 Open Access Article
Open Access ArticleMagnetic control of cellular processes using biofunctional nanoparticles
Cornelia
Monzel
 a,
Chiara
Vicario
a,
Jacob
Piehler
b,
Mathieu
Coppey
a and
Maxime
Dahan
*a
a,
Chiara
Vicario
a,
Jacob
Piehler
b,
Mathieu
Coppey
a and
Maxime
Dahan
*a
aInstitut Curie, PSL Research University, Laboratoire Physico Chimie, CNRS UMR168, UPMC, F-75005 Paris, France. E-mail: maxime.dahan@curie.fr
bUniversity of Osnabrück, Department of Biology/Chemistry, Division of Biophysics, 49076 Osnabrück, Germany
First published on 9th August 2017
Abstract
Remote control of cellular functions is a key challenge in biomedical research. Only a few tools are currently capable of manipulating cellular events at distance, at spatial and temporal scales matching their naturally active range. A promising approach, often referred to as ‘magnetogenetics’, is based on the use of magnetic fields, in conjunction with targeted biofunctional magnetic nanoparticles. By triggering molecular stimuli via mechanical, thermal or biochemical perturbations, magnetic actuation constitutes a highly versatile tool with numerous applications in fundamental research as well as exciting prospects in nano- and regenerative medicine. Here, we highlight recent studies, comment on the advancement of magnetic manipulation, and discuss remaining challenges.
1 Introduction
Remote activation of cellular processes constitutes an important challenge in nanotechnology and bioengineering. This challenge can now be addressed with techniques based on advanced physical (optical, magnetic or electrical) and chemical modalities which go well beyond conventional genetic or pharmacological approaches. These novel techniques enable systematic activation at the single cell level but within environments as complex as a living organism. Recent studies, for example, succeeded in the control of oriented migration,1–3 intracellular transport,4 gene expression,5,6 and differentiation.7 Thus, it becomes possible to probe specific biological mechanisms, to quantitatively interrogate the complex molecular circuitries that mediate signaling events, or even to control cell behavior for regenerative medicine applications.Optical techniques based on photocontrol of protein activity, using photoactivatable reagents8,9 or optogenetics,2,10,11 have been very beneficial, with a broad range of applications in cell biology and neuroscience. Yet, light-based methods also present some limitations, such as the need to express multiple genetically-modified photo-reactive proteins, the potential phototoxicity of optical stimuli in the biological specimen, or the difficulty to apply sustained spatially-patterned illumination, which can be challenging in thick and scattering tissues.
An emerging and complementary approach to remotely control biological processes is based on magnetic stimulation. So far, the effect of magnetic fields on cellular response has been investigated in different contexts. A first instance is the field of ‘magnetosensation’, i.e. the ability of some organisms to detect magnetic fields, e.g. for the purpose of navigation.12–14 A second area of research focuses on the influence of magnetic fields on biological processes in general.15–17 Here, usually strong magnetic fields (>1 T) have been applied to show that biopolymers with high diamagnetic anisotropy (e.g. microtubules and nucleic acid chains) can respond to the external magnetic field. While both of these fields are active and interesting areas of research, this review elaborates on a third approach, in which the magnetic control of biological processes is mediated by functionalized magnetic nanoparticles (MNPs). Recent studies have indeed demonstrated how MNPs can serve to convert the external signal of a static or oscillating magnetic field of moderate strength (∼100 mT, field gradient ∼ 101 to 104 T m−1) into biological events (see Table 1 for an overview). These studies, which go well beyond the conventional use of MNPs as imaging contrast agents18,19 or drug-delivery carriers,20–24 demonstrated how different magnetic field conditions effectuate mechanical, thermal or biochemical stimuli that trigger a specific cellular event. The benefits of such flexibility are further reinforced by the fact that magnetic fields can non-invasively penetrate deep into tissues, wherefore cellular functions and circuits can be probed in live cultured cells, tissues and organisms. Altogether, these novel manipulation tools have laid the ground for a new field, sometimes termed ‘magnetogenetics’. Here, we review recent advances in magnetogenetics and highlight its potential for both fundamental and applied biomedical research.
| Application | Mechanism | Force F/magnetic flux density B/amplitude magnetic flux density B0/field gradient ∇B | MNP | Reference |
|---|---|---|---|---|
| Magneto-mechanical stimulation | ||||
| Control of Notch & E-cadherin receptor activity | Stretching/pulling by magnetic tip | F = 1–47 pN | Core: 10–30 nm zinc-doped iron oxide, coating: silica & gold shell; thiolated DNA | 5 |
| Stretching of chromatin for gene transcription upregulation | Stretching/twisting by 3D magnetic multipoles | B ∼ 250 mT, B < 2.5 mT (twisting field) | Core: 4 μm ferromagnetic bead, coating: RGD peptides | 34 |
| Control of TRPV4 ion channel gating | Pulling/clustering by electromagnet (EM) or permanent magnet (PM) | B = 50 mT (EM & PM), 500 mT (PM) | Endoferritin particle | 39 |
| Control of Wnt-frizzled receptor activity | Stretch/pulling by oscillatory motion on magnetic arrays | B = 25–120 mT | Core: 300 nm iron oxide, coating: antibodies or RGD tri-peptide | 40 |
| Stimulation of filopodia formation & oriented cell division | Attraction/pulling by magnetic array |
F ∼ 100 nN, B = 25–100 mT, ∇B = 2500–70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 T m−1 000 T m−1 |
Coating: dextran | 47 |
| Control of stereocilia tilt for ion channel gating | Attraction/pulling by magnetic tip | F = 0.1 pN, ∇B = 103 T m−1 | Core: ∼50 nm zinc-doped iron oxide, coating: 3.8 nm SiO2 shell | 35 |
| Control of Drosophila embryonic tissue deformation for gene expression | Magnetic tweezer induced mechanical tissue deformation | F = 60 nN, ∇B = 120 T m−1 | Core: 7.5 nm maghemite, coating: citrate molecules | 44 |
| Modulation of cell endocytosis | Pulling/clustering by permanent magnet | F = 1–100 pN, ∇B ∼ 104 T m−1 | Core: 30 nm iron oxide | 81 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Magneto-thermal stimulation | ||||
| Control of TRPV1 ion channel gating | Heat activation via radio-frequency waves | f = 40 MHz, B0 = 1.3 mT | Core: 6 nm manganese ferrite, coating: streptavidin & PEG-phospholipids, SLP (specific loss power) = 2.5 W g−1 | 55 |
