Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ezetimibe alleviates non-alcoholic fatty liver disease through the miR-16 inhibiting mTOR/p70S6K1 pathway
Xiang Wang *,
Yunbing Meng and
Junrong Zhang
*,
Yunbing Meng and
Junrong Zhang
Department of Endocrinology and Metabolism, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450052, China. E-mail: wangxiang801028@163.com; Tel: +86-371-66295042
First published on 1st August 2017
Abstract
Emerging studies have indicated the role of ezetimibe, miR-16 and mTOR signaling in non-alcoholic fatty liver disease (NAFLD). However, few studies have demonstrated their correlation and regulation functions in NAFLD. Therefore, the protective effect of ezetimibe in NAFLD and the potential mechanisms were explored in the current study. A NAFLD in vivo rat model was established. In addition, a rat hepatocyte cell line, BRL3A, was treated with 1 mM free fatty acids (FFA) to induce NAFLD in vitro. Hepatic triglyceride and lipid metabolism genes (ACC, Fas) and serum biochemical parameters (cholesterol, ALT) were measured. miR-16 and mTOR/p70S6K1 pathway protein expression in both rat livers and cultured cells was also assessed. The direct targeting relationship between miR-16 and mTOR was determined by the dual luciferase reporter gene assay. BRL3A cells were transfected with an miR-16 mimic for in vitro functional studies, and HFD-fed rats were tail vein injected with miR-16 inhibitor and in vivo experimental validation was carried out. NAFLD models both in vivo and in vitro were established successfully. The expression of hepatic triglyceride and lipid metabolism genes was increased and serum biochemical parameters, miR-16 and mTOR/p70S6K1 pathway proteins, were unusually expressed in NAFLD rats and BRL3A cells. Ezetimibe was found to alleviate abnormal expression of these molecules. Through the dual luciferase reporter gene assay, we found that mTOR was a target gene of miR-16. In addition, miR-16 over-expression could reverse the up-regulation of lipid metabolism genes in BRL3A cells, while this effect could be attenuated by pcDNA-mTOR. Ezetimibe significantly reduced the expression of ACC and Fas. After rapamycin treatment, the action of ezetimibe in regulating Fas and AAC was not reversed; however, rapamycin further reduced the expression of ACC and Fas. Silencing miR-16 reversed the protections given by ezetimibe in HFD rats, which showed up-regulated hepatic triglyceride, lipid metabolism genes, serum biochemical parameters and mTOR/p70S6K1 pathway proteins. Ezetimibe alleviated NAFLD induced by a HFD, at least partly, through inhibition of the mTOR/p70S6K1 pathway by miR-16 and affecting the regulation of lipid metabolism, which provides a therapeutic method for NAFLD.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is emerging as the most common liver disease worldwide.1 A population screening using ultrasonography estimates that the incidence rate of NAFLD is approximately 15% in China.2 The disease is characterized by accumulation of triglycerides and free cholesterol in the liver tissues.3 This condition mainly results from a lipid metabolism pathway imbalance, mainly including triacylglycerol delivery, synthesis, export or oxidation.4 It is believed that the special disorder is closely associated with cryptogenic cirrhosis and might lead to hepatocellular carcinoma (HCC).5 Nevertheless, the underlying molecular mechanism and therapeutic medicines for NAFLD remain largely unknown. Recently, a new class of RNA regulatory genes, known as microRNAs (miRNAs), have received great attention because of numerous findings of their dysregulation effect in various diseases6,7 and their potential as diagnostic and therapeutic targets.MiRNAs are a type of small, non-coding RNA molecule that function in post-transcriptional regulation of gene expression.8 Increasing amount of evidence supports their role in different physiological progressions and human diseases, particularly in NAFLD,9 cirrhosis and HCC. Numerous miRNAs have been demonstrated to participate in the process of NAFLD, for example, decreasing expression of miR-34a potentially contributes to altered lipid metabolism in NAFLD, which may be a therapeutic strategy against NAFLD by regulating its target PPARα and SIRT1.10 miR-185 could regulate the expression of lipid metabolism genes and improve insulin sensitivity in mice with NAFLD, shown through in vivo and in vitro experiments.11 However, whether miR-16 is also involved in the progression of NAFLD is unclear and its exact role of in the regulation of lipid metabolism is yet to be determined. In addition, the accuracy of bioinformatic predictions needs to be experimentally confirmed.
