Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Catalysis of a 1,3-dipolar reaction by distorted DNA incorporating a heterobimetallic platinum(II) and copper(II) complex†
Iván
Rivilla
 a,
Abel
de Cózar
a,
Abel
de Cózar
 ab,
Thomas
Schäfer
ab,
Thomas
Schäfer
 bc,
Frank J.
Hernandez
bc,
Frank J.
Hernandez
 c,
Alexander M.
Bittner
c,
Alexander M.
Bittner
 cd,
Aitziber
Eleta-Lopez
cd,
Aitziber
Eleta-Lopez
 d,
Ali
Aboudzadeh
d,
Ali
Aboudzadeh
 c,
José I.
Santos
c,
José I.
Santos
 e,
José I.
Miranda
e,
José I.
Miranda
 e and
Fernando P.
Cossío
e and
Fernando P.
Cossío
 *a
*a
aDepartment of Organic Chemistry I, Centro de Innovación en Química Avanzada (ORFEO-CINQA), Universidad del País Vasco/Euskal Herriko Unibertsitatea (UPV/EHU), Donostia International Physics Center (DIPC), Po Manuel Lardizabal 3, E-20018 Donostia/San Sebastián, Spain. E-mail: fp.cossio@ehu.es
bIkerbasque, Basque Foundation for Science, Ma Díaz de Haro 3, E-48013 Bilbao, Spain
cNanoBioSeparations Group, POLYMAT University of the Basque Country (UPV/EHU), Avda. Tolosa 72, E-20018 Donostia/San Sebastián, Spain
dCIC NanoGUNE, Consolider. Tolosa Hiribidea, 76, E-200018 Donostia/San Sebastián, Spain
eSGIker NMR Facility, Universidad del País Vasco/Euskal Herriko Unibertsitatea (UPV/EHU), Avda. Tolosa 72, E-20018 Donostia/San Sebastián, Spain
First published on 22nd August 2017
Abstract
A novel catalytic system based on covalently modified DNA is described. This catalyst promotes 1,3-dipolar reactions between azomethine ylides and maleimides. The catalytic system is based on the distortion of the double helix of DNA by means of the formation of Pt(II) adducts with guanine units. This distortion, similar to that generated in the interaction of DNA with platinum chemotherapeutic drugs, generates active sites that can accommodate N-metallated azomethine ylides. The proposed reaction mechanism, based on QM(DFT)/MM calculations, is compatible with thermally allowed concerted (but asynchronous) [π4s + π2s] mechanisms leading to the exclusive formation of racemic endo-cycloadducts.
Introduction
The main role of DNA is the storage of genomic information leading to the biosynthesis of proteins via diverse forms of RNA.1 In turn, proteins play multiple roles in living systems, catalysis being among the most important ones. This general pattern has been refined as a consequence of the discovery of catalytic RNAs (ribozymes)2 and DNAs (deoxyribozymes).3 However, since DNA is structurally less versatile than RNA and proteins, additional molecules and functional groups are required to expand the catalytic space of DNA. Roelfes and Feringa4 brilliantly demonstrated the feasibility of this concept by intercalation of aza-chalcones in the double helix and subsequent coordination to copper salts. Further work5 demonstrated that various intercalating heterocycles and metallic salts in the presence of DNA are able to promote, among other transformations, Diels–Alder reactions,4,6 Friedel–Crafts alkylations7 and Michael8,9 (including oxa-Michael)10,11 additions. It is remarkable that all these reactions were carried out in water, although in several cases the system tolerated organic co-solvents.7 Other DNA activation methods include covalent attachment of active catalytic sites such as proline organocatalysts,12 as well as Ir(I)13 and Pt(II)14 complexes.Despite the relevance of (3 + 2) cycloadditions in the chemical synthesis of five-membered rings,15 the enzymatic version of this reaction has not been identified in living systems.16,17 Only a very recent example of a possible enzymatic 1,3-dipolar reaction has been reported to date.18 In addition, nonenzymatic 1,3-dipolar reactions have been postulated in the biosynthesis of several alkaloids19 and natural products such as furanocembranoids20,21 and santolin Y.22 Therefore, to the best of our knowledge a biomolecule-assisted bona fide (3 + 2) cycloaddition between azomethine ylides and alkenes to produce unnatural proline derivatives has not been reported to date. Within this context, we herein describe the first example of a DNA-assisted 1,3-dipolar reaction in water.