| Control of TRPV1 ion channel gating for neuronal cell excitation | Heat activation via radio-frequency waves | f = 500 kHz, B0 = 18 mT | Core: 22 nm iron oxide, coating: PEG, SLP = 660 W g−1 | 56 |
| Control of TRPV1 ion channel gating for neuronal cell excitation & glucose homeostasis | Heat activation via radio-frequency waves (Stanley 2012/2015/2016), static magnetic fields (Stanley 2015/2016) | f = 465 kHz; BRF ∼ 5 mT (Stanley 2012) & ∼30 mT (Stanley 2015/2016); B ∼ 0.1–1 T (Stanley 2016) | Core: 20–25 nm iron oxide (Stanley 2012), coating: carboxylic acids (Stanley 2012), endoferritin particle (Stanley 2015/2016), SAR (specific absorption rate) = 0.63 W g−1 (Stanley 2012) | 6, 57 and 58 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Magneto-biochemical stimulation | ||||
| Control of FcεRI receptor activity for inflammatory responses | Clustering by electromagnetic tip | F ∼ 0.01 fN, B ∼ 100 mT | Core: 5 nm iron, coating: 10 nm polymer shell, with amine groups | 59 |
| Control of DR4 receptor activity for cell apoptosis | Attraction/clustering by magnetic arrays | F ∼ 30 fN, B ∼ 200 mT (magnetic arrays), B ∼ 500 mT (zebrafish) | Core: 15 nm zinc-doped iron oxide, coating: thiols | 61 |
| In vitro control of microtubule nucleation | Attraction/pulling by magnetic tip | F ∼ 10 fN, B ∼ 150 mT, ∇B = 50 T m−1 | Core: 100 nm iron oxide, coating: 10 nm polymer shell with carboxylic acids | 65 |
| Control of intracellular Rac-GTPase signalling | Attraction/pulling by magnetic tip | F ∼ 10–30 pN, B ∼ 200 mT, ∇B = 103 to 104 T m−1 | Core: 500 nm iron oxide, coating: 10 nm polymer shell with streptavidin | 64 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Forces in nature | ||||
| Thermal energy | Thermal energy per degree of freedom: kT = 4 × 10−21J = 4 pNnm, (can be compared to the interaction energy, E, between the MNP magnetic moment, m, and the magnetic flux density, B: E = −mB) | 80 | ||
| Cell forces | Ion channel gating force (force-sensitive channels in auditory hair cells): F = 2 × 10−13 N (Howard 1988), traction forces/pulling forces by actin retraction fibres/actin polymerization forces: F ∼ 1–100 nN (Sniadecki 2007, Tan 2003, Tseng 2012, Prass 2006, and Fink 2011), tissue deformation forces (during snail-dependent apex pulsations in mesoderm cells/of stomodeal cells in Drosophila embryo): F ∼ 0.5 nN/60 nN (Mitrossilis 2017/Desprat 2008) | 44, 47, and 82–87 | ||
2 Magnetic control of cellular processes
Magneto-mechanical stimulation
The use of MNPs in cell biology has long been associated with the study of mechano-transduction, that is, the conversion of a mechanical stimulus into an electrical or biochemical signal.25 Attaching a bead at a cell surface to bend, stretch, or twist the cell membrane has proven very useful to analyze cell mechanical features26–28 and the transduction of mechanical constraints by membrane-associated molecular complexes, such as integrins3,29 or cadherins.27,30 In recent years, the scope of these experiments has been largely expanded by means of carefully designed and targeted MNPs.31–33 Combined with advanced magnetic systems, these MNPs enabled the manipulation of various biological processes with spatio-temporal resolution in the submicrometer and millisecond range, and down to the level of single proteins.34One such example is the work by Seo et al.,5 where mono-functionalized iron-oxide nanoparticles capped with a gold shell (with ∼50 nm total diameter) were used to manipulate individual Notch receptors or E-cadherin at the cell surface. Utilizing a calibrated tweezer setup, they found that individual Notch receptors require a critical loading force in the 2–9 pN range to activate downstream signaling. For E-cadherin, they identified the importance of cooperative spatial aggregation and mechanical loading to achieve actin and vinculin adaptor recruitment (Fig. 1a).
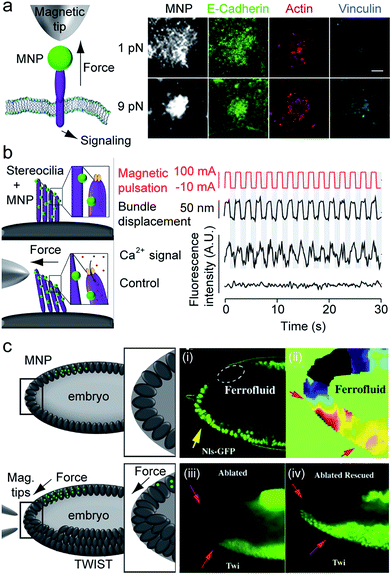 | ||
| Fig. 1 Magneto-mechanical stimulation. (a) Left: Mechanoreceptor activation via pulling. A magnetic tip in close proximity of an MNP bound receptor mechanically loads the MNP–receptor complex and activates intracellular signaling. Right: Immunofluorescence staining for MNPs, E-cadherin and the signal activation reporters, actin and vinculin. 9 pN forces result in enhanced molecular colocalization and recruitment compared to 1 pN forces. Scale bar, 2 μm. Taken from Seo et al.5 with permission from Elsevier. (b) Left: Gating ion channels of the inner ear via mechanical deflection. A magnetic field gradient tilts stereocilia with attached MNPs. Bundle displacement opens the tip link at the channel top (yellow) resulting in ion influx. Right: Traces of pulsed magnetic stimulation, the corresponding hair bundle displacement, the fluorescence signal of a Ca2+-sensitive dye inside the bundle, and the control experiment, where channel opening was prevented. Reprinted with permission from Lee et al.35 Copyright 2014 American Chemical Society. (c) Left: TWIST expression correlates with mechanical cell compression in a Drosophila embryo. To probe this, the natural compression of endodermal cells is blocked and mimicked instead by magnetically pulling on cells containing a ferrofluid. These magnetized cells are adjacent to endodermal cells and compress the tissue upon magnetic field application. Right: (i) cell compression induced by magnetic manipulation (yellow arrow). (ii) Particle imaging velocimetry indicates compression changes – lowest in black and highest in red. (iii) Inhibition of TWIST expression in the uncompressed cells in the ablated embryo (between the red arrows). (iv) Recovery of TWIST expression in the cells of the ablated embryo by inducing physiological compression with magnetic fields. Taken from Desprat et al.44 with permission from Elsevier. | ||
Remote actuation of signaling by magneto-mechanical stimulation may also be realized through direct gating of ion channels via deflection or stretching. For instance, Lee et al.35 demonstrated ultrafast (sub-ms) mechanical control of the deflection-relaxation dynamics of stereocilia bundles bound to cubic MNPs (size ∼50 nm) and submitted to oscillating fields in inner ear hair cells (Fig. 1b). Wheeler et al. used the ‘transient receptor potential cation channel subfamily V member 4’ (TRPV4), a membrane receptor known to gate in response to osmotic, chemical or mechanical cues.36–39 By fusing TRPV4 to genetically-encoded ferritin nanoparticles and applying a static magnetic gradient, they triggered Ca2+ transients, which elicited action potential in neurons, enhanced tactile behavior in zebrafish, and affected reward behavior in mice.39 It should be noted though that the physical mechanism underlying the actuation of TRPV4 by endogenous ferritin is a subject of current debate (see discussion).
