Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein–inorganic hybrid system for efficient his-tagged enzymes immobilization and its application in L-xylulose production†
Sanjay K. S. Patel a,
Sachin V. Otari
a,
Sachin V. Otari a,
Yun Chan Kang
a,
Yun Chan Kang *b and
Jung-Kul Lee
*b and
Jung-Kul Lee *a
*a
aDepartment of Chemical Engineering, Konkuk University, 1 Hwayang-Dong, Gwangjin-Gu, Seoul, 143-701, South Korea. E-mail: jkrhee@konkuk.ac.kr
bDepartment of Materials Science and Engineering, Korea University, Anam-Dong, Seongbuk-Gu, Seoul, 136-713, South Korea. E-mail: yckang@korea.ac.kr
First published on 13th January 2017
Abstract
The facile synthesis of protein–inorganic hybrid nanoflowers was evaluated for the efficient immobilization of recombinant his-tagged enzymes, which have a broad range of potential applications. In this study, we report the preparation of a metal–protein hybrid nanoflower system for efficient immobilization of the recombinant enzymes L-arabinitol 4-dehydrogenase from Hypocrea jecorina (HjLAD) and NADH oxidase from Streptococcus pyogenes (SpNox). Compared with free enzymes, synthesized hybrid nanoflowers exhibited enhanced enzymatic activities of 246 and 144% for HjLAD and SpNox, respectively. We have demonstrated that immobilized enzymes retained high catalytic activity and improved the tolerance towards pH and temperature changes. Synthesized nanoflowers also retained high storage stability and reusability. In addition, the immobilized enzymes exhibited significantly enhanced L-xylulose production under co-factor regeneration conditions than the free enzyme combination. These results demonstrate that variations in the concentration of metals and synthesis conditions of nanoflowers can be extended to efficiently immobilize recombinant his-tagged enzymes.
1. Introduction
Recently, enzyme immobilization using a hybrid system with metals as protein–inorganic hybrid has received significant attention for its ability to overcome the primary limitations associated with free enzyme efficiency and stability.1–8 The process of enzyme immobilization is useful for improving these properties of the enzymes. However, there are still problems associated with low loading, efficiency, and stability of the enzymes immobilized on various supports.9–12 Therefore, suitable supports to achieve high loading and immobilization efficiency using different methods, including adsorption, covalent bonding, and cross-linking have been discussed.9,11,12 A simple method of enzyme immobilization using an enzyme-embedded metal hybrid system appears to be more efficient for retaining high enzyme efficiency than most immobilization processes performed on solid support materials, including various nano-particles.1,13–17 Ge et al. developed a unique protein–inorganic encapsulation hybrid system with enzyme–Cu(PO4)2·3H2O, including α-lactalbumin, laccase, carbonic anhydrase, and lipase for efficient immobilization.1 The formation of the enzyme–Cu hybrid as a nanoflower occurred in a three-step process of nucleation, aggregation, and anisotropic growth as described previously.1 Initially, enzymes interact with primary nanocrystal of Cu3(PO4)2 to start the nucleation. Thereafter, a large aggregation of enzyme and nanocrystals is formed and finally, controlled anisotropic growth results in a branched flower like structure. Similarly, a Cu-based enzyme–metal hybrid nanoflower system was extended to different enzymes including catalase,18 α-chymotrypsin,19 trypsin,20 horseradish peroxidase,2 and lactoperoxidase.21 This type of system is also suitable for protein encapsulation using other inorganic metal components such as CaHPO4,22 Co3(PO4)2,3 Zn3(PO4)2,23 and ZnO.24 The encapsulation of enzymes using CaCO3 porous particles also contributed to better loading of enzymes and other applications.25–27 However, the synthesis of enzyme–metal hybrids using unstable enzymes at 25 °C is limited due to 3 days of long incubation.1,3,28 Therefore, modification of synthesis conditions is required to improve the efficiency of unstable enzymes after immobilization.Rare sugars are unique monosaccharides that play a significant role as recognition elements in bio-active molecules with a broad range of applications.29–31 L-Xylulose is a rare sugar used primarily as a potential inhibitor of α-glucosidases and indicator of liver cirrhosis disease.32 The conversion of carbohydrates by metal catalysts are known due their high selectivity and stability. Still, the purity of low cost based metal catalyst, their leaching and compatibility are major issues during the reaction conditions. The enzymatic production of L-xylulose is primarily limited by the cost of the co-factors such as nicotinamide adenine dinucleotide (NAD+), which is a necessary component.10,33,34 Thus, the use of a co-factor regeneration system for efficient L-xylulose production may be a cost-effective approach.