Results and discussion
The design of the covalent modification of DNA was based on the ability of Pt(II) chemotherapeutic drugs to bind mainly 1,2-intrastrand GpG units,23,24 thus providing a concave-convex distortion of the double helix that could mimic the active sites of metalloenzymes. In previous work25 we reported new chiral ligands that can bind Cu(II) salts26 and efficiently catalyze (3 + 2) cycloadditions involving azomethine ylides. Therefore, we reasoned that a DNA–Pt(II)–Cu(II) heterobimetallic complex similar to that depicted in Scheme 1 could catalyze this reaction.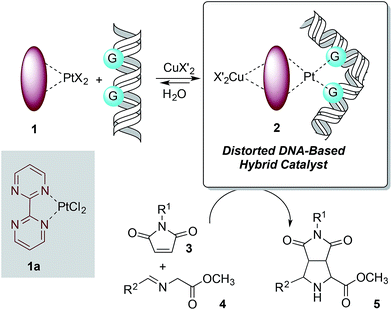 | ||
| Scheme 1 Basic design of DNA-based catalysts for 1,3-dipolar reactions. The distortion of DNA by coordination of G bases with Pt(II) is shown. | ||
First, we tested the feasibility of this basic design using computational methods. As starting point we took the crystal structure reported by Takahara et al.27 for a cisplatin-double-stranded oligodeoxynucleotide complex (pdb code: 1aio), where G*G* denotes a 1,2-intrastrand cis-[(H3N)2Pt-d(GpG)] crosslink. To this structure 2,2′-bipyrimidin (bipym) was added and the azomethine ylide derived from methyl (E)-2-(benzylidene-amino)acetate 4a was coordinated to a copper(II) metallic centre. The resulting structure was stabilized by incorporating 509 water molecules and 22 sodium cations (Fig. 1a). The whole ensemble was optimized using a hybrid QM/MM ONIOM28–30 scheme (ESI†). The full structure thus optimized was found to keep the folded geometry of the distorted double helix, where the Cu(II)–bipym–Pt(II)–G*G* system generated a cavity to which the azomethine ylide was coordinated.
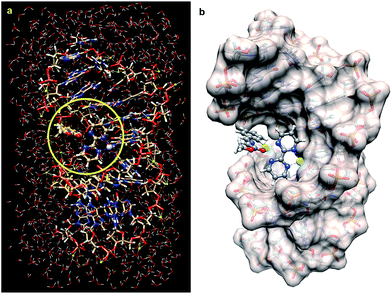 | ||
| Fig. 1 (a) Fully optimized (B3LYP/LanL2DZ:UFF level of theory) structure of a DNA double strand containing the d(CCTCTG*G*TCTCC)-d(GGAGACCAGAGG) sequence, in which the complex 1a is bound to the G*pG* unit and to the N–Cu(II) azomethine ylide derived from imine 4a (R2 = Ph, Scheme 1), surrounded by 509 water molecules and 22 sodium cations. The QM part of the optimization is within the circle highlighted in yellow. (b) The same optimized structure but showing the solvent accessible surface of the DNA fragment. The water molecules have been removed for clarity. Pt(II) and Cu(II) atoms are represented in light blue and green, respectively. | ||
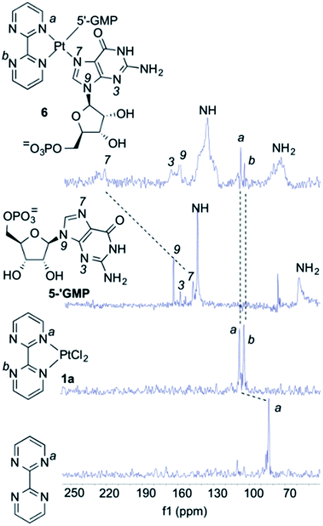
As it can be seen by inspection of Fig. 1b, the resulting ensemble closely resembles the active site of a metalloenzyme. As a proof-of-concept experiment, we next examined the ability of (bipym)PtCl2 complex 1a to bind two equivalents of guanosine. Since 1a and its derivatives posed solubility problems, we monitored the different species in the solid state using Cross Polarization-Magic Angle Spinning (CP-MAS) spectroscopy.31 The NMR spectrum of bipym showed only one 15N-NMR signal, as expected from its D2h symmetry (Fig. 2). In contrast, the two nitrogen atoms of 1a coordinated to Pt(II) could be readily distinguished. Then, we used 5′-GMP as a suitable equivalent of G units in DNA and analysed the 1H–15N CP-MAS spectra at different mixing times to assign the five nitrogen atoms of the G-unit. Combination of 1a and 5′-GMP resulted in the formation of an adduct whose 15N chemical shift associated with the N7 atom of the purine system was considerably deshielded with respect to the signal recorded for 5′-GMP (Fig. 2).
We concluded that Pt(II) complex interacts with guanine units yielding a square planar complex, in which two equivalent G units bind the Pt(II) centre by means of the respective N7 atoms of the purine bicycle. We next studied the interaction between 1a and oligodeoxynucleotides in order to assess the binding abilities of different DNA sequences.