Other studies have aimed at controlling developmental processes, such as stem cell differentiation or tissue organization.7,40–43 For instance, transmembrane ion channels TREK-1, which are responsible for setting the resting membrane potential and intracellular Ca2+-concentrations, and which influence differentiation processes, were labeled in human mesenchymal stem cells with 250 nm MNPs and were stimulated by applying a slowly oscillating (∼1 Hz) magnetic field over a period of 7 or 21 days in vitro and in vivo.7 The resulting increase in the proteins Sox9, osteopontin and collagen provided evidence that a mechanical stimulation of TREK-1 is sufficient to induce the differentiation of osteoprogenitor cell populations toward an osteogenic lineage. Next to the overall change in gene expression, an interesting point raised by these studies is that long-term magnetic stimulation over a day and up to several weeks showed a significant effect on the targeted gene expression. The expression of the reporter molecules of TREK-1, for example, exhibited a 2-fold increase after 7 days of magnetic field exposure7 and a similar increase was observed in a study of the Wnt signaling pathway, where expression levels were compared after 6 h and 1 day.40 Hence, investigating the temporal effects of magnetic stimulation is important to achieve full control of a biological process.
Desprat et al. demonstrated how mechanical forces during Drosophila embryo development are coupled to the regulation of TWIST, a transcription factor vitally involved in early anterior endoderm cell differentiation:44 first, the natural compression movement occurring at the onset of Drosophila gastrulation was suppressed and TWIST expression remained low. Thereafter, the natural deformation of cells was mimicked by pulling on ferrofluid loaded cells in the neighborhood of the otherwise naturally compressed cells. This mechanical stimulus rescued TWIST up-regulation and resulted in gene expression profiles similar to those occurring naturally (Fig. 1c). More recently, a similar approach was used to show how mechanical pressure contributes to the onset of tumorigenesis in a mouse model for colon tumor development.45
Finally, directional magnetic pulling of MNPs internalized in endosomes was used by the group of Di Carlo in different biological contexts – e.g. to induce coordinated filopodia formation, to bias spindle orientation in dividing cells,46,47 as well as to mechanically stimulate calcium influx48 and polarity of neurons.49 A breakthrough in these studies was to establish a massively parallelized assay, where microfabricated magnetic arrays, with cells plated in close proximity to one micromagnet, enabled high-throughput measurements with unprecedented control. With this assay, the group provided an elegant solution for one of the major challenges in many magnetic manipulation studies, namely, the application of well-defined magnetic forces over a large population of cells.
Magneto-thermal stimulation
A second magnetic actuation modality is based on the thermal response of magnetic nanoparticles. When placed in a radio-frequency (∼1 MHz) magnetic field, some MNPs (depending on their size, shape, and magnetic content) are able to convert the field stimulation into heat.50,51 Thus, they can be used as local hotspots to stimulate thermo-responsive molecules – several of which are naturally found in mammalian cells. The most prominent examples are proteins of the ‘transient receptor potential’ (TRP) channel family, ion channels which are located in the plasma membrane of cells. TRPM8 and TRPA1, for example, are ion channels sensitive to cold temperatures (active between 25–28 °C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 52 and <17 °C,53 respectively) and TRPV1, TRPV3, and TRPV4 (active >42 °C, >38 °C and >35 °C, respectively) are activated when heated. Note, that the most suitable activation temperature can vary between different cell types due to their different thermosensitivity.54 The studies discussed in the following, report the control of TRPV1 cation gating in different cells and this gating in turn was used to trigger specific signaling pathways (Fig. 2a).
52 and <17 °C,53 respectively) and TRPV1, TRPV3, and TRPV4 (active >42 °C, >38 °C and >35 °C, respectively) are activated when heated. Note, that the most suitable activation temperature can vary between different cell types due to their different thermosensitivity.54 The studies discussed in the following, report the control of TRPV1 cation gating in different cells and this gating in turn was used to trigger specific signaling pathways (Fig. 2a).