Commercially available stable enzymes have been widely studied for immobilization with Cu(PO4)2·3H2O because the synthesis can be conducted at room temperature (25 °C) and incubated for up to 3 days.1 Thus, the methods must be modified to extend the application of nanoflowers and improve enzyme stability at 25 °C. In this study, we adapted a modified nanoflower synthesis method using 2 mM of Cu for incubation of 24 h at 4 °C to improve the properties of the recombinant his-tagged enzymes L-arabinitol 4-dehydrogenase from Hypocrea jecorina (HjLAD) and NADH oxidase from Streptococcus pyogenes (SpNox). The resulting immobilized enzymes were used to produce L-xylulose from L-arabinitol.
2. Experimental section
2.1. Materials and reagents
Escherichia coli cultivation medium (Luria-Bertani, LB) was purchased from Duchefa – Postbus (Haarlem, The Netherlands). NAD+, NADH, fluorescein isothiocyanate (FITC), rhodamine B isothiocyanate (RBITC), copper sulphate, and L-arabinitol were purchased from Sigma-Aldrich (St. Louis, MO, USA). UltraPure water and phosphate-buffered saline (pH 7.4, 1×) were purchased from Life Technologies (Carlsbad, CA, USA). All other reagents were of analytical grade.2.2. Cell culture and protein purification
Recombinant E. coli BL21 (DE3) strains harboring pET28a-HjLAD,34 or pET28a-SpNox were cultured at 37 °C in LB medium supplemented with kanamycin (50 g mL−1).35 HjLAD and SpNox expression was induced with isopropyl-β-D-thiogalactopyranoside (0.1 M). The induced cells were harvested by centrifugation at 4 °C for 15 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, followed by rinsing with phosphate-buffered saline, and the cell pellet was stored at −20 °C until use. To purify recombinant HjLAD or SpNox, the cell pellets were suspended in binding buffer (50 mM NaH2PO4 and 300 mM NaCl, pH 8.0). The cell suspension was incubated on ice for 30 min in the presence of 1 mg mL−1 lysozyme. The cells were disrupted by sonication at 4 °C for 5 min and the lysate was centrifuged at 14
000 × g, followed by rinsing with phosphate-buffered saline, and the cell pellet was stored at −20 °C until use. To purify recombinant HjLAD or SpNox, the cell pellets were suspended in binding buffer (50 mM NaH2PO4 and 300 mM NaCl, pH 8.0). The cell suspension was incubated on ice for 30 min in the presence of 1 mg mL−1 lysozyme. The cells were disrupted by sonication at 4 °C for 5 min and the lysate was centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 20 min at 4 °C to remove the cell debris. The resulting crude extract was retained at 4 °C for purification. The cell-free extract was then applied onto an Ni-NTA Super flow column (3.4 × 13.5 cm, QIAGEN, Hilden, Germany) that had been previously equilibrated with binding buffer. Unbound proteins were washed from the column with washing buffer (50 mM NaH2PO4, 300 mM NaCl, and 60 mM imidazole, pH 8.0). Next, recombinant HjLAD or SpNox was eluted from the column with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). Both crude and purified enzyme fractions were analyzed by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and visualized by staining with Coomassie blue R250 (Fig. S1†).
000 × g for 20 min at 4 °C to remove the cell debris. The resulting crude extract was retained at 4 °C for purification. The cell-free extract was then applied onto an Ni-NTA Super flow column (3.4 × 13.5 cm, QIAGEN, Hilden, Germany) that had been previously equilibrated with binding buffer. Unbound proteins were washed from the column with washing buffer (50 mM NaH2PO4, 300 mM NaCl, and 60 mM imidazole, pH 8.0). Next, recombinant HjLAD or SpNox was eluted from the column with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). Both crude and purified enzyme fractions were analyzed by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and visualized by staining with Coomassie blue R250 (Fig. S1†).