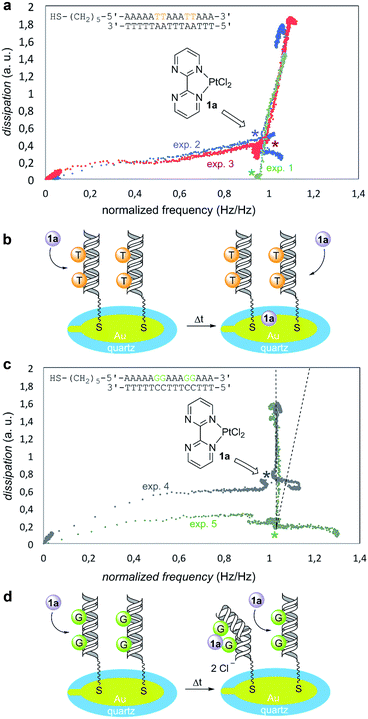
To this end, we used a quartz-crystal microbalance with dissipation monitoring (QCM-D) device.32 These QCM-D experiments measured changes in frequency, which correlate with mass adsorption on the sensor surface, and the dissipation of energy of the adsorbed film, which in turn correlates with its viscoelasticity.33,34 The dissipation vs. frequency points were collected for various times and ordered graphically from left to right and from bottom to top (Fig. 3). In a reference experiment denoted as “exp. 1” in Fig. 3a, the Pt(II) complex 1a was injected on bare gold (no DNA immobilized) in PBS buffer. Under these conditions, dissipation increased linearly with frequency, thus resulting in an incremental dissipation entirely correlated with the mass adsorbed on the surface. To carry out the experiments in the presence of DNA, we selected two different model oligodeoxynucleotides on the basis of the well-known binding ability of GpG pairs with platinum drugs.23,24 We tested in duplicate the oligomer containing 5′-thiol-AAAAATTAAATTAAA-3′ binding sequence (Fig. 3a, experiments 2 and 3). An overlay with the reference experiment 1 revealed that this response was indeed non-specific as both experiments showed practically identical profiles with respect to the reference. We interpreted these results as negative experiments in which there was no significant interaction between Pt(II) complex 1a and G-free deoxyoligonucleotide (Fig. 3b). We next examined the behavior of the oligomer containing 5′-thiol-AAAAAGGAAAGGAAA-3′ binding sequence (Fig. 3c, experiments 4 and 5). In this case, upon addition of 1a both plots showed an identical increase in dissipation while simultaneously no frequency change was observed, in sharp contrast with the previous blank and negative experiments 1–3. This indicated that first, there was no measurable non-specific binding occurring as it would be detected as a change in frequency (see the dotted lines in Fig. 3c); second, injection of 1a induced a strong change in dissipation, which can be only attributed to conformational changes taking place in the GpG pairs-containing immobilized and hybridized deoxyoligo-nucleotide. From these results, we concluded that GpG-containing DNA interacts specifically with Pt(II) complex 1a resulting in an increasingly more dissipative film, which in turn correlates with additive conformational changes of DNA upon addition of 1a (Fig. 3d).
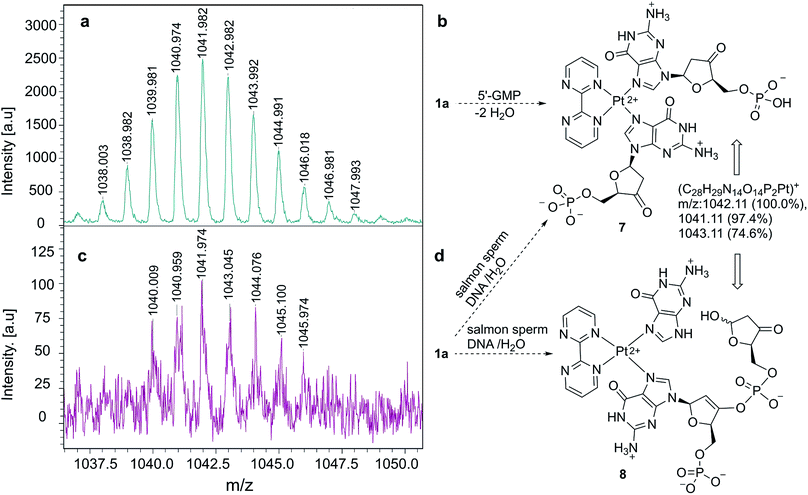
Once we verified that complex 1a binds selectively G-containing oligonucleotides, we tested its binding ability to DNA strands. As a control experiment, we mixed 1a with 5′-GMP and the resulting complex was analyzed by MALDI-TOF mass spectrometry.35,36 We observed an ensemble of high mass-to-charge (m/z) signals distributed around a value of m/z ≈ 1042 a.u., as shown in Fig. 4a. These signals correlate satisfactorily with complex 7 (Fig. 4b), closely related to 6 (Fig. 2), in which two ketone units have been generated via dehydration-tautomerization of one hydroxy group of each ribose unit. After this control experiment, the same protocol was followed, but instead of 5′-GMP we used DNA sodium salt from salmon sperm (salmon sperm DNA in Fig. 4) with a % G-C content of 41.2% and a molecular mass of 1.3 × 106 Da (ca. 2000 bp). In this case, the same profile was observed in the corresponding MALDI-TOF mass spectrum (Fig. 4c), with a high m/z ensemble centered at ca. 1042 a.u. This response was interpreted in terms of ion 7 or, for two consecutive GpG units in the starting salmon sperm DNA, as the keto-enol phosphoric ester depicted as ion 8 in Fig. 4d. On the basis of these experiments, we concluded that the 1a–G2 complexes observed in monomeric and oligomeric G-containing species can be extended to double strain DNA.