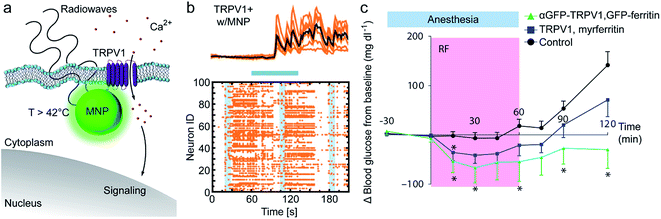 | ||
| Fig. 2 Magneto-thermal stimulation. (a) Controlling membrane ion channels via MNP heating. An MNP in close proximity to the temperature-sensitive ion channel TRPV1 is locally heated to >42 °C with a radio-frequency (RF) magnetic field. Subsequently, TRPV1 opens, enabling Ca2+ influx. The ion concentration increase is used to activate selected signaling processes. (b) Magneto-thermal TRPV1 stimulation for the controlled activation of hippocampal neurons. The ion channel gating evokes correlated and repeated trains of action potentials in TRPV1 expressing neurons. Top: 10 fluorescence traces (orange) with an average overlay (black) before, during, and after magnetic stimulation (blue bar). Bottom: Raster plots of 100 randomly selected neurons exhibiting repetitive calcium spikes. Shaded blue bars represent alternating magnetic field pulses. Taken from Chen et al.56 with permission from AAAS. (c) Remote regulation of glucose homeostasis in mice by TRPV1 activation. During channel activation in a RF magnetic field (pink area) a calcium-dependent transgene expression of insulin is initiated. Enhanced insulin expression followed by reduced blood glucose levels in mice are predominantly observed for ferritin–MNPs directly coupled to TRPV1 via the GFP–antiGFP nanobody interaction (αGFP–TRPV1/GFP–ferritin), as well as for ferritin–MNPs associated to the cell plasma membrane (TRPV1/myrferritin). *P < 0.05. Data are mean ± s.e.m. Taken from Stanley et al.57 with permission from AAAS. | ||
The magneto-thermal stimuli reported in recent studies enabled the control of molecular activity states in single cells, tissue and animals.6,55–58 More precisely, calcium influx into cells was controlled via TRPV1 gating and evidenced at the single cell level, either in HEK293 cells55–57 or in hippocampal neuronal cells leading to the firing of action potentials.55,56 In animals, magneto-thermal actuation was first applied in C. elegans worms where heating of MNPs near sensory neurons triggered a thermal avoidance, i.e. a spatial retraction response.55 In mice, activation of TRPV1 in ventral tegmental brain regions evoked considerable neuronal excitation (Fig. 2b) and highlighted the potential of magneto-thermal tools for remote and local deep-brain stimulation without the need for implants and connectors.56 The remote gating of TRPV1 was achieved by means of a high concentration (∼mg ml−1) of synthetic iron-oxide MNPs (size 10–30 nm) either freely diffusing in the cell cytoplasm56 or targeted to the cell plasma membrane via biotin–streptavidin interactions to increase the efficiency of ion channel heating as well as to reduce side effects.55
In a different set of experiments, Stanley et al. followed a similar strategy in order to control insulin expression and, consequently, the level of blood glucose in mice (Fig. 2c).6,57,58 To this end, they used endogenous ferritin as heat generating nanoparticles targeted to TRPV1 to thermally gate the ion channel. Channel gating resulted in elevated intracellular calcium levels, which in turn induced the expression of bioengineered insulin. An increase in insulin followed by a decrease in blood glucose levels was achieved in mice, either by transplanting mesenchymal stem cells expressing the genetic constructs, or by adenoviral delivery of transgenes.6,57 In a subsequent study, the same ferritin–TRPV1 molecular system was used in targeted hypothalamic glucose-sensing neurons to activate or inhibit (using mutated chloride-permeant TRPV1 channels) neuronal activity and thereby to regulate metabolism and control insulin levels in mice.58 Yet, as commented above for ref. 39, it remains uncertain which physical mechanisms can account for the magnetic actuation of channel gating (see discussion).
Magneto-molecular stimulation
A third magnetic actuation modality relies on the control of biomolecular activity patterns in the cell by modulation of the concentration and spatial distribution of signaling molecules. An early example is the use of MNPs specifically bound to membrane receptors, to control the oligomerization-dependent activation mechanisms in the cell plasma membrane.59–61 Here, a static field is applied to reversibly cluster receptor–MNP complexes through dipole–dipole interactions between the nanoparticles, which subsequently triggers an intracellular signaling response. By doing so, Mannix et al. probed the immune surveillance as performed by mast cells and found that aggregation of FcεRI–dinitrophenyl receptor–ligand complexes was sufficient to increase cytosolic calcium and to initiate a local inflammatory response59 (Fig. 3a). Similarly, the magnetically-induced dimerization of EGF receptors led to the activation of downstream signaling cascades, as evidenced by the phosphorylation of the receptors.60 Building on this approach, Cho et al. targeted iron-oxide MNPs to the death receptor 4 (DR4) of colon cancer cells to mimic the natural apoptosis signaling pathway via MNP-DR4 aggregation in vitro. When injected into a zebrafish embryo, these MNPs targeted a DR4 related receptor and aggregated upon magnetic field stimulation in the zebrafish tail, altering its morphology.61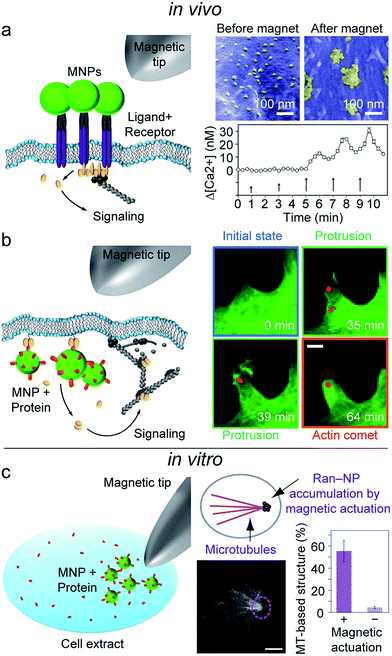 | ||
| Fig. 3 Magneto-molecular stimulation. (a) and (b) In vivo control of receptor signal transduction via functionalized MNP accumulation. (a) Left: magnetic field induced aggregation of MNPs bound to plasma membrane receptors. Right: (top) scanning electron microscopy images of antibody–MNP-treated cells before and after magnetic field application. Antibody–MNPs shown in yellow are individually distributed before and clustered after magnetic field treatment. Taken from Cho et al.61 with permission from NPG. (bottom) Oligomerization of FcεRI–DNP receptor–ligand complexes triggers calcium signaling. Graph depicting successively enhanced intracellular calcium levels (Δ[Ca2+]) for cells subjected to five electromagnetic pulses (arrows). Taken from Mannix et al.59 with permission from NPG. (b) Left: remote control of Rac-GTPase signaling via cytosolic accumulation of functionalized MNPs. Right: Guanine nucleotide exchange factors TIAM1 coupled to MNP (orange) are attracted to distinct subcellular sites in a magnetic field gradient. TIAM1–MNPs localize into an inactive area of the cell border and activate Rac1, which in turn triggers actin branched polymerization and protrusion formation. An actin comet is observed at 64 min. Scale bar, 1 μm. Taken from Etoc et al.64 with permission from NPG. (c) Left: in vitro formation of microtubule asters from cellular extract via magnetic accumulation of MNPs conjugated with Ran, a molecular switch regulating microtubule self-assembly during mitosis. Middle: (top) schematic representation and (bottom) fluorescence image of microtubule nucleation and assembly at the point of Ran-NP accumulation (pink dotted circle). Scale bar, 10 μm. Right: quantification of microtubule-based structures, with and without magnetically accumulated Ran-NP. Taken from Hoffmann et al.65 with permission from NPG. | ||
Besides the ability to activate signaling pathways at the plasma membrane receptor level, magnetogenetics also permits the investigation and control of the intracellular signaling machinery, or, more generally, of the subcellular organization. This approach faces the challenges of nanoparticle internalization in the cytosol (escaping endosomal pathways) and surface passivation to ensure colloidal stability inside the complex cytoplasmic environment. In the case of subcellular manipulation, MNPs tagged with signaling proteins and in the fluid phase of the cytoplasm can function as signaling nanoplatforms, able to interact with downstream effectors and to activate signaling pathways. With magnetic gradients, these nanoplatforms can further be displaced through the cytoplasm and accumulate at specific locations in order to modulate the local concentration of signaling molecules.62,63 Etoc et al. used this approach with 500 nm-MNPs coupled to Tiam1, a GEF molecule for the small GTPase Rac1.64 Once brought to the plasma membrane, MNPs triggered actin cytoskeleton remodeling and cell protrusion formation (Fig. 3b). Another experiment in droplets of Xenopus egg extract probed the spatial arrangement of the Ran/RCC1 signaling pathway involved in cell cytoskeleton and mitosis regulation.65 When accumulating the particles in a field gradient, RanGTP coupled MNPs were found to act as a bioinspired switch where microtubule aster growth is only initiated if a distinct concentration threshold is exceeded (Fig. 3c).