The mammalian target of rapamycin (mTOR), member of the phosphatidylinositol 3 kinase (PI3K)-related serine/threonine kinase, plays a central part in the biological process of cell growth, proliferation and survival.12 Moreover, the mTOR/p70S6K1 pathway has been described as having an important role in various diseases. A recent study presents that miR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1 signaling, which indicates that mTOR/P70S6K1 might be used as a therapeutic target in response to cisplatin in ovarian cancer treatment.13 In addition, angiotensin II (ANG II)-stimulated DNA synthesis is mediated by EGF receptor-dependent PI3K/Akt/mTOR/p70S6K1 signal pathways in mouse embryonic stem cells.14 Taking these results together, it is likely that the mTOR/p70S6K1 pathway may also be an effective treatment for NAFLD.
To develop a better therapeutic method for NAFLD it is urgently necessary to explore effective pharmacological agents, because of the increasing prevalence of the disease. In clinical settings, ezetimibe remains the most widely used first-line drug for the treatment of hypercholesteremia by inhibiting cholesterol absorption.15 Unexpectedly, it is found that ezetimibe treatment can induce a complete resistance to cholesterol gallstone formation and NAFLD.16 In addition, it has been shown that ezetimibe may exert these above effects by reducing Srebp-1c expression in mice fed a high-fat diet.17 Another study indicates that ezetimibe is effective in reducing serum low-density lipoprotein cholesterol levels resistant to lifestyle intervention in patients with NAFLD.18 Despite these notable findings, mechanisms by which ezetimibe administration ameliorates NAFLD remain largely unexplored.
In the present study we investigated whether ezetimibe could alleviate the development of NAFLD and elaborate on the potential mechanism for inhibition of the development of NAFLD.
2. Materials and methods
2.1 NAFLD model establishment
A total of 18 male SD rats (4 weeks of age) weighing about 100–120 g were selected and used for NAFLD model establishment. The rats were acclimated for 1 week before the experiment started, housed under controlled environmental conditions with free access to standard laboratory diet and water for acclimatization. After one week of acclimatization to laboratory conditions these animals were randomly divided into three groups, with 6 rats in each group. These groups were either fed the standard chow diet (SCD group) for 7 weeks, a high-fat diet (HFD group) for 7 weeks, or a high-fat diet for 7 weeks followed by gavaging ezetimibe (0.0064% w/w, 7 mg per kg per day) for further 7 weeks (HFD + ezetimibe group). The period of procedures lasted for 14 weeks in total. At the end of experiments all rats were sacrificed, and the damaged liver lesions were disassociated for real time PCR or western blotting analysis. Human subjects were not used. The animals were maintained in accordance with the guidelines of the National Institute of Nutrition, Hyderabad, India. All animal experiments were approved by the Experimental Animal Ethics Committee of The First Affiliated Hospital of Zhengzhou University.2.2 Tail vein injection of lentiviral vector
Male SD rats of 4 weeks of age, each weighing about 100–120 g, were chosen and acclimated for 1 week before they were used for experiments. The sequence of the miR-16 inhibitor was built into a pLenti6.3/TO/V5 vector (Invitrogen, Carlsbad, USA). The rats were given a HFD for 4 weeks and then divided into three groups: the ezetimibe group were gavaged with ezetimibe (n = 10); the ezetimibe + NC group were treated with ezetimibe and 100 μl negative control siRNA (NC) via the tail vein (n = 10); the ezetimibe + miR-16 inhibitor group were maintained on ezetimibe injected with an equal volume of pLenti6.3/TO/V5 vector for 4 more weeks (n = 10). The animals were then sacrificed. Blood was collected just before they were sacrificed for serum biochemical analysis. Liver tissues were quickly excised, cleaned completely with ice-cold PBS, and preserved in liquid nitrogen for further analysis.2.3 Cell culture
The rat normal hepatocyte cell line BRL3A cells used in the current study were obtained from Shanghai Institute of Cell Biology (Shanghai, China). BRL3A cells were maintained in Dulbecco's modified eagle medium (DMEM) cell culture media supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Sigma), 2 mM glutamine, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U ml−1 penicillin and 100 μg ml−1 streptomycin under a humidified atmosphere of 5% CO2 at 37 °C.
U ml−1 penicillin and 100 μg ml−1 streptomycin under a humidified atmosphere of 5% CO2 at 37 °C.
