and electrode blockage is already observed at a surface concentration of 3 ML. Error bars in all figures represent three times the standard deviation.
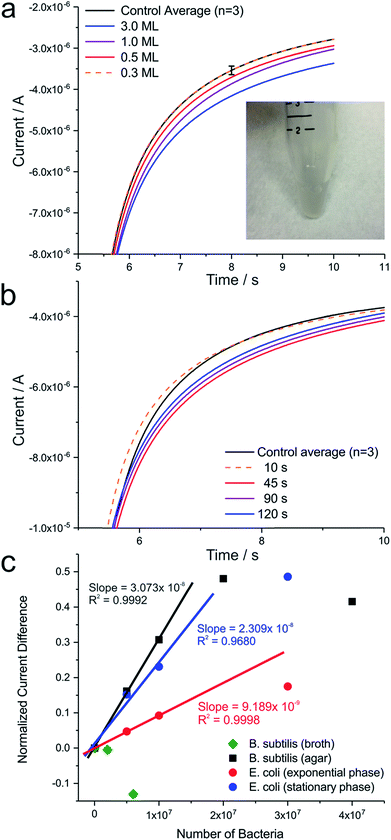
To transfer the previously established proof of concept to more pertinent, pathogenic bacteria, E. coli, cultured under aerobic conditions, were investigated. In cell culture, standard oxidase tests performed under these conditions result in a negative outcome, missing the characteristic blue colour indication of TMPD+˙ (Fig. 4a inset). Although E. coli bacteria do possess a ccm gene cluster in the aeg-46.5 operon region, which becomes important for the functional pathway involved in cytochrome c maturation,31 amounts of cytochrome c synthesised under aerobic bacteria growth cannot be detected by techniques in molecular biology, such as SDS-PAGE.32 However, the genomic information of E. coli strongly suggests that cytochrome c maturation in this organism follows a pathway similar to the one that has been shown for other bacteria.31 Furthermore, gradual gene activity levels as well as protein expression up and down regulation under conditions, such as oxygen deprivation have been reported in literature,33 in contrast to straight forward on or off switch mechanisms. In fact, cytochrome c-heme lyase activity was found to be present in aerobe E. coli,34 which gives reason to suspect a minimal expression of cytochrome c in E. coli, even in the presence of oxygen. Interestingly, as it can be seen in Fig. 4a, a current increase in the presence of E. coli at the macroelectrode was observed despite a negative classical colorimetric oxidase test, demonstrating how electrochemical techniques have the potential to outperform other methods in molecular biology.
Furthermore, Fig. 4b illustrates the dependency of the electrochemical current signal on an experimental delay time. After immobilization of E. coli bacteria, the electrode was placed in solution and kept for various delay times before a potential step was applied. At a delay time of 45 s, a maximum current response was achieved (Fig. 4b, red line), indicating that bacteria present require time to generate sufficient amounts of TMPD+˙ to be recognized at the electrode. If a delay time of 90 s or greater is applied, the concentration differences are thought to equilibrate and the current response decreases. The enhanced electrochemical signal in the presence of E. coli can also be recorded during cyclic voltammetry (Fig. S2†). However, due to the small current range detected, scan rates as low as 2 mV s−1 need to be applied in order to visualize the oxidation process of TMPD by the bacteria. At these slow scan rates contribution to the current signals by convective effects have to be assumed. Consequently, chronoamperometry measurements were chosen for quantitative analyses.