3 Conclusion and perspectives
All of the above examples attest to the great variety of biological questions that can be addressed using magnetic manipulation mediated by MNPs (see Table 1 for an overview). A key factor is the unique spectrum of magnetic actuation modalities (mechanical, thermal, and biochemical) by which quantitative and spatio-temporally modulated stimuli can be generated.So what can be expected from magnetogenetics in the future? An exciting prospect is the possibility to control cellular behavior and tissue engineering directly in organisms where, contrary to light illumination, magnetic fields can penetrate easily and without inducing damage. Conceivably, remote activation of cellular processes, such as cell differentiation or oriented migration, might contribute to the success of cell-based therapies, for instance to target and differentiate stem cells at sites of injury, during lesion repair, or for the treatment of neurological disorders. Magnetic stimulation might also become a valuable and non-invasive technique for the control of hormone release and metabolic activity during pathologies.58 Interestingly, MNPs can serve to hijack signalling pathways and exploit the ability of individual cells to sense signalling cues with great sensitivity, to process signalling information (including its amplification), and to mobilize complex molecular machineries to ensure a proper response. Thus, magnetic stimuli provide a means to non-invasively instruct cells to carry out tasks (such as differentiation and growth) that they are already programmed to perform, but do not necessarily accomplish at the right time and the right place. In view of realizing such control of vital biological functions, magnetogenetics will also benefit from further studies in the fields of ‘magnetosensation’12–14 and magneto-responsive biomolecules,15,17 where the effect of magnetic fields in the absence of MNPs is investigated.
While much work is still needed before magnetogenetics becomes a versatile tool in the arsenal of regenerative medicine and nanomedicine, several stimulating applications can already be envisaged in the short term. First, the ability to displace or heat MNPs at sub-micrometer scales, and thereby to apply forces, activate molecular functions or trigger biochemical reactions, should prove invaluable to get novel insights into the mechanisms of biological processes. Second, as is well known in engineering, applying a variable input and measuring the output is a powerful means to dissect the molecular circuits underlying the cellular response to external cues. By quantitatively modulating the amplitude, frequency, and spatial localization of the stimulus, one can identify key aspects of the cue processing, such as amplification, filtering or the existence of thresholds. These important systems-level features are usually difficult to infer from the genetic manipulation of molecular components. Finally, magnetic manipulation involving MNPs might serve for the design of (semi-)synthetic molecular machines inside the cell, i.e. the engineering of biological oscillators and switches based on MNPs coupled to molecular nanoplatforms.
Several substantial challenges remain to take full advantage of the promises of magnetogenetics. Novel MNPs will surely be needed, with physical (size and shape), chemical (composition) and biofunctional surface properties tailored to ensure optimized magnetic response and efficient biotargeting as well as passivation against non-specific interactions. An obvious challenge and well-identified issue in bionanosciences is the delivery of MNPs within cells.66 Here, development of cell loading protocols alternative to injection, endocytosis, or permeabilizing reagents is needed, in particular, when anticipating magnetic manipulation with MNPs inside organisms.
A solution might reside in the use of genetically-encoded MNPs, in the form, for instance, of natural ferritin cage proteins67–71,79 or viral capsids.39,57,72 Yet, a difficulty with endogenous ferritin is that they store iron as ferrihydrite, which has reduced magnetic properties compared to iron oxides.77,78 Several strategies can be envisaged to enhance the magnetic response of fully genetically encoded systems. For ferritin-expressing eukaryotic cells, Kim et al.72 established protocols to elevate intracellular iron levels by combining ferritin expression with DMT1-based iron import and ferrous ammonium sulfate-supplemented culture medium. Another approach might be to exploit the ability of magnetotactic bacteria to synthesize biomineralized Fe3O4 crystals, so-called magnetosomes. Recently, Kolinko et al. have transferred this biomineralization potential from magnetotactic bacteria to a foreign non-magnetic prokaryotic organism.73 This successful endeavor is an exciting step towards the more challenging goal of enabling eukaryotic cells to synthesize tailored magnetic nanostructures.