2.4 Cellular steatosis and ezetimibe treatment
BRL3A cell lines were plated to six well plates and pre-treated with ezetimibe (50 μM, dissolved in DMSO) for 16 h. Fat over-loading incubation of cells was performed by exposing BRL3A cells to a mixture of FFA (oleate and palmitate) at a final ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and final concentration of 1 mM. Afterwards, cells were washed and collected for further real time PCR or western blotting analysis.
1 and final concentration of 1 mM. Afterwards, cells were washed and collected for further real time PCR or western blotting analysis.
2.5 miRNA transfection and over-expression of mTOR
BRL3A cells were pre-treated with FFA and cultured in six well plates for 24 h. miR-16 mimic, inhibitor and mTOR over-expression (pcDNA-mTOR) plasmid, along with their negative controls (pre-NC, NC or pcDNA), were transfected into cells using lipofectamine 2000 reagent (Invitrogen, USA) following the manufacturer's instructions when cells were at 50–70% confluence. The mimic, inhibitor, pcDNA-mTOR and NCs were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).2.6 Quantitative real-time PCR
Real time PCR was preformed to detect the transcription of miR-16, ACC and Fas. Total RNA was extracted from liver tissues and BRL3A cells using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. First, cDNA was reverse-transcribed by a reverse-transcription kit (Takara, Dalian, China). Real-time PCR analysis was conducted on an Applied Bio-system 7500 instrument using SYBR Green PCR Master Mix (Qiagen, China). The relative expression of miR-16, ACC and Fas was calculated using the 2−△△CT method. The CT values were normalized using GAPDH as the internal control.2.7 Western blotting analysis
After different treatments, rat livers or BRL3A cells were rinsed in ice-cold phosphate buffer and lysed in RIPA lysis buffer to collect proteins. Samples were then centrifuged for 20 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. The protein concentration was determined by Coomassie brilliant blue protein assay. An equal amount of protein was electrophoresed on 12% SDS-PAGE and transferred to PVDF membranes (Millipore, USA). Membranes were then blocked with 5% nonfat milk at room temperature for 1 h. Subsequently, the membranes were incubated with each primary antibody from Abcam (MA, USA), including anti-p70S6K1 antibody, anti-mTOR antibody, and anti-β-actin, overnight at 4 °C. Next, the membranes were washed and incubated with HRP-conjugated secondary antibody (Santa Cruz Biotechnology, USA) for 1 h. Band intensity was quantified using the software Quantity one. All total protein expression values were normalized to the β-actin level.
000g. The protein concentration was determined by Coomassie brilliant blue protein assay. An equal amount of protein was electrophoresed on 12% SDS-PAGE and transferred to PVDF membranes (Millipore, USA). Membranes were then blocked with 5% nonfat milk at room temperature for 1 h. Subsequently, the membranes were incubated with each primary antibody from Abcam (MA, USA), including anti-p70S6K1 antibody, anti-mTOR antibody, and anti-β-actin, overnight at 4 °C. Next, the membranes were washed and incubated with HRP-conjugated secondary antibody (Santa Cruz Biotechnology, USA) for 1 h. Band intensity was quantified using the software Quantity one. All total protein expression values were normalized to the β-actin level.
2.8 Luciferase activity assay