To promote an increased cytochrome c maturation in E. coli, bacteria were cultured to the stationary phase, which resulted in a positive oxidase test in E. coli (Fig. S3† inset), probably due to the competition for oxygen in solution. As expected, the increased cytochrome c oxidase activity under these growth conditions can be seen electrochemically and is presented in Fig. S3.†
As during dropcasting both living and dead bacteria of a solution are deposited at an electrode. Flow cytometry measurements were performed to estimate the percentage of living cells in solution, which contribute to the electrochemical signal (Fig. S4†). Analysis of B. subtilis revealed a cell viability of about 90%, whereas E. coli bacteria showed about 97% cell viability, which had to be respected for the calculation of a quantitative turnover number. Taking the electrochemical current during chronoamperometry (measurement point at 8 s), a turnover number was determined. At a bacteria count ranging from zero up to 107, this turnover number was observed to scale linearly with the number of bacteria (Fig. 4c). As such, a turnover number per single bacterium (Tsb) could be calculated using the equation
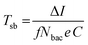 | (3) |
As a summary, Fig. 4c consolidates measurements in B. subtilis and E. coli under different growth conditions, representing the normalized current difference (current difference divided by the current recorded at the unmodified electrode) as a function of the number of bacteria immobilized at the macroelectrode. A linear relationship for currents obtained for samples containing up to 1 × 107 bacteria can be seen, before electrode blockage is observed at higher bacteria concentrations.
Experimental
Chemical reagents
All chemicals were purchased from Sigma-Aldrich, if not indicated otherwise. Phosphate buffered saline (PBS, 0.17 M) solution consists of 8 g sodium chloride (≥99%), 0.2 g potassium chloride (≥99%), 1.44 g sodium phosphate dibasic (≥99%), 0.24 g potassium phosphate dibasic (≥99%) and was completed to 1 L using nanopure water with a resistivity not less than 18.2 MΩ cm at 25 °C (Millipore water purification system).Synthesis of TMPD-BF4
The radical cation salt TMPD-BF4 was prepared following the method of Yamauchi et al.23 In short, TMPD (99%) was dissolved in 18 mL nanopure H2O and 24 mL methanol, containing 9 g sodium tetrafluoroborate (Alfa Aesar, UK, 97%). Aqueous bromine solution (32 mL, 0.252 mol kg−1) was added dropwise to the cooled TMPD solution (−10 °C). Resulting crystals were washed repeatedly with ice-cold methanol, followed by dry ether, and were recrystallized from methanol. Crystals appeared brownish purple, in accordance with literature,23 and a melting point of 125–127 °C was determined. Please see ESI for details.†Bacteria culture and oxidase test
For the oxidase test, 200 μL of a 1% (wt) N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD-2HCl) solution was added to 1 mL of cell suspension (1 × 107 cells μL−1). Images were taken within 30 s of solution mixture.
Electrochemical measurements
Electrochemical measurements were carried out using a modular potentiostat (PGSTAT302N, Autolab, UK). All experiments involving bacteria cultures were conducted at 37 °C inside a Faraday cage. An in-house fabricated gold microelectrode with a diameter of 6.9 μm was employed to determine the concentration of TMPD-BF4 in solution by cyclic voltammetry. Information about calculations can be found in the ESI.† Working electrodes were polished prior to experiments using a water–alumina mix (1.0, 0.3 and 0.05 μm, 30 seconds for each grade) on microcloth polishing pads (Buehler, USA).38 In all experiments, a standard calomel electrode and a platinum mesh were employed as reference and counter electrodes, respectively.Bioelectrochemical measurements
A 3 mm (diameter) gold macroelectrode (Alvatek Ltd, UK) was used as working electrode for all bioelectrochemical measurements. After polishing, sonication of the macroelectrode was applied for 2 minutes to assure removal of alumina powder from the electrode surface. Bacteria were immobilized onto the electrode by drop casting. For this purpose, the macroelectrode was placed in an electrode holder inside a 50 mL falcon tube, keeping it in upright position. Bacteria suspension was diluted so that all depositions consisted of 3 μL bacteria suspension representing monolayer concentrations of 0.5 to 30. The falcon tube was closed and bacteria suspension dried under N2 flow. Evaporation was monitored by eye and N2 flow was stopped the moment all liquid was evaporated. The electrode was removed from the holder and was placed in the electrochemical set up. The electrochemical cell remained thermostatted to 37 °C throughout the experiments.Chronoamperometry was carried out immediately after dropcasting of bacteria to minimize cell death at the electrode. In case of E. coli organisms, a delay time of 45 s was applied after the electrode was brought in contact with the solution, holding the electrode at open circuit potential. An oxidative potential of 250 mV was applied for 5 s, followed by a potential step to a reductive regime of −150 mV, which was held for 5 s also. All potentials are indicated vs. a saturated calomel reference electrode (SCE).