To increase the magnetic response, a complementary strategy is to improve our ability to generate strong magnetic gradients.74 Yet, the main difficulty is that applying strong gradients implies working at a short distance, which is particularly challenging in tissues and organisms where distances between the magnetic device and the nanoparticles are in the millimeter/centimeter range. One interesting way to overcome this challenge might be the use of locally magnetizable implants.75,76
Importantly, the development of magnetogenetics requires a careful and quantitative examination of the physical mechanisms of action. In particular, the activation mechanisms proposed in some of the recent studies, notably using endogenous ferritin nanoparticles,6,39 have been heavily questioned. Indeed, based on physical arguments and the experimental conditions reported, it was estimated that the force and/or torque exerted by the endogenous ferritin NP on the attached TRPV4 channel are 4 to 9 orders of magnitude lower than those due to thermal agitation.80 Hence, it is difficult to comprehend how mechanical actuation could be achieved. Similarly, the heating response in a RF field, even with a ferritin optimized with cobalt doping, is about ten orders of magnitude too small.80 Thus there is a need for clarifying the exact nature of the physical or biochemical stimuli induced by the magnetic field and MNPs in these experiments.6,39
To conclude, we anticipate that, in view of its benefits as well as numerous challenges and open questions, the magnetic control of cellular behavior will become an active field of research in the forthcoming years. With its successful advancement, magnetogenetics should be established as a key technology, complementary to optogenetic tools, and will find multidisciplinary applications in chemistry, bioengineering, bionanosciences, biology, biophysics, or neurosciences.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
C. M. acknowledges financial support from the German Academic Exchange Service (DAAD) and Fonds der Chemischen Industrie. M. D acknowledges funding from the French National Research Agency (ANR) Paris-Science-Lettres Program (ANR-10-IDEX-0001-02 PSL). M. D. acknowledge funding from Labex CelTisPhyBio (No. ANR-10-LBX-0038). This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 686841 (MAGNEURON).References
- M. Gautier, I. Dhennin-Duthille, A. S. Ay, P. Rybarczyk, I. Korichneva and H. Ouadid-Ahidouch, Br. J. Pharmacol., 2014, 171, 2582–2592 CrossRef CAS PubMed.
- Y. I. Wu, D. Frey, O. I. Lungu, A. Jaehrig, I. Schlichting, B. Kuhlman and K. M. Hahn, Nature, 2009, 461, 104–108 CrossRef CAS PubMed.
- Z. Liu, Y. Liu, Y. Chang, H. R. Seyf, A. Henry, A. L. Mattheyses, K. Yehl, Y. Zhang, Z. Huang and K. Salaita, Nat. Methods, 2015, 13, 143–146 CrossRef PubMed.
- P. van Bergeijk, M. Adrian, C. C. Hoogenraad and L. C. Kapitein, Nature, 2015, 518, 111–114 CrossRef CAS PubMed.
- D. Seo, K. M. Southard, J. Kim, H. J. Lee, J. Farlow, J. Lee, D. B. Litt, T. Haas, A. P. Alivisatos, J. Cheon, Z. J. Gartner and Y. Jun, Cell, 2016, 165, 1507–1518 CrossRef CAS PubMed.
- S. A. Stanley, J. Sauer, R. S. Kane, J. S. Dordick and J. M. Friedman, Nat. Med., 2015, 21, 92–98 CrossRef CAS PubMed.
- J. M. Kanczler, H. S. Sura, J. Magnay, D. Green, R. O. C. Oreffo, J. P. Dobson and A. J. El Haj, Tissue Eng., Part A, 2010, 16, 3241–3250 CrossRef CAS PubMed.
- G. Mayer and A. Heckel, Angew. Chem., Int. Ed., 2006, 45, 4900–4921 CrossRef CAS PubMed.
- N. Umeda, T. Ueno, C. Pohlmeyer, T. Nagano and T. Inoue, J. Am. Chem. Soc., 2011, 133, 12–14 CrossRef CAS PubMed.
- A. Levskaya, O. D. Weiner, W. A. Lim and C. A. Voigt, Nature, 2009, 461, 997–1001 CrossRef CAS PubMed.
- K. Zhang and B. Cui, Trends Biotechnol., 2015, 33, 92–100 CrossRef CAS PubMed.
- S. Johnsen and K. J. Lohmann, Nat. Rev. Neurosci., 2005, 6, 703–712 CrossRef CAS PubMed.
- J. L. Kirschvink, M. M. Walker and C. E. Diebel, Curr. Opin. Neurobiol., 2001, 11, 462–467 CrossRef CAS PubMed.
- T. Ritz, M. Ahmad, H. Mouritsen, R. Wiltschko and W. Wiltschko, J. R. Soc., Interface, 2010, 7(suppl. 2), S135–S146 CrossRef CAS PubMed.
- L. Zhang, Y. Hou, Z. Li, X. Ji, Z. Wang, H. Wang, X. Tian, F. Yu, Z. Yang, L. Pi, T. J. Mitchison, Q. Lu and X. Zhang, eLife, 2017, 6, e28212 Search PubMed.
- T. Higashi, A. Yamagishi, T. Takeuchi, N. Kawaguchi, S. Sagawa, S. Onishi and M. Date, Blood, 1993, 82, 1328–1334 CAS.
- D. L. Worcester, Biophysics, 1978, 75, 5475–5477 CAS.
- J. W. Bulte, T. Douglas, B. Witwer, S. C. Zhang, E. Strable, B. K. Lewis, H. Zywicke, B. Miller, P. van Gelderen, B. M. Moskowitz, I. D. Duncan and J. A. Frank, Nat. Biotechnol., 2001, 19, 1141–1147 CrossRef CAS PubMed.
- N. Lee and T. Hyeon, Chem. Soc. Rev., 2012, 41, 2575–2589 RSC.
- Q. A. Pankhurst, J. Connolly, S. K. Jones and J. Dobson, J. Phys. D: Appl. Phys., 2003, 36, R167–R181 CrossRef CAS.
- A. K. Gupta and A. S. G. Curtis, J. Mater. Sci.: Mater. Med., 2004, 15, 493–496 CrossRef CAS PubMed.
- N. Griffete, J. Fresnais, A. Espinosa, C. Wilhelm, A. Bée and C. Ménager, Nanoscale, 2015, 7, 18891–18896 RSC.
- T. Georgelin, S. Bombard, J.-M. Siaugue and V. Cabuil, Angew. Chem., Int. Ed., 2010, 49, 8897–8901 CrossRef CAS PubMed.
- J. Dobson, Nanomedicine, 2006, 1, 31–37 CrossRef CAS PubMed.
- J. G. Añoveros and D. P. Corey, Curr. Biol., 1996, 6, 541–543 CrossRef.
- B. Fabry, G. N. Maksym, S. A. Shore, P. E. Moore, R. A. Panettieri, J. P. Butler and J. J. Fredberg, J. Appl. Physiol., 2001, 91, 986–994 CAS.