The sequences of rat mTOR from GeneBank (NC_000001.11) were used. First, luciferase cDNA fused with the rat 3′ UTR of the mTOR gene was amplified and cloned into the psi-CHECK-2 vector to generate the luciferase reporter plasmid psi-CHECK-mTOR-3′ UTR. When BRL3A cells grew to 70% confluence, miR-16 mimic or inhibitor or psi-CHECK-mTOR-3′ UTR plasmid were co-transfected into cells using lipofectamine 2000 (Invitrogen, USA). The relative luciferase activity of mTOR 3′ UTR was measured using the Dual-Luciferase Reporter Assay (Promega, USA).2.9 Statistical analysis
All the results are presented as mean ± S.D. values. Statistical significance of difference was determined using the student's t-test in Graphad Prism 5 (Graphad, San Diego, CA, USA), and the results were considered significant when the P-values were less than 0.05.3. Results
3.1 Determination of biochemical parameters and lipid metabolism-related genes in NAFLD rats
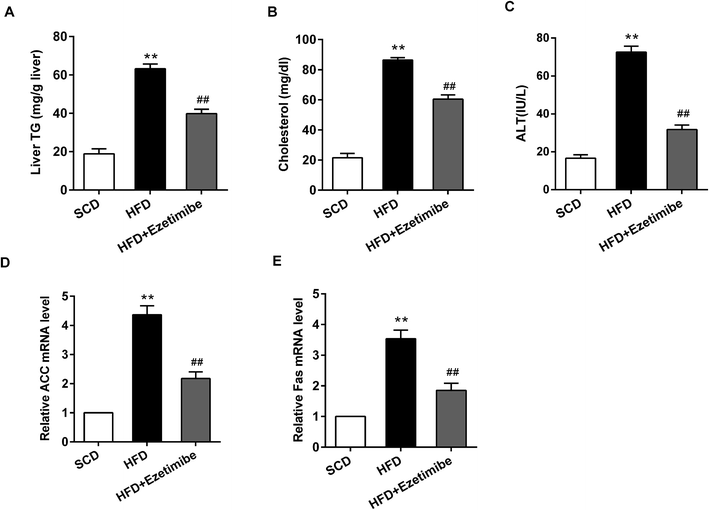
After a HFD induced NAFLD in rats, the serum and damaged livers were collected and analyzed. The production of biochemical parameters (triglycerides, cholesterol, ALT) and lipid metabolism related genes (ACC, Fas) was detected using ELISA and the quantitative RT-PCR method, respectively. In this study, we found that when compared with that in the SCD group the liver triglyceride expression (Fig. 1A), serum cholesterol (Fig. 1B) and ALT activity (Fig. 1C) in the HFD group were significantly increased. Moreover, qRT-PCR analysis showed that relative ACC (Fig. 1D) and Fas (Fig. 1E) mRNA levels were significantly elevated in the HFD group. However, after the animals were treated with HFD and gavaged with ezetimibe at a concentration of 7 mg per kg per day, differences in the expression of the above indicators could be significantly reversed compared to the HFD group.3.2 Abnormal expression of miR-16 and mTOR pathway proteins in livers of NAFLD rats
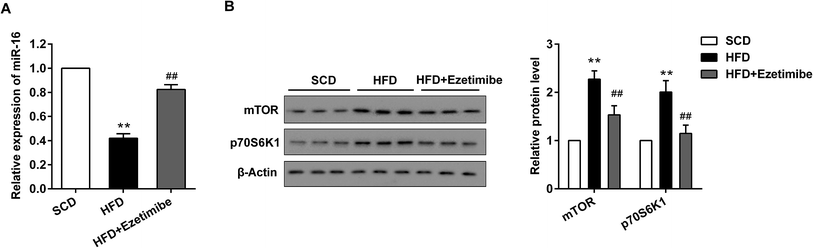
The expression of miR-16 and mTOR pathway proteins was assessed by qRT-PCR and western blotting. As exhibited in Fig. 2A, down-regulated miR-16 expression induced by HFD was strongly reversed by ezetimibe treatment, to a level close to the basal expression. Further analysis for total mTOR and p70S6K1 proteins showed that they were significantly increased in the HFD group when compared with that in SCD group, and treatment of NAFLD animals with ezetimibe caused a significant decrease (Fig. 2B).3.3 Alteration of miR-16 and mTOR pathway proteins in BRL3A cells in vitro
BRL3A cell lines were pre-treated with ezetimibe (50 μM) for 16 h and exposed to a mixture of FFA (1 mM, oleate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) palmitate = 2
palmitate = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
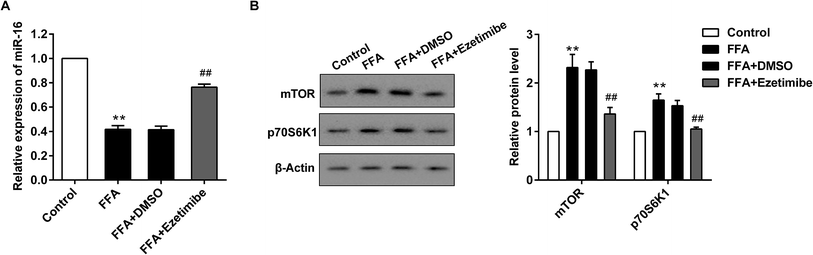
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to establish an NAFLD model in vitro. After various treatments cells were collected for qRT-PCR and western blotting analysis. Fig. 3A shows a significant reduction of the miR-16 level in the presence of FFA treatment compared with control cells, while supplementation with FFA + ezetimibe significantly elevated the expression of miR-16 when compared with that of the FFA + DMSO group. Moreover, the expression of total mTOR and p70S6K1 proteins was detected by western blotting and results showed that the two proteins were apparently up-regulated in FFA-processed BRL3A cells compared with that in controls. However, they were visibly reduced after ezetimibe and FFA were introduced together as compared to those in the FFA + DMSO group (Fig. 3B).