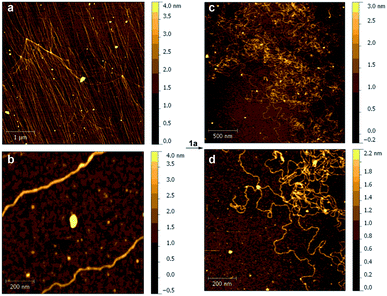
We also studied the effect of Pt(II) complex 1a on the structure of DNA strands. Thus, samples of DNA (ca. 48 kb), from λ phage-infected E. coli were analyzed by atomic force microscopy (AFM).37 The corresponding DNA strands were unambiguously identified on oxidized silicon38 by the corresponding AFM images (Fig. 5a and b). When 1a was added, the AFM scans showed large morphological changes consisting of local kinks and crosslinks (Fig. 5c and d). On the basis of the previously presented experiments, these changes were attributed to the formation of intra- and interstrand cis-{(bipym)Pt(d[GpG + GpXpG + GpA])} adducts.39
These advanced analytical studies permitted us to establish the main features of the Pt(II)-mediated binding between molecule 1a and DNA. We concluded that the thus generated hybrid system could catalyse 1,3-dipolar reactions provided that (i) the heterobimetallic Cu(II)–Pt(II) centre is able to generate in situ the required N-metallated azomethine ylide derived from the corresponding imine 4; and (ii) the active site can accommodate the dipolarophile 3.
In order to test the catalytic ability in (3 + 2) cycloadditions of the DNA–heterobimetallic complex, we prepared catalyst 2 (Scheme 1) by using salmon sperm DNA in a buffered solution of (N-morpholino)propane-sulfonic acid (MOPS), to which 1a was added, followed by copper(II) triflate, triethylamine and the corresponding maleimide 3 and imine 4 (Scheme 2). After six days of reaction at room temperature, nine distinct endo-(3 + 2)-cycloadducts (5aa–5bf, Table 1) could be generated. This endo stereochemistry was secured on the basis of the spectroscopic data and, in two cases, by X-ray diffraction analysis (ESI†).
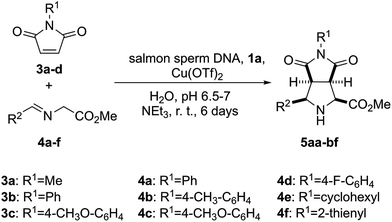 | ||
| Scheme 2 Synthesis of (3 + 2) endo-cycloadducts 5aa–5bf catalysed by the heterobimetallic complex formed by salmon sperm DNA and 1a in the presence of copper(II) triflate. | ||
| Entry | R1 (3) | R2 (4) | 5 | Yielda (mmol × 10−3) [%] |
|---|---|---|---|---|
| a Yields of isolated pure products correspond to averaged values obtained after three experiments. The limiting reactant was the corresponding maleimide 3 (0.05 mmol). b Structure confirmed by X-ray diffraction analysis (ESI).40 | ||||
| 1 | Me (3a) | Ph (4a) | 5aa | 10.5 ± 0.4 [21] |
| 2 | Ph (3b) | Ph (4a) | 5ba | 18.0 ± 0.5 [36] |
| 3 | 4-MeO–C6H4 (3c) | Ph (4a) | 5ca | 10.0 ± 0.8 [20] |
| 4 | Ph (3b) | 4-Me–C6H4 (4b) | 5bb | 17.9 ± 0.4 [23] |
| 5 | Ph (3b) | 4-MeO–C6H4 (4c) | 5bc | 17.5 ± 0.3 [35] |
| 6 | Ph (3b) | 4-F-C6H4 (4d) | 5bd | 12.5 ± 0.2 [25] |
| 7 | Ph (3b) | Cyclohexyl (4e) | 5be | 13.5 ± 0.4 [27] |
| 8b | Ph (3b) | 2-Thyenyl (4f) | 5bf | 16.0 ± 0.9 [32] |
In order to assess the relevance of each component of the catalytic system we tested 17 possibilities resulting from the combination of all the reagents except at least one (see Table S1 of the ESI†). In all these control experiments no 1,3-dipolar reaction was observed. In particular, when different combinations of triethylamine and the Cu(II) and Pt(II) salts were tested in the absence of salmon sperm DNA, the reaction did not proceed. Similarly, different combinations in the presence of salmon sperm DNA but in the absence of base, 2,2′-bipyrimidine and/or one of the metals did not produce any (3 + 2) cycloadduct. It is interesting to note that the combination of DNA, 2,2′-bipyrimidine and Cu(OTf)2 in the absence of Pt(II) was also unproductive (see Table S1 of the ESI,† entry 13), thus indicating that the powerful Roelfes–Feringa method consisting of a metallated intercalating heterocycle cannot catalyse this challenging 1,3-dipolar cycloaddition. In addition, these experiments demonstrate that there is no background reaction (see in particular entry 1 of Table S1 of the ESI†). In summary, these control experiments demonstrated that combination of 2,2′-bipyrimidine, Pt(II), Cu(II) and DNA, most likely by bonding to consecutive GG units, is required to achieve moderate yields of (3 + 2) racemic endo-cycloadducts 5 (see Table S1 of the ESI,† entry 10).