- M. L. Gardel, J. H. Shin, F. C. MacKintosh, L. Mahadevan, P. Matsudaira and D. A. Weitz, Science, 2004, 304, 1301–1305 CrossRef CAS PubMed.
- P. Janmey and C. Schmidt, in Cytoskeletal Mechanics: Models and Measurements, ed. M. R. K. Mofrad and R. D. Kamm, Cambridge University Press, Cambridge, 2006, pp. 18–49 Search PubMed.
- N. Wang, J. P. Butler and D. E. Ingber, Science, 1993, 260, 1124–1127 CAS.
- G. F. Weber, M. A. Bjerke and D. W. DeSimone, Dev. Cell, 2012, 22, 104–115 CrossRef CAS PubMed.
- W. Wang, X. Ji, H. Bin Na, M. Safi, A. Smith, G. Palui, J. M. Perez and H. Mattoussi, Langmuir, 2014, 30, 6197–6208 CrossRef CAS PubMed.
- A. K. Gupta and M. Gupta, Biomaterials, 2005, 26, 3995–4021 CrossRef CAS PubMed.
- S. Bamrungsap, T. Chen, M. I. Shukoor, Z. Chen, K. Sefah, Y. Chen and W. Tan, ACS Nano, 2012, 6, 3974–3981 CrossRef CAS PubMed.
- A. Tajik, Y. Zhang, F. Wei, J. Sun, Q. Jia, W. Zhou, R. Singh, N. Khanna, A. S. Belmont and N. Wang, Nat. Mater., 2016, 15, 1287–1296 CrossRef CAS PubMed.
- J.-H. Lee, J. Kim, M. Levy, A. Kao, S.-H. Noh, D. Bozovic and J. Cheon, ACS Nano, 2014, 8, 6590–6598 CrossRef CAS PubMed.
- E. Cao, M. Liao, Y. Cheng and D. Julius, Nature, 2013, 504, 113–118 CrossRef CAS PubMed.
- M. Liao, E. Cao, D. Julius and Y. Cheng, Nature, 2013, 504, 107–112 CrossRef CAS PubMed.
- C. E. Paulsen, J.-P. Armache, Y. Gao, Y. Cheng and D. Julius, Nature, 2015, 525, 552 CrossRef CAS PubMed.
- M. A. Wheeler, C. J. Smith, M. Ottolini, B. S. Barker, A. M. Purohit, R. M. Grippo, R. P. Gaykema, A. J. Spano, M. P. Beenhakker, S. Kucenas, M. K. Patel, C. D. Deppmann and A. D. Güler, Nat. Neurosci., 2016, 19, 756–761 CrossRef CAS PubMed.
- M. Rotherham and A. J. El Haj, PLoS One, 2015, 10, e0121761 Search PubMed.
- J. de Boer, R. Siddappa, C. Gaspar, A. van Apeldoorn, R. Fodde and C. van Blitterswijk, Bone, 2004, 34, 818–826 CrossRef CAS PubMed.
- C. Y. Logan and R. Nusse, Annu. Rev. Cell Dev. Biol., 2004, 20, 781–810 CrossRef CAS PubMed.
- B. Hu, A. Haj and J. Dobson, Int. J. Mol. Sci., 2013, 14, 19276–19293 CrossRef PubMed.
- N. Desprat, W. Supatto, P.-A. Pouille, E. Beaurepaire and E. Farge, Dev. Cell, 2008, 15, 470–477 CrossRef CAS PubMed.
- M. E. Fernández-Sánchez, S. Barbier, J. Whitehead, G. Béalle, A. Michel, H. Latorre-Ossa, C. Rey, L. Fouassier, A. Claperon, L. Brullé, E. Girard, N. Servant, T. Rio-Frio, H. Marie, S. Lesieur, C. Housset, J.-L. Gennisson, M. Tanter, C. Ménager, S. Fre, S. Robine and E. Farge, Nature, 2015, 523, 92–95 CrossRef PubMed.
- P. Tseng, D. Di Carlo and J. W. Judy, Nano Lett., 2009, 9, 3053–3059 CrossRef CAS PubMed.
- P. Tseng, J. W. Judy and D. Di Carlo, Nat. Methods, 2012, 9, 1113–1119 CrossRef CAS PubMed.
- A. Tay, A. Kunze, C. Murray and D. Di Carlo, ACS Nano, 2016, 10, 2331–2341 CrossRef CAS PubMed.
- A. Kunze, P. Tseng, C. Godzich, C. Murray, A. Caputo, F. E. Schweizer and D. Di Carlo, ACS Nano, 2015, 9, 3664–3676 CrossRef CAS PubMed.
- A. Riedinger, P. Guardia, A. Curcio, M. A. Garcia, R. Cingolani, L. Manna and T. Pellegrino, Nano Lett., 2013, 13, 2399–2406 CrossRef CAS PubMed.
- T. T. T. N’Guyen, H. T. T. Duong, J. Basuki, V. Montembault, S. Pascual, C. Guibert, J. Fresnais, C. Boyer, M. R. Whittaker, T. P. Davis and L. Fontaine, Angew. Chem., Int. Ed., 2013, 52, 14152–14156 CrossRef PubMed.
- D. D. McKemy, W. M. Neuhausser and D. Julius, Nature, 2002, 416, 52–58 CrossRef CAS PubMed.
- L. S. Premkumar and M. Abooj, Life Sci., 2013, 92, 415–424 CrossRef CAS PubMed.
- C. W. Song, H. Park and R. J. Griffin, Thermotherapy for Neoplasia, Inflammation, and Pain, Springer, Japan, Tokyo, 2001 Search PubMed.
- H. Huang, S. Delikanli, H. Zeng, D. M. Ferkey and A. Pralle, Nat. Nanotechnol., 2010, 5, 602–606 CrossRef CAS PubMed.
- R. Chen, G. Romero, M. G. Christiansen, A. Mohr and P. Anikeeva, Science, 2015, 347, 1477–1480 CrossRef CAS PubMed.
- S. A. Stanley, J. E. Gagner, S. Damanpour, M. Yoshida, J. S. Dordick and J. M. Friedman, Science, 2012, 336, 604–608 CrossRef CAS PubMed.
- S. A. Stanley, L. Kelly, K. N. Latcha, S. F. Schmidt, X. Yu, A. R. Nectow, J. Sauer, J. P. Dyke, J. S. Dordick and J. M. Friedman, Nature, 2016, 531, 647–650 CrossRef CAS PubMed.