1) to establish an NAFLD model in vitro. After various treatments cells were collected for qRT-PCR and western blotting analysis. Fig. 3A shows a significant reduction of the miR-16 level in the presence of FFA treatment compared with control cells, while supplementation with FFA + ezetimibe significantly elevated the expression of miR-16 when compared with that of the FFA + DMSO group. Moreover, the expression of total mTOR and p70S6K1 proteins was detected by western blotting and results showed that the two proteins were apparently up-regulated in FFA-processed BRL3A cells compared with that in controls. However, they were visibly reduced after ezetimibe and FFA were introduced together as compared to those in the FFA + DMSO group (Fig. 3B).
3.4 miR-16 targeted mTOR to regulate pathway related factors expression
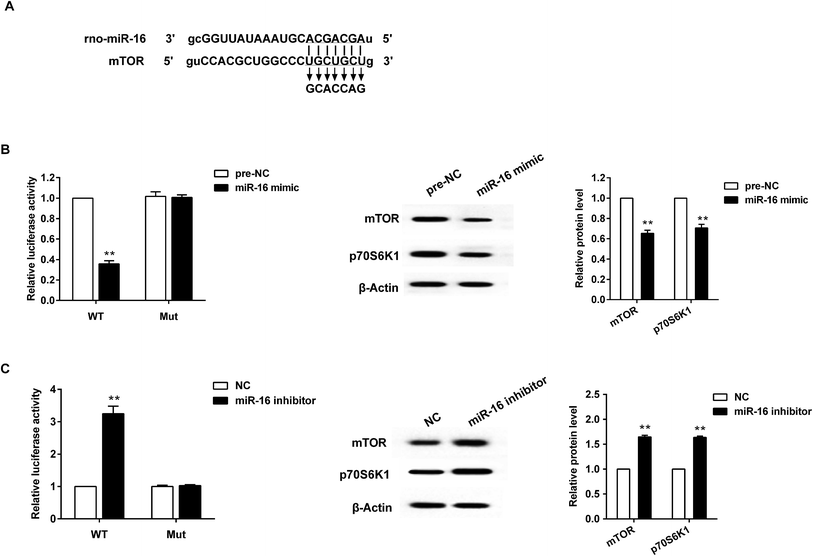
A bioinformatics database (TargetScan) was used to identify miR-16 target genes. Among hundreds of predicted targets, mTOR was chosen for further validation in BRL3A cells using luciferase reporter assays. Fig. 4A shows the predicted miR-16-binding sites in 3′UTR of mTOR. The luciferase reporter assay was then conducted, and it demonstrated that over-expression of miR-16 decreased mTOR 3′UTR-WT but not mTOR 3′UTR-Mut activity. In addition, miR-16 mimic significantly reduced the total p70S6K1 and mTOR expression at the protein level (Fig. 4B). On the other hand, down-regulation of miR-16 increased mTOR 3′UTR-WT but not mTOR 3′UTR-Mut activity, along with the p70S6K1 and mTOR expression level (Fig. 4C).3.5 miR-16 mediating mTOR involved in synthesis of lipid metabolism related genes in BRL3A cells
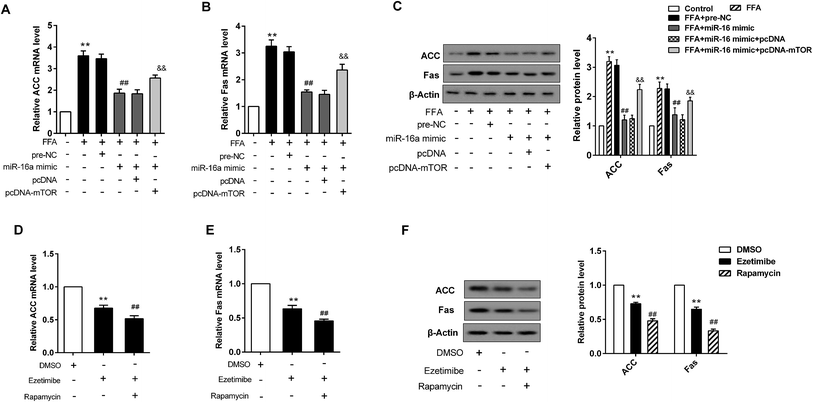
BRL3A cells were pre-treated with FFA for 24 h. Following this, miR-16 mimic and pcDNA-mTOR plasmid, along with their negative controls (pre-NC or pcDNA), were transfected into the cells. When BRL3A cells were exposed to FFA the relative ACC mRNA level was elevated, whereas it was decreased after the introduction of miR-16a mimic. However, the reduction could be reversed by mTOR over-expression (Fig. 5A). miR-16a mimic also prominently reduced the expression of Fas in BRL3A cells exposed to FFA. However, its effect was strongly attenuated by over-expression of mTOR (Fig. 5B). The corresponding protein expression and ACC and Fas protein production histogram is presented in Fig. 5C. Moreover, ezetimibe treatment significantly decreased relative ACC and Fas mRNA level, while mTOR inhibitor further exacerbated this trend (Fig. 5D and E). Similarly, the protein expression of ACC and Fas was also visibly reduced in cells treated with ezetimibe; however, the downward trend was more prominent when cells were treated with ezetimibe plus rapamycin (Fig. 5F).3.6 Knockdown of miR-16 promoted the levels of lipid metabolism related genes and mTOR pathway proteins in vivo