Conclusions
We have successfully demonstrated that the colorimetric oxidase test, commonly employed in cell and microbiology, can be transferred to an electrochemical set up and the expression of cytochrome c oxidase was quantified in the model organism B. subtilis, resulting in a turnover number Tsb of 9.8 (±0.3) × 108 L mol−1 s−1 for single bacteria. Furthermore, we conclude that although grown aerobically, E. coli bacteria show trace amounts of cytochrome c oxidase, whose activity can be detected electrochemically, revealing a Tsb of 3.8 (±0.06) × 108 L mol−1 s−1 (about 39% compared to B. subtilis). The electrochemical recognition of the TMPD oxidation by bacterial oxidases can be applied to a variety of pathogens. Improving the experimental set up to lower the LOD to a competitive range and transferring the presented principle onto a selective device, the detection of pathogenic bacteria, such as Neisseria meningitidis, or bacteria related to sexually transmitted infections, is anticipated.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Prof. Dr C. J. Schofield for providing the E. coli bacteria cultures, as well as Adam Hardy and Dr Matteo Ferla for technical assistance. The authors thank Prof. Dr Kayla King for providing lab space and materials for the bacteria culture of B. subtilis. S. K. thanks the support from the European Commission under the Marie Curie Programme (grant number 702009). The contents reflect only the authors' views and not the views of the European Commission. The research leading to these results has received partial funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement no. 320403.References
- K. L. Adams, M. Puchades and A. G. Ewing, Annu. Rev. Anal. Chem., 2008, 1, 329–355 CrossRef CAS PubMed
.
- D. Ivnitski, I. Abdel-Hamid, P. Atanasov, E. Wilkins and S. Stricker, Electroanalysis, 2000, 12, 317–325 CrossRef CAS
.
- E. Y. Jomma and S.-N. Ding, Curr. Anal. Chem., 2016, 12, 5–21 CrossRef CAS
.
- C. Batchelor-McAuley, J. Ellison, K. Tschulik, P. L. Hurst, R. Boldt and R. G. Compton, Analyst, 2015, 140, 5048–5054 RSC
.
- L. Yang and R. Bashir, Biotechnol. Adv., 2008, 26, 135–150 CrossRef CAS PubMed
.
- S. Bergner, P. Vatsyayan and F.-M. Matysik, Anal. Chim. Acta, 2013, 775, 1–13 CrossRef CAS PubMed
.
- A. J. Bard, X. Li and W. Zhan, Biosens. Bioelectron., 2006, 22, 461–472 CrossRef CAS PubMed
.
-
A. Lehninger, D. Nelson and M. Cox, Lehninger Biochemie, Springer-Verlag, Berlin, Heidelberg, New York, 3rd edn, 2001 Search PubMed
.
- M. Nebel, S. Grützke, N. Diab, A. Schulte and W. Schuhmann, Angew. Chem., Int. Ed., 2013, 52, 6335–6338 CrossRef CAS PubMed
.
- X. Li, J. Dunevall and A. G. Ewing, Acc. Chem. Res., 2016, 49, 2347–2354 CrossRef CAS PubMed
.
- A. Yakushenko, E. Kätelhön and B. Wolfrum, Anal. Chem., 2013, 85, 5483–5490 CrossRef CAS PubMed
.
- S. Kuss, D. Polcari, M. Geissler, D. Brassard and J. Mauzeroll, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9249–9254 CrossRef CAS PubMed
.
- C. Chan, L. Sepunaru, S. V. Sokolov, E. Kätelhön, N. P. Young and R. G. Compton, Chem. Sci., 2017, 8, 2303–2308 RSC
.
-
Industrial Catalysis: A Practical Approach, ed. J. Hagen, Wiley-VCH, Weinheim, 2nd edn, 2006 Search PubMed
.