- R. J. Mannix, S. Kumar, F. Cassiola, M. Montoya-Zavala, E. Feinstein, M. Prentiss and D. E. Ingber, Nat. Nanotechnol., 2008, 3, 36–40 CrossRef CAS PubMed.
- A. A. Bharde, R. Palankar, C. Fritsch, A. Klaver, J. S. Kanger, T. M. Jovin and D. J. Arndt-Jovin, PLoS One, 2013, 8, e68879 CAS.
- M. H. Cho, E. J. Lee, M. Son, J.-H. Lee, D. Yoo, J. Kim, S. W. Park, J.-S. Shin and J. Cheon, Nat. Mater., 2012, 11, 1038–1043 CrossRef CAS PubMed.
- F. Etoc, C. Vicario, D. Lisse, J.-M. Siaugue, J. Piehler, M. Coppey and M. Dahan, Nano Lett., 2015, 15, 3487–3494 CrossRef CAS PubMed.
- D. Liße, C. Monzel, C. Vicario, J. Manzi, I. Maurin, M. Coppey, J. Piehler and M. Dahan, Adv. Mater. Search PubMed , in press.
- F. Etoc, D. Lisse, Y. Bellaiche, J. Piehler, M. Coppey and M. Dahan, Nat. Nanotechnol., 2013, 8, 193–198 CrossRef CAS PubMed.
- C. Hoffmann, E. Mazari, S. Lallet, R. Le Borgne, V. Marchi, C. Gosse and Z. Gueroui, Nat. Nanotechnol., 2013, 8, 199–205 CrossRef CAS PubMed.
- J. B. Delehanty, H. Mattoussi and I. L. Medintz, Anal. Bioanal. Chem., 2009, 393, 1091–1105 CrossRef CAS PubMed.
- E. Skoropata, R. D. Desautels, E. Falvo, P. Ceci, O. Kasyutich, J. W. Freeland and J. van Lierop, Phys. Rev. B: Condens. Matter Mater. Phys., 2014, 90, 174424 CrossRef.
- K. Fan, C. Cao, Y. Pan, D. Lu, D. Yang, J. Feng, L. Song, M. Liang and X. Yan, Nat. Nanotechnol., 2012, 7, 459–464 CrossRef CAS PubMed.
- M. Uchida, M. L. Flenniken, M. Allen, D. A. Willits, B. E. Crowley, S. Brumfield, A. F. Willis, L. Jackiw, M. Jutila, M. J. Young and T. Douglas, J. Am. Chem. Soc., 2006, 128, 16626–16633 CrossRef CAS PubMed.
- Z. Zhen, W. Tang, C. Guo, H. Chen, X. Lin, G. Liu, B. Fei, X. Chen, B. Xu and J. Xie, ACS Nano, 2013, 7, 6988–6996 CrossRef CAS PubMed.
- D. He and J. Marles-Wright, New Biotechnol., 2015, 32, 651–657 CrossRef CAS PubMed.
- T. Kim, D. Moore and M. Fussenegger, J. Biotechnol., 2012, 162, 237–245 CrossRef CAS PubMed.
- I. Kolinko, A. Lohße, S. Borg, O. Raschdorf, C. Jogler, Q. Tu, M. Pósfai, É. Va Tompa, J. M. Plitzko, A. Brachmann, G. Wanner, R. Müller, Y. Zhang and D. Schüler, Nat. Nanotechnol., 2014, 9, 193–197 CrossRef CAS PubMed.
- V. Zablotskii, T. Polyakova, O. Lunov and A. Dejneka, Sci. Rep., 2016, 6, 1–13 CrossRef PubMed.
- B. Polyak and G. Friedman, Expert Opin. Drug Delivery, 2009, 6, 53–70 CrossRef CAS PubMed.
- B. Polyak, I. Fishbein, M. Chorny, I. Alferiev, D. Williams, B. Yellen, G. Friedman and R. J. Levy, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(2), 698–703 CrossRef CAS PubMed.
- N. Gálvez, B. Fernandez, E. Valero, P. Sánchez, R. Cuesta and J. M. Domínguez-Vera, C. R. Chim., 2008, 11, 1207–1212 CrossRef.
- A. García-Prieto, J. Alonso, D. Muñoz, L. Marcano, A. Abad Díaz de Cerio, R. Fernández de Luis, I. Orue, O. Mathon, A. Muela and M. L. Fdez-Gubieda, Nanoscale, 2016, 8, 1088–1099 RSC.
- E. Fantechi, C. Innocenti, M. Zanardelli, M. Fittipaldi, E. Falvo, M. Carbo, V. Shullani, L. Di Cesare Mannelli, C. Ghelardini, A. M. Ferretti, A. Ponti, C. Sangregorio and P. Ceci, ACS Nano, 2014, 8, 4705–4719 CrossRef CAS PubMed.
- M. Meister, eLife, 2016, 5, 589–599 Search PubMed.
- V. Zablotskii, O. Lunov, A. Dejneka, L. Jastrabík, T. Polyakova, T. Syrovets and T. Simmet, Appl. Phys. Lett., 2011, 99, 183701 CrossRef.
- J. L. Tan, J. Tien, D. M. Pirone, D. S. Gray, K. Bhadriraju and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1484–1489 CrossRef CAS PubMed.
- M. Prass, K. Jacobson, A. Mogilner and M. Radmacher, J. Cell Biol., 2006, 174(6), 767–772 CrossRef CAS PubMed.
- D. Mitrossilis, J.-C. Röper, D. Le Roy, B. Driquez, A. Michel, C. Ménager, G. Shaw, S. Le Denmat, L. Ranno, F. Dumas-Bouchiat, N. M. Dempsey and E. Farge, Nat. Commun., 2017, 8, 13883 CrossRef CAS PubMed.
- N. J. Sniadecki, A. Anguelouch, M. T. Yang, C. M. Lamb, Z. Liu, S. B. Kirschner, Y. Liu, D. H. Reich and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(37), 14553–14558 CrossRef CAS PubMed.
- J. Fink, N. Carpi, T. Betz, A. Bétard, M. Chebah, A. Azioune, M. Bornens, C. Sykes, L. Fetler, D. Cuvelier and M. Piel, Nat. Cell Biol., 2011, 13(7), 771–778 CrossRef CAS PubMed.
- J. Howard and A. J. Hudspeth, Neuron, 1988, 1, 189–199 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |