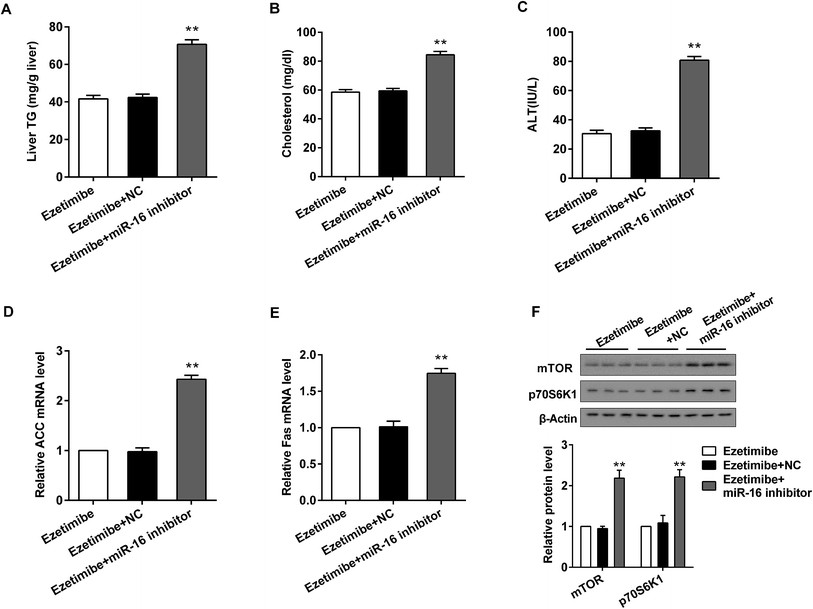
To verify the effects of miR-16 on the expression of lipid metabolism-related genes and mTOR pathway proteins in vivo, the HFD-fed rats were divided into three groups: ezetimibe, ezetimibe + NC and ezetimibe + miR-16 inhibitor. miR-16 inhibitor tail vein injection dramatically increased the expression of liver triglycerides (Fig. 6A), serum cholesterol (Fig. 6B) and ALT (Fig. 6C) in NAFLD rats. Using qRT-PCR analysis we confirmed that the knockout of miR-16 prominently elevated both the relative ACC (Fig. 6D) and Fas (Fig. 6E) mRNA levels. The mTOR pathway proteins (p70S6K1 and mTOR) of NAFLD rats were also significantly improved by miR-16 inhibitor treatment (Fig. 6F).4. Discussion
In the present study, we found that mTOR pathway proteins and lipid metabolism related genes were up-regulated, and the miR-16 relative level was down-regulated in NAFLD model rats and in BRL3A cells treated with FFA. Ezetimibe could alleviate abnormal expression of these molecules. With the presence of miR-16 mimic in rat hepatocyte BRL3A cells, the expression of lipid metabolism-related genes (ACC and Fas) was significantly decreased compared to that in controls; however, this effect could be reversed by mTOR over-expression.Although the pathogenesis of NAFLD is not entirely explored, the vital role of miRNAs as major mediators of fatty acid and lipid metabolism deserves attention. Moreover, emerging evidence supports a key role for miRNAs in regulating lipid metabolism and insulin sensitivity, e.g. miR-29 fine-tuning the expression of key FOXA2-activated lipid metabolism genes which is dysregulated in animal models of insulin resistance and diabetes.19 Silencing of endogenous miR-26a in mice fed with a conventional diet impairs insulin sensitivity, enhances glucose production and increases fatty acid synthesis.20 A recent article reported that miR-16 participates in the progression of liver steatosis, a type of NAFLD.21 Similarly, in the current study, we found that miR-16 is down-regulated in NAFLD rats and BRL3A cells, which indicates that it may be associated with the development of NAFLD.
To investigate the regulatory mechanism of miR-16, we also detected the role of the mTOR/p70S6K1 pathway in NAFLD. Through luciferase activity assay analysis, we found that miR-16 can target mTOR to inhibit its down-stream gene expression, also regulating hepatic lipid metabolism and the development of NAFLD. In addition, mTOR represents an exciting target to offer new clinical avenues for the treatment of fatty liver disease and the complications of this disease.22 Thus far, it has been demonstrated that there is a role of mTOR action in metabolism, and it has been highlighted that there exists a potent mTOR/ERRα regulatory axis with significant clinical impact.23 However, in the field of liver diseases, reports about miRNAs and mTOR are extremely rare. Uesugi's finding identified that miR-218 targeted the mTOR component Rictor and inhibited AKT phosphorylation in oral cancer.24 Moreover, miR-7 regulated the mTOR pathway and proliferation in adult pancreatic β-cells.25 In the present study, over-expression of miR-16 contributes to reduced fatty acids and cholesterol biosynthesis, which parallels the observed reduction in relative levels of the lipid metabolism-related genes ACC and Fas. Therefore, there might be some correlations between miR-16, the mTOR/p70S6K1 pathway and lipid metabolism, which needs to be investigated further.
Currently, no standard treatments are used to reverse NAFLD, and effective medical intervention is particularly critical. Previous studies showed that various pharmacological agents were applied in the treatment of NAFLD. For example, betulinic acid alleviates NAFLD by inhibiting SREBP1 activity via the AMPK–mTOR–SREBP signaling pathway.26 Gao et al. demonstrated the beneficial effects of the traditional Chinese medicine sea buckthorn (SBT) on patients with NAFLD, which may be further developed as a promising therapy for the treatment of NAFLD.27 Currently, our study indicated that ezetimibe alleviated NAFLD progression through inhibition of the mTOR/p70S6K1 pathway by miR-16.
In summary, our study showed increased mTOR pathway proteins and lipid metabolism related genes, as well as decreased miR-16 relative levels, in NAFLD rats and BRL3A cells. Moreover, the finding that ezetimibe mitigated NAFLD through inhibition of the mTOR/p70S6K1 pathway by miR-16 added to our understanding of the mechanisms. However, further studies should be established to explore the deep mechanism underlying NAFLD for new therapies.
Conflict of interest
None.Acknowledgements
We are sincerely thankful to every member involved in the project, and this study was supported by Science Foundation for Young Scientists of The First Affiliated Hospital of Zhengzhou University.References
- M. F. Karim, et al., Non-alcoholic Fatty Liver Disease (NAFLD) – A Review, Mymensingh Med. J., 2015, 24(4), 873–880 CAS.
- F. Montazerifar, M. Karajibani and A. R. A. Moghaddam, Relationship between fatty liver disease and biochemical factors in obese adolescents, Rawal Med. J., 2014, 39(1), 15–18 Search PubMed.
- Y. Y. Seo, et al., Tumor Necrosis Factor-α as a Predictor for the Development of Nonalcoholic Fatty Liver Disease: A 4-Year Follow-Up Study, Endocrinol. Metab., 2013, 28(1), 41–45 CrossRef PubMed.
- A. Mühlroth, et al., Pathways of Lipid Metabolism in Marine Algae, Co-Expression Network, Bottlenecks and Candidate Genes for Enhanced Production of EPA and DHA in Species of Chromista, Mar. Drugs, 2013, 11(11), 4662–4697 CrossRef PubMed.
- R. Kumar, Hard clinical outcomes in patients with NAFLD, Hepatol. Int., 2013, 7(2), 790–799 CrossRef PubMed.
- G. Reid, et al., Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer, Epigenomics, 2017, 8(8), 1079–1085 CrossRef PubMed.
- Y. Qi, et al., Prognostic value of the microRNA-29 family in multiple human cancers: a meta-analysis and systematic review, Clin. Exp. Pharmacol. Physiol., 2017, 44(4), 441–454 CrossRef CAS PubMed.