We interpreted our results as follows: Previously formed DNA-1a adducts bound Cu(OTf)2 and the resulting heterobimetallic complex 2a (Fig. 5a) coordinated imine 4 to form intermediate species INT1, from which the corresponding N-metallated azomethine ylide INT2 was formed via triethylamine-assisted deprotonation. This 1,3-dipole interacted with dipolarophile 3 to form the corresponding (3 + 2) cycloadduct and regenerating INT1via interaction with another equivalent of imine, thus completing the catalytic cycle. In order to understand the origins of the endo selectivity we optimized the possible endo- and exo-transition structures under the same computational framework used to optimize the structure of INT2 (Fig. 5b and c). Both saddle points were found to be quite asynchronous but still associated with a concerted [π4s + π2s] symmetry allowed mechanism, as indicated by the bond distances corresponding to the formation of the two new σ-bonds. We also found that endo-TS was stabilized by a strong electrostatic interaction between the nitrogen atom of the imide moiety and the metallic centre (Fig. 5b). As a result, exo-TS was calculated to be ca. 13 kcal mol−1 higher in energy than its endo congener, thus predicting the preferential formation of cycloadduct endo-5aa under kinetic control, in nice agreement with the experimental data.
We performed similar calculations in the absence of DNA. In these studies, the features of the computational model (including surrounding water molecules, see the ESI†) remained identical. The results are gathered in Fig. 6d and e. We observed that in the presence of Cu(II) and bipyrimidine, the chief features of the transition structure leading to endo-5aa are similar to those found for the 3a + 4a → 5aa reaction in the presence of DNA. However, the activation energy was found to be ca. 12 kcal mol−1 higher, which corresponds to a k(DNA–Cu–Pt)/k(Cu) ratio of ca. 4.8 × 108 (Fig. 6d). When the square planar diaqua–Pt(II) moiety was incorporated to the reaction coordinate (Fig. 6e), the shape of the corresponding saddle point did not change significantly and the activation energy was slightly lower than in the previous case, with a calculated k(DNA–Cu–Pt)/k(Cu–Pt) ratio of ca. 2.7 × 107. It is interesting to note that the DNA-free simulations also predict the preferential formation of the endo-cycloadduct (see Fig. S3 and Table S3 of the ESI†). These results are in agreement with the absence of reactivity observed when DNA was not present. We interpret the lower activation energies in the presence of DNA in terms of the destabilization of the substrate by restriction of the conformational freedom and by the electrostatic repulsion between the anionic polyphosphate environment and the anionic part of the starting 1,3-dipole, which is alleviated along the reaction coordinate leading to the non-zwitterionic cycloadduct.
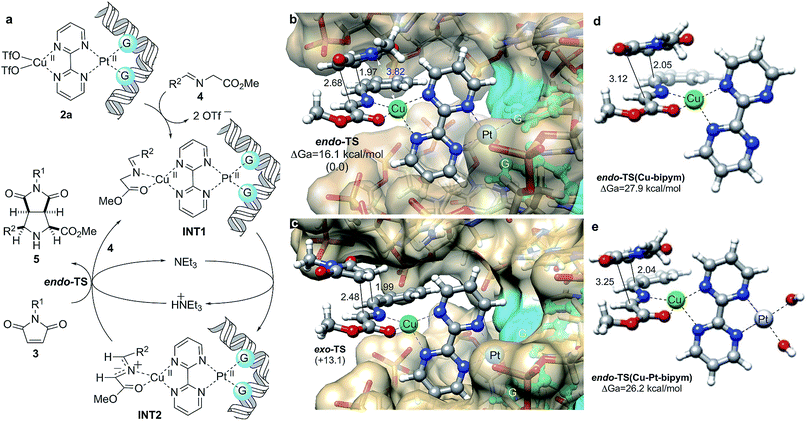 | ||
| Fig. 6 (a) Proposed catalytic cycle for (3 + 2) cycloadditions between azomethine ylides INT1 and maleimides 3 catalyzed by in situ formed DNA–bipym–Pt(II)–Cu(II) complexes. (b, c) Fully optimized geometries and relative energies of endo- and exo- transition structures (TS) associated with the 3a + 4a → 5aa reaction. Numbers in parentheses are the relative energies, in kcal mol−1. Bond distances are given in Å. Guanine units bound to Pt(II) are highlighted in blue. Calculations were performed at the B3LYP/LanL2DZ:UFF level of theory as indicated in Fig. 1b. Water molecules and sodium cations were included in the calculations (MM level, UFF force field) but are not shown for clarity. (d, e) Same transition structures associated with the formation of cycloadduct endo-5aa in the absence of DNA and Pt(II) (d), and in the absence of DNA (e). In the latter case, two binding water ligands have been included. Also in (d) and (e), water molecules were included in the calculations (MM level, UFF force field, not shown for clarity). | ||
Conclusions