2.3. Synthesis of metal–enzyme hybrid nanoflowers
The synthesis of enzyme–Cu(PO4)2·3H2O hybrid nanoflowers was performed as reported previously with some modifications.30 Briefly, 5 mL of phosphate-buffered saline solution (10 mM, pH 7.4) containing different concentrations of recombinant enzymes from 0.05–1.0 mg mL−1 were mixed with 50 μL of CuSO4 in water (200 mM) at 4 °C for 24 h to form enzyme–Cu(PO4)2·3H2O hybrid nanoflowers. In addition, the synthesis was also performed as previously described at 4 °C and 25 °C using 6.0 and 0.8 mM CuSO4 for comparison, respectively (Table S1†).3,28 The encapsulation yield (EY) and relative activity (RA) were calculated as follows (eqn (1) and (2)):| EY (%) = 100 × amount of enzyme immobilized/total initial content of protein | (1) |
| RA (%) = 100 × total activity of the immobilized enzyme/total activity of the free enzyme | (2) |
2.4. Measurements of activity
The activity of purified and nanoflowers of HjLAD or SpNox enzymes was determined spectrophotometrically by monitoring the change in A340 upon reduction and oxidation of NAD+ and NADH at 25 °C for HjLAD and SpNox, respectively. The HjLAD assay mixture for oxidation consisted of NAD+ (1.5 mM), arabinitol (200 mM), and HjLAD in Tris–glycine–NaOH (100 mM, pH 9.5). The SpNox assay mixture consisted of NADH (200 μM) and enzyme in potassium–phosphate buffer (50 mM, pH 7.0). One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of NADH or NAD+ per minute for HjLAD and SpNox under the assay conditions, respectively.2.5. Characterization of synthesized nanoflowers
The effects of pH on the activities of the purified and nanoflowers of HjLAD or SpNox enzymes were determined using standard assay conditions in various buffers (50 mM): sodium-acetate (pH 4.0–5.5), potassium-phosphate (pH 6.0–7.5), Tris–glycine–NaOH (8.0–9.5) and glycine NaOH (10.0). Further, the effect of temperature was analyzed by assaying the enzyme samples over the range of 25–70 °C. To determine the apparent kinetic parameters (Km and Vmax), the samples were incubated at 25 °C with varying concentrations of L-arabinitol (25–400 mM) and NADH (5–400 μM) for purified and nanoflowers of HjLAD or SpNox under standard assay conditions, respectively. Kinetic parameters were obtained using non-linear regression fitting of the Michaelis–Menten equation using Prism 5 (Graphpad, Inc., La Jolla, CA, USA).2.6. Stability and reusability
The stabilities of both free and nanoflowers of HjLAD or SpNox enzymes were investigated by incubation under optimum conditions in 50 mM buffer at 25 °C. Samples were withdrawn to measure the residual activity at different intervals. The denaturation kinetics parameters were determined using non-linear regression of data analysis as described previously.10 The storage stability was monitored at 4 °C over a period of two months. The reusability of the HjLAD or SpNox nanoflowers was assessed at 25 °C under standard assay conditions for 10 cycles of reuse. After each cycle, the immobilized enzyme was removed by centrifugation for 5 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. Further, immobilized enzyme was washed with buffer and used for the second cycle. The initial activity of the free enzymes or nanoflowers was considered as 100%.
000 × g. Further, immobilized enzyme was washed with buffer and used for the second cycle. The initial activity of the free enzymes or nanoflowers was considered as 100%.
2.7. L-Xylulose production
The production of L-xylulose by free or nanoflowers combinations of HjLAD and SpNox enzymes were analyzed in a 5 mL thermoreactor with a stir bar under optimum conditions.10 The reaction mixture contained 32.8 mM of L-arabinitol, 20 U of each free or nanoflowers enzyme, and 2 mM of NAD+ at pH 8.0. L-Xylulose production was performed at 25 °C for incubation up to 6 h. Samples were withdrawn at regular time intervals and analyzed using a colorimetric assay and an Ultimate 3000 high-pressure liquid chromatography as described previously.30,332.8. Instrumentation