- M. Azimzadeh, et al., An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer, Biosens. Bioelectron., 2016, 77, 99–106 CrossRef CAS PubMed.
- J. M. Estep, et al., Adipocytokine expression associated with miRNA regulation and diagnosis of NASH in obese patients with NAFLD, Liver Int., 2014, 35(4), 1367–1372 CrossRef PubMed.
- J. Ding, et al., Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease, Sci. Rep., 2015, 5, 13729 CrossRef PubMed.
- X. C. Wang, et al., Micro RNA-185 regulates expression of lipid metabolism genes and improves insulin sensitivity in mice with non-alcoholic fatty liver disease, World J. Gastroenterol., 2014, 20(47), 17914–17923 CAS.
- J. Moore, et al., Rosemary extract reduces Akt/mTOR/p70S6K activation and inhibits proliferation and survival of A549 human lung cancer cells, Biomed. Pharmacother., 2016, 83, 725–732 CrossRef CAS PubMed.
- X. Shaohua, et al., MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1, Oncotarget, 2011, 6(28), 26457–26471 Search PubMed.
- H. J. Han, et al., ANG II-stimulated DNA synthesis is mediated by ANG II receptor-dependent Ca(2+)/PKC as well as EGF receptor-dependent PI3K/Akt/mTOR/p70S6K1 signal pathways in mouse embryonic stem cells, J. Cell. Physiol., 2007, 211(3), 618–629 CrossRef CAS PubMed.
- S. Oswald, et al., Drug Interactions Between the Immunosuppressant Tacrolimus and the Cholesterol Absorption Inhibitor Ezetimibe in Healthy Volunteers, Clin. Pharmacol. Ther., 2011, 89(4), 524 CrossRef CAS PubMed.
- O. D. Bari, et al., Ezetimibe: Its Novel Effects on the Prevention and the Treatment of Cholesterol Gallstones and Nonalcoholic Fatty Liver Disease, J. Lipids, 2012, 2012(2), 302847 Search PubMed.
- T. Muraoka, et al., Ezetimibe decreases SREBP-1c expression in liver and reverses hepatic insulin resistance in mice fed a high-fat diet, Metab., Clin. Exp., 2011, 60(5), 617–628 CrossRef CAS PubMed.
- N. Oza, et al., Efficacy of ezetimibe for reducing serum low-density lipoprotein cholesterol levels resistant to lifestyle intervention in patients with non-alcoholic fatty liver disease, Hepatol. Res., 2014, 44(7), 812–817 CrossRef CAS PubMed.
- C. L. Kurtz, et al., MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes, Diabetes, 2014, 63(9), 3141 CrossRef PubMed.
- X. Fu, et al., MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids, J. Clin. Invest., 2015, 125(6), 2497–2509 Search PubMed.
- J. H. Kim, et al., Reverse Expression of Aging-Associated Molecules through Transfection of miRNAs to Aged Mice, Mol. Ther.–Nucleic Acids, 2017, 6, 106–115 CrossRef CAS PubMed.
- C. Wang, et al., Functional crosstalk between AKT/mTOR and Ras/MAPK pathways in hepatocarcinogenesis: implications for the treatment of human liver cancer, Cell Cycle, 2013, 12(13), 1999–2010 CrossRef CAS PubMed.
- C. Chaveroux, et al., Molecular and genetic crosstalks between mTOR and ERRα are key determinants of rapamycin-induced nonalcoholic fatty liver, Cell Metab., 2013, 17(4), 586–598 CrossRef CAS PubMed.
- A. Uesugi, et al., The Tumor Suppressive MicroRNA miR-218 Targets the mTOR Component Rictor and Inhibits AKT Phosphorylation in Oral Cancer, Cancer Res., 2011, 71(17), 5765–5778 CrossRef CAS PubMed.
- Y. Wang, et al., MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells, Diabetes, 2012, 62(3), 887–895 CrossRef PubMed.
- H. Y. Quan, et al., Betulinic acid alleviates non-alcoholic fatty liver by inhibiting SREBP1 activity via the AMPK–mTOR–SREBP signaling pathway, Biochem. Pharmacol., 2013, 85(9), 1330–1340 CrossRef CAS PubMed.
- Z. Gao, et al., Efficacy of Sea Buckthorn Therapy in Patients with Nonalcoholic Fatty Liver Disease, Chin. Med., 2014, 5(4), 223–230 CrossRef.
| This journal is © The Royal Society of Chemistry 2017 |