In this work we have shown that (2,2′-bipym)PtCl2 complex 1a binds 5′-GMP by its N7 atom. In addition, oligomers containing GpG pairs can interact selectively with 1a, in line with the major formation of cis-[Pt(II){GpG}] complexes in platinum drugs. Moreover, 1a promotes significant structural changes in λ-DNA, resulting in highly distorted microenvironments around the metallic centres, most likely because of the formation of cis-[Pt(II){GpG} + {GpXpG} + {GpA}] adducts. On the basis of these experiments, we conclude that a suitable bimetallic complex based on Pt(II) can distort the double helix of DNA to generate an active site similar to those found in well-known metalloenzymes, as supported by QM/MM calculations. Finally, we have shown that the system formed by 1a, salmon sperm DNA and Cu(OTf)2 catalyses (3 + 2) cycloadditions between azomethine ylides (formed in situ from the corresponding imines) and maleimides. Only the racemic endo-cycloadducts are obtained, probably because of competing geometries of the intermediate N-metallated azomethine ylides linked to the 1a–DNA hybrid system. This result demonstrates that modified biomolecules can catalyse chemical reactions in water, for which there is no equivalent in living systems.Experimental section
Computational methods
All the stationary points were fully optimized and characterized (harmonic analysis) within the QM/MM scheme using B3LYP hybrid functional.29 LANL2DZ basis set and effective core potential,24 as well as UFF force field25 were used within the ONIOM28 framework as implemented in the Gaussian09 (ref. 30) suite of programs.NMR experiments
Liquid NMR spectra were recorded on either a Bruker Avance 500 MHz or 400 MHz spectrometer at standard temperature and pressure, equipped with an z gradient BBO probe. The 15N spectra were recorded using an inverse gated sequence at 40.54 MHz, with a time domain of 32k, a spectral width of 16 kHz, an interpulse delay of 5 s and an acquisition time of 1 s. DMSO-d8 was used as internal standard for 1H and MeNO2 (376.86 ppm “Bruker scale”) as external reference for 15N NMR spectra. Solid State NMR spectra were recorded on a Bruker 400 AVANCE III WB spectrometer 9.40 T (1H = 400 MHz). 13C CP-MAS and 1H spectra were collected by using a 4 mm CP-MAS probe at a spinning of 10 kHz. These experiments were carried out using the standard pulse sequence at 100.6 MHz, with a time domain of 2 K, a spectral width of 29 kHz, a contact time of 1.5 ms and an interpulse delay of 5 s for 13C spectra. A nominal frequency of 400 MHz was used for 1H, employing the DUMBO pulse sequence, a time domain of 2 K, a spectral width of 20 kHz and an interpulse delay of 5 s. 15N solid state spectra were collected by using a 7 mm MASDVT probe at a spinning of 5 KHz. 15N CP-MAS NMR spectra were recorded using a standard pulse sequence at 40.56 MHz, a time domain of 1k, a spectral width of 96 kHz, a contact time of 2 ms and an interpulse delay of 5 s.MALDI-TOF mass spectrometry
Matrix Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) measurements were performed on a Bruker Autoflex Speed system (Bruker, Germany) equipped with a 355 nm NdYAG laser. Spectra were acquired in the positive reflection mode. trans-2-[3-(4-tert-Butylphenyl)-2-methyl-2-propenylidene] malonitrile (DCTB, Fluka) was used as a matrix. Sodium trifluoroacetate and silver trifluoroacetate (Aldrich) were added as the cationic ionization agents (10 g L−1 dissolved in THF). The matrix was also dissolved in THF at a concentration of 20 g L−1. All the samples were dissolved in water at a concentration. The matrix and salt were premixed in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Approximately 0.5 μL of the sample were hand spotted on the ground steel target plate and 0.5 μL of the matrix/salt obtained mixture were quickly added to the hand spotted samples. For each spectrum 5000 laser shots were accumulated.
1 ratio. Approximately 0.5 μL of the sample were hand spotted on the ground steel target plate and 0.5 μL of the matrix/salt obtained mixture were quickly added to the hand spotted samples. For each spectrum 5000 laser shots were accumulated.
AFM experiments
DNA samples were prepared from lyophilized lambda phage DNA, methylated from E. coli host strain W3110 (Mw. 31.5 × 103 kDa, 48 kb, from sigma Sigma-Aldrich). DNA solutions (0.02 mg mL−1 in (N-morpholino)propanesulfonic acid, MOPS, 10 mM) were prepared and in each experiment 10 μL were deposited on the freshly cleaned silicon oxide surface. Then 10 μL of DNA solution was deposited on a silicon wafer, freshly hydrolyzed with oxygen plasma. Immediately afterwards the solution droplet was softly blown with a nitrogen stream. A saturated solution of compound 1a in Millipore water (18 Mohm cm, TOC < 5 ppb) was prepared and a droplet of 20 μL was deposited on DNA/silicon oxide sample. The Pt(II) salt was incubated for 30 minutes and dried with nitrogen stream. DNA surface topographies were imaged with an atomic force microscopy (AFM 5500, Agilent Technologies/Keysight Technologies) in air, in AC mode with an oscillation frequency of 63 kHz. The images were obtained at 512 and 1024 points per lines. All the data were processed with Gwyddion 2.31 program.41QCM-D experiments
The experiments were carried out using a quartz-crystal microbalance with dissipation monitoring from Biotin Scientific and respective quartz sensors QSX 301 Gold (Biolin Scientific). For each experiment, a new sensor was used. Each experiment started with obtaining a stable baseline by passing PBS–buffer (Sigma Aldrich) at a flow rate of 100 μL min−1 through the sensor module (E1. Biolin Scientific) using a peristaltic pump (Ismatec, Reglo Digital). Frequency and dissipation data were recorded using the acquisition software QSoft 401 (Biolin Scientific). The 5th harmonic of the frequency was used for further data processing as this harmonic has previously proven to yield most reliable signals.34 Stability was assumed when the average frequency signal was less than 0.05 Hz during five min. Subsequently, each thiol-containing oligomer (Biomers) was injected (5 μM) at the same feed flow velocity for 10 min, followed by rinsing with PBS buffer during additional 10 min in order to wash-off any loosely bound DNA. The complementary strand (5 mM) was then injected at the same flow rate and duration, followed again by injection of PBS buffer. Finally, the Pt(II) complex 1a was injected at a concentration of 0.1 mg mL−1, followed by rinsing with buffer. For better visualization, in Fig. 3 the frequency data were normalized for the steady-state frequency (Fmax) measured when the DNA duplex had formed after injection of the complementary strand and before injection of the Pt(II) complex. In this way, the effect of the Pt(II) complex on the dissipation of the DNA was comparable between measurements irrespective of the absolute DNA coverage of the sensor.Reagents and catalysis
[Pt(DMSO)2Cl2] and [(bipym)PtCl2] were prepared following the procedure described in the literature.42 The α-imino glycinate esters R2HC = NCH2CO2Me 4, with R2 = Ph (4a), p-Me(C6H4) (4b), cyclohexyl (4e), 3′-thienyl (4f), p-F(C6H4) (4d), and p-OMe(C6H5) (4c), were prepared using the synthetic procedures described in the literature.25Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Spanish MINECO (grants CTQ2013-45415-P, CTQ2016-80375-P MAT2013-46006-R and CTQ2016-81797-REDC, PCIN-2015-240) and by the Basque Government (grants IT673-13 and PI-2013 57). We thank the SCI/IZO-SGIker, the UPV/EHU (UFI QOSYC) and the DIPC for allocation of computational and analytical resources and. T. S. gratefully acknowledges ERC Starting-Grant St 209842 (MATRIX) and ERC PoC 713641 (ESSENS).Notes and references
- F. Crick, Nature, 1970, 227, 561–563 CrossRef CAS PubMed.
- K. Kruger, P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling and T. R. Cech, Cell, 1982, 31, 147–157 CrossRef CAS PubMed.
- S. K. Silvermann, Org. Biomol. Chem., 2007, 2, 2701–2706 Search PubMed.
- G. Roelfes and B. L. Feringa, Angew. Chem., Int. Ed., 2005, 44, 3230–3232 CrossRef CAS PubMed.
- A. J. Boersma, R. P. Megens, B. L. Feringa and G. Roelfes, Chem. Soc. Rev., 2010, 39, 2083–2092 RSC.
- A. J. Boersma, B. L. Feringa and G. Roelfes, Org. Lett., 2007, 9, 3647–3650 CrossRef CAS PubMed.
- R. P. Megens and G. Roelfes, Org. Biomol. Chem., 2010, 8, 1387–1393 CAS.
- D. Coquière, B. L. Feringa and G. Roelfes, Angew. Chem., Int. Ed., 2007, 46, 9308–9311 CrossRef PubMed.
- E. Benedetti, N. Duchemin, L. Bethge, S. Vonhoff, S. Klussmann, J.-J. Vasseur, J. Cossy, M. Smietana and S. Arseniyadis, Chem. Commun., 2015, 51, 6076–6079 RSC.
- R. P. Megens and G. Roelfes, Chem. Commun., 2012, 48, 6366–6368 RSC.
- A. Boersma, D. Coquière, D. Geerdink, F. Rosati, B. L. Feringa and G. Roelfes, Nat. Chem., 2010, 2, 991–995 CrossRef CAS PubMed.
- Z. Tang and A. Marx, Angew. Chem., Int. Ed., 2007, 46, 7297–7300 CrossRef CAS PubMed.
- P. Fournier, R. Fiammengo and A. Jäschke, Angew. Chem., Int. Ed., 2009, 48, 4426–4429 CrossRef CAS PubMed.
- L. Gjonaj and G. Roelfes, ChemCatChem, 2013, 5, 1718–1721 CrossRef CAS.
- Synthetic Applications of 1,3-Dipolar Reaction Chemistry Toward Heterocycles and Natural Products, ed. A. Padwa and W. H. Pearson, J. Wiley & Sons, Hoboken, 2003 Search PubMed.
- M. Baunach and C. Hertweck, Angew. Chem., Int. Ed., 2015, 54, 12550–12555 CrossRef CAS PubMed.
- E. H. Krenske, A. K. Patel and N. Houk, J. Am. Chem. Soc., 2013, 135, 17638–17642 CrossRef CAS PubMed.
- K. A. P. Payne, M. D. White, K. Fisher, B. Khara, S. S. Bailey, D. Parker, N. J. W. Rattray, D. K. Trivedi, R. Goodacre, R. Reveridge, P. Barran, S. E. J. Rigby, N. S. Crutton, S. Hay and D. Leys, Nature, 2015, 522, 497–501 CrossRef CAS PubMed.
- P. Painter, R. P. Pemberton, B. M. Wong, K. C. Ho and D. J. Tantillo, J. Org. Chem., 2014, 79, 432–435 CrossRef CAS PubMed.
- P. A. Roethle, P. T. Hernandez and D. Trauner, Org. Lett., 2006, 8, 5901–5904 CrossRef CAS PubMed.
- B. Tang, C. D. Bray and G. Pattenden, Tetrahedron Lett., 2006, 47, 6401–6404 CrossRef CAS.
- S. Strych, G. Journot, R. P. Pemberton, S. C. Wang, D. J. Tantillo and D. Trauner, Angew. Chem., Int. Ed., 2015, 54, 5079–5083 CrossRef CAS PubMed.
- L. Kelland, Nat. Rev. Cancer, 2007, 7, 573–584 CrossRef CAS PubMed.
- E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed.
- E. Conde, D. Bello, A. de Cózar, M. Sánchez, M. A. Vázquez and F. P. Cossío, Chem. Sci., 2012, 3, 1486–1491 RSC.
- E. E. Maroto, S. Filippone, M. Suárez, R. Martínez-Álvarez, A. de Cózar, F. P. Cossío and N. Martín, J. Am. Chem. Soc., 2014, 136, 705–712 CrossRef CAS PubMed.
- P. M. Takahara, A. C. Rosenzweig, C. A. Frederick and S. J. Lippard, Nature, 1995, 377, 649–652 CrossRef CAS PubMed.
- L. W. Chung, W. M. C. Sameera, R. Ramozzi, A. J. Page, M. Hatanaka, G. P. Petrova, T. V. Harris, X. Li, Z. Ke, F. Liu, H.-B. Li, L. Ding and K. Morokuma, Chem. Rev., 2015, 115, 5678–5796 CrossRef CAS PubMed.
- T. Vreven, K. S. Byun, I. Komáromi, S. Dapprich, J. A. Montgomery Jr, K. Morokuma and M. J. Frisch, J. Chem. Theory Comput., 2006, 2, 815–826 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision D.01, Gaussian, Inc., Wallingford CT, 2013 Search PubMed.
- W. Kolodziejski and J. Klinowski, Chem. Rev., 2002, 102, 613–628 CrossRef CAS PubMed.
- C. L. Cheng, Y. P. Chang and Y. H. Chu, Chem. Soc. Rev., 2012, 41, 1947–1971 RSC.
- F. Caruso, E. Rodda, D. N. Furlong, K. Niikiura and Y. Okahata, Anal. Chem., 1997, 69, 2043–2049 CrossRef CAS PubMed.
- M. B. Serrano-Santos, E. Llobet, V. C. Özalp and T. Schäfer, Chem. Commun., 2012, 48, 10087–10089 RSC.
- C. Jurinke, P. Oeth and D. Van den Boom, Mol. Biotechnol., 2004, 26, 147–163 CrossRef CAS PubMed.
- L. Y. Hemeryck, S. A. Moore and L. Vanhaecke, Anal. Chem., 2016, 88, 7436–7446 CrossRef CAS PubMed.
- X. M. Hou, X. H. Zhang, K. J. Wei, C. Ji, S. X. Dou, W. C. Wang, M. Li and P. Y. Wang, Nucleic Acids Res., 2009, 37, 1400–2141 CrossRef CAS PubMed.
- Y. Kan, Q. Tan, G. Wu, W. Si and Y. Chen, Sci. Rep., 2014, 5, 8442 CrossRef PubMed.
- M. Kartalou and J. M. Essigmann, Mutat. Res., 2001, 478, 1–21 CrossRef CAS PubMed.
- CCDC 1411373 and 1411372 contain the supplementary crystallographic data for compounds 5bb and 5bf, respectively.†.
- D. Nečas and P. Klapetek, Cent. Eur. J. Phys., 2012, 10, 181–188 Search PubMed.
- I. Bar-Nahum, M. A. Khenkin and R. Neumann, J. Am. Chem. Soc., 2004, 126, 10236–10237 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, characterization of cycloadducts and intermediates, computational data. CCDC 1411372 and 1411373. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7sc02311a |
| This journal is © The Royal Society of Chemistry 2017 |