The morphological and elemental mapping analysis of synthesized nanoflowers was investigated using scanning electron microscopy (FE-SEM, Jeol JSM-6060, Tokyo, Japan).36,37 Absorption spectra of catalytic products and reactions were recorded on a spectrophotometer (Varian Cary 100 Bio UV-Vis spectrophotometer, Palo Alto, CA, USA).38 Dynamic light scattering (DLS, Wyatt Technology, CA, USA) measurement was used to determine the average size of synthesized nanoflowers.39 HjLAD and SpNox enzymes were labeled with FITC and RBITC, respectively.12 Confocal laser scanning microscope (CLSM) images synthesized nanoflowers with labeled enzymes were acquired using an FV-1000 Olympus confocal microscope (Olympus, Tokyo, Japan).123. Results and discussion
3.1. Synthesis and characterization of Cu(PO4)2·3H2O–enzyme hybrid nanoflower
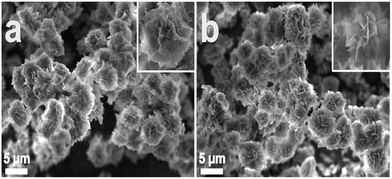
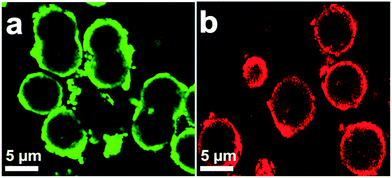
A diagram for the synthesis of metal–protein hybrid nanoflowers using Cu(PO4)2·3H2O and recombinant enzymes is presented in Fig. 1. Different concentrations of enzymes (0.05–1.00 mg mL−1) were used to synthesize nanoflowers using 2 mM of Cu(PO4)2·3H2O for incubation for 24 h at 4 °C (Table 1). The structural morphology of the synthesized Cu(PO4)2·3H2O–enzyme nanoflowers with 0.25 mg protein mL−1 of purified HjLAD and SpNox was confirmed by SEM analysis (Fig. 2). The synthesized nanoflowers were obtained in fine regular structures. DLS analysis suggested that the synthesized HjLAD and SpNox nanoflowers were homogeneously distributed with average particle sizes of 6.6 and 6.3 μm, respectively (Fig. S2†). When the morphological changes of the synthesized nanoflowers of his-tagged enzymes were evaluated at additional pH 6.5 and 8.0, similar diameters of the synthesized nanoflowers were observed with an increase in petal density at higher pH values of 7.4 and 8.0 (Fig. S3†). Elemental mapping analysis was performed to verify the components of the synthesized nanoflower HjLAD (Fig. S4†). The presence of Cu and P in the synthesized nanoflower suggests the formation of the metal and enzyme hybrid system as enzyme–Cu(PO4)2·3H2O. Further, efficient immobilization of HjLAD and SpNox was determined using CLSM images of synthesized nanoflowers of labeled proteins as FITC and RBITC, respectively (Fig. 3). The high intensity of the FITC and RBITC colors in the synthesized nanoflowers confirmed the efficient immobilization.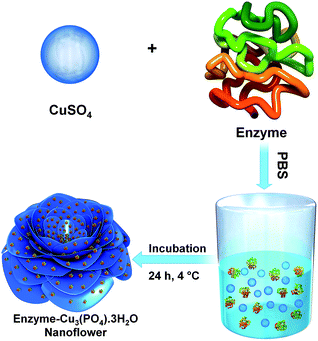 | ||
| Fig. 1 Schematic representation of his-tagged recombinant enzyme–Cu3(PO4)·3H2O hybrid nanoflower synthesis. | ||
| Protein (mg mL−1) | HjLAD | SpNox | ||
|---|---|---|---|---|
| EYa (%) | RAb (%) | EY (%) | RA (%) | |
| a Encapsulation yields.b Relative activity. | ||||
| 0.05 | 78.6 ± 6.9 | 212 ± 18 | 77.5 ± 6.8 | 127 ± 13 |
| 0.10 | 71.2 ± 6.5 | 229 ± 20 | 66.4 ± 6.1 | 138 ± 14 |
| 0.25 | 63.4 ± 5.8 | 246 ± 21 | 58.9 ± 5.7 | 144 ± 16 |
| 0.50 | 24.6 ± 2.5 | 187 ± 16 | 21.3 ± 2.0 | 108 ± 12 |
| 1.00 | 9.10 ± 0.8 | 94.8 ± 7.0 | 8.30 ± 0.8 | 63.3 ± 6.1 |
The EY of both enzymes, HjLAD and SpNox, was reduced with increasing concentrations of purified protein from 0.05 to 1.0 mg mL−1. The EYs were in the range of 9.1–78.6 and 8.3–73.5% for the HjLAD and SpNox, respectively. The synthesized nanoflowers exhibited enhanced RAs of 246 and 144% at a protein concentration of 0.25 mg mL−1 for HjLAD and SpNox with EYs of 63.4% and 58.9%, respectively. Further increases in the concentration of HjLAD from 0.25 to 1.0 mg mL−1 resulted in a significant reduction in both the EYs and RAs of enzymes to 9.1% and 94.8%, respectively. Under similar conditions, SpNox showed much lower EY and RA values of 8.3% and 63.3%, respectively. The synthesized Cu(PO4)2·3H2O his-tagged enzymes HjLAD and SpNOx hybrid nanoflowers retained 141- and 6-fold higher RAs than alcohol dehydrogenases from Geobacillus stearothermophilus and Lactococcus lactis immobilized using similar metal-his-tagged protein hybrid systems.3 Here, a decreased incubation time of 24 h showed better results than 3 day incubation. In a previous study, a lower RA was observed for alcohol dehydrogenases after immobilization, likely because of the low stability of enzymes during the synthesis procedure at 25 °C.3 To confirm these results, the immobilization of HjLAD and SpNox was performed at 25 °C for 3 days. Here, synthesized HjLAD and SpNox nanoflowers showed significantly lower RA values of 14.4% and 17.1%, respectively (Table S1†). These results suggest that enzyme stability is important for the efficient synthesis of nanoflowers.
Further, the concentration of metals also significantly influenced the EY and RA of immobilized enzymes during the nanoflower synthesis processes (Table S1†). A concentration of Cu (2 mM) appeared to be more effective for immobilizing HjLAD and SpNox compared to the higher concentration of 6 mM reported for the more stable enzymes glucose oxidase and horseradish peroxidase under similar conditions.28 At a Cu concentration of 6 mM, both HjLAD- and SpNox-synthesized nanoflowers showed lower RAs of 86.0% and 65.8%, respectively (Table S1†). These results suggest that his-tagged recombinant enzymes can be efficiently immobilized by modifying metal–protein hybrid systems, including metal concentration, incubation period, and temperature. In addition, this system retained higher RA values of 246% and 144% for HjLAD and SpNox after immobilization compared to those of 44% and 94% for dehydrogenases from G. stearothermophilus and L. lactis, respectively, obtained using Co3(PO4)2-his-tagged protein hybrid systems.3 This system is more efficient than protein–inorganic capsules system of his-tagged epoxide hydrolase from Sphingomonas sp. HXN-200 and Fmoc-diphenylalanine–polyethyleneimine–silicate with RA of 60%.40 Similarly, his-tagged enzymes ADP-dependent glucokinase, cellulase, and glucose-6-phosphate dehydrogenase immobilized through biomimetic layer-by-layer mineralization on diatom silica resulted in lower efficiency of 75–90%.41 A comparison analysis of his-tagged enzymes immobilization through different protein–inorganic hybrid systems are presented in Table S2.†
3.2. Characterization of synthesized nanoflowers
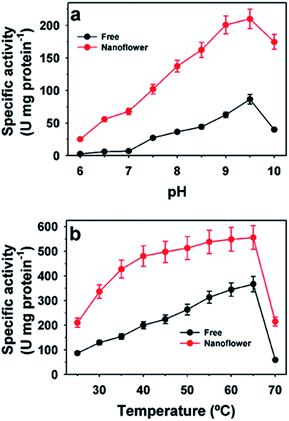
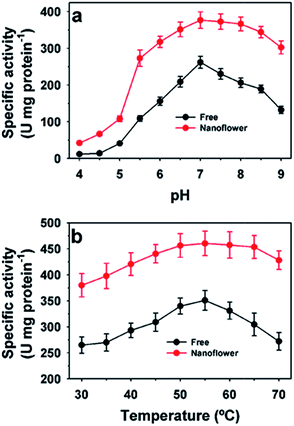
The activity profiles of free and immobilized HjLAD nanoflowers at different pH and temperature values are presented in Fig. 4. The maximum activity of the free and HjLAD nanoflowers was observed at pH 9.5 (Fig. 4a). HjLAD nanoflowers retained high residual activities over a broad range of pH 6.0–9.0 and at 10.0. Immobilization may provide a suitable buffering environment to retain high residual activity under both acidic and basic conditions. At temperatures of 25–60 °C, the HjLAD nanoflowers retained much higher residual activity than the free enzyme. The wider temperature profile of synthesized HjLAD nanoflowers suggested its lower sensitivity than the free form of the enzyme. Similar results in the activities profiles were observed with synthesized SpNox nanoflowers at different pH and temperature values (Fig. 5). Both optimum pH and temperature were similar for the free and SpNox nanoflowers with values of 7.0 and 55 °C. SpNox nanoflowers showed a broader range of pH profile and were more stable than the free enzyme. In both acid and basic pH ranges of 4.0–6.5 and 7.5–9.0, SpNox nanoflowers retained higher residual activity than the free enzyme. At a temperature range of 60–70 °C, SpNox nanoflowers retained higher stability than the free enzyme. These his-tagged recombinant enzymes immobilized through protein–Cu(PO4)2·3H2O hybrid systems showed better stability as compared with that of alcohol dehydrogenase immobilized to Co3(PO4)2.3 The different immobilization process may have influenced the properties of the his-tagged enzymes.The Michaelis–Menten model was applied to determine kinetic parameters for both free and synthesized nanoflowers (Table 2). Apparent Km and Vmax values for HjLAD nanoflowers were 21.3 mM and 235 μmol min−1 mg protein−1, respectively, compared with 18.0 mM and 93.8 μmol min−1 mg protein−1 for free HjLAD, respectively.10 The Km and Vmax values of the SpNox nanoflowers were 28.0 μM and 495 μmol min−1 mg protein−1, respectively, compared with values of 27.2 μM and 344 μmol min−1 mg protein−1 for free SpNox, respectively.35 Here, the enhanced activity of immobilized enzymes may be related to the high surface area of the synthesized nanoflower or a co-operative effect of the encapsulated enzymes to overcome mass-transfer limitations or a favorable enzyme conformation.1,16,17
| Enzyme | Km (mM) | Vmax (μmol min−1 mg protein−1) |
|---|---|---|
| Free HjLAD | 18.0 ± 2.2 | 93.8 ± 7.6 |
| HjLAD nanoflower | 21.3 ± 2.0 | 235 ± 21 |
| Free SpNox | 27.2 ± 2.4 | 344 ± 32 |
| SpNox nanoflower | 28.0 ± 2.5 | 495 ± 41 |
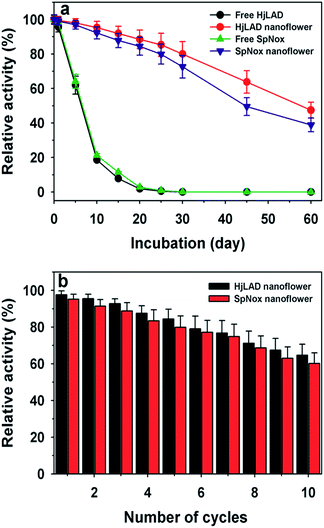
3.3. Stability and reusability
The stability parameters at 25 °C of the free and immobilized enzymes are presented in Table 3. The lower deactivation constant (kd) for the immobilized enzymes compared to that of the free enzymes suggests that HjLAD and SpNox nanoflowers are more stable than their free enzyme forms. The t1/2 of HjLAD and SpNox nanoflowers were approximately 61.0 and 88.7 h, respectively, whereas the respective free enzyme values were 3.2 and 7.6 h. Immobilization of enzymes HjLAD and SpNox as nanoflowers resulted in 18- and 12-fold higher stability under similar conditions compared to that of the free enzymes. The storage stabilities of the HjLAD and SpNox nanoflowers were analyzed over a period 60 days at 4 °C (Fig. 6a). HjLAD and SpNox nanoflowers retained excellent residual activities of 80.0% and 72.6% after 30 days of incubation, respectively. Both free enzymes completely lost their activities under the same conditions. After 60 days of incubation, both HjLAD and SpNox nanoflowers retained residual activities of 47.4% and 38.2%, respectively. The high storage stabilities of the unstable enzymes HjLAD and SpNox after immobilization were improved compared to those in previous reports of the more stable laccase nanoflowers.1,42| Enzyme | Kd (h−1) | t1/2 (h) |
|---|---|---|
| Free HjLAD | 0.218 ± 0.020 | 3.20 ± 0.2 |
| HjLAD nanoflower | 0.011 ± 0.001 | 61.0 ± 5.8 |
| Free SpNox | 0.091 ± 0.008 | 7.60 ± 0.7 |
| SpNox nanoflower | 0.008 ± 0.001 | 88.7 ± 7.6 |
The reusability of both HjLAD and SpNox synthesized nanoflowers was examined under standard assay conditions for 10 cycles of reuse (Fig. 6b). HjLAD and SpNox nanoflowers retained residual activities of 64.7% and 60.2% after 10 cycles of reuse. Here, both synthesized HjLAD and SpNox nanoflowers showed much higher residual activities of 80% as compared to that of 50% for glucose oxidase43 and 60% for laccase after five cycles of reuse.42 Additionally, the reusability of HjLAD and SpNox nanoflowers were much better than papain immobilized as Cu(PO4)2·3H2O–enzyme hybrid nanoflowers.44 Papain nanoflowers exhibited only 15% residual activity after six cycles of reuse. Similarly, encapsulated yeast alcohol dehydrogenase as calcium-mineralized chitosan beads retained only 43% residual activity after eight cycles of reuse.45
3.4. Production of L-xylulose