- G. N. Nobre, M. J. Charrua and M. M. Silva, J. Med. Microbiol., 1987, 23, 359–361 CrossRef CAS PubMed
.
-
H. D. Isenberg, Clinical Microbiology Procedures Handbook, American Society for Microbiology, 2004 Search PubMed
.
-
J. F. MacFaddin, Biochemical Tests for Identification of Medical Bacteria, Lippincott Williams and Wilkins, 3rd edn, 2000 Search PubMed
.
- L. Michaelis, M. P. Schubert and S. Granick, J. Am. Chem. Soc., 1939, 61, 1981–1992 CrossRef CAS
.
- D. Menshykau, I. Streeter and R. G. Compton, J. Phys. Chem. C, 2008, 112, 14428–14438 CAS
.
-
B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Molecular Biology of the Cell, Taylor & Francis Books, Inc., 4th edn, 2002 Search PubMed
.
- O. Tenaillon, D. Skurnik, B. Picard and E. Denamur, Nat. Rev. Microbiol., 2010, 8, 207–217 CrossRef CAS PubMed
.
- J. Gordon and J. W. McLeod, J. Pathol. Bacteriol., 1928, 31, 185–190 CrossRef CAS
.
- J. Yamauchi and H. Fujita, Bull. Chem. Soc. Jpn., 1990, 63, 2928–2932 CrossRef CAS
.
-
D. A. C. Brownson and C. E. Banks, in The Handbook of Graphene Electrochemistry, Springer-Verlag London Ltd., London, 2014 Search PubMed
.
- M. Rudolph, J. Electroanal. Chem. Interfacial Electrochem., 1991, 314, 13–22 CrossRef CAS
.
- M. Rudolph, J. Electroanal. Chem., 1992, 338, 85–98 CrossRef CAS
.
- M. Rudolph, J. Electroanal.
Chem., 1994, 375, 89–99 CrossRef CAS
.
- M. Rudolph, D. P. Reddy and S. W. Feldberg, Anal. Chem., 1994, 66, 589A–600A CrossRef CAS
.
-
M. Rudolph, in Physical electrochemistry: Principles, methods and applications, ed. I. Rubinstein, Marcel Dekker, New York, 1995 Search PubMed
.
- S. Kuss and R. G. Compton, Electrochim. Acta, 2017, 242, 19–24 CrossRef CAS
.
- L. Thony-Meyer, F. Fischer, P. Kunzler, D. Ritz and H. Hennecke, J. Bacteriol., 1995, 177, 4321–4326 CrossRef CAS PubMed
.
- C. Iobbi-Nivol, H. Crooke, L. Griffiths, J. Grove, H. Hussain, J. Pommier, V. Mejean and J. A. Cole, FEMS Microbiol. Lett., 1994, 119, 89–94 CrossRef CAS PubMed
.
- J. S. Chang, T. Wendt, W. Qu, L. Kong, Y. S. Zou, A. M. Schmidt and S.-F. Yan, Circ. Res., 2008, 102, 905 CrossRef CAS PubMed
.
- C. von Wachenfeldt and L. Hederstedt, FEBS Lett., 1990, 270, 147–151 CrossRef CAS PubMed
.
- Agilent Technologies, E. coli Cell Culture Concentration from OD600, http://www.genomics.agilent.com/biocalculators/calcODBacterial.jsp, accessed 1 June 2017.
- A. J. Fisher, T. N. Rosenstiel, M. C. Shirk and R. Fall, Anal. Biochem., 2001, 292, 272–279 CrossRef CAS PubMed
.
- M. C. Arrieta, B. K. Leskiw and W. R. Kaufman, Exp. Appl. Acarol., 2006, 39, 297–313 CrossRef CAS PubMed
.
- T. J. Cardwell, J. Mocak, J. H. Santos and A. M. Bond, Analyst, 1996, 121, 357–362 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc03498a |
| This journal is © The Royal Society of Chemistry 2017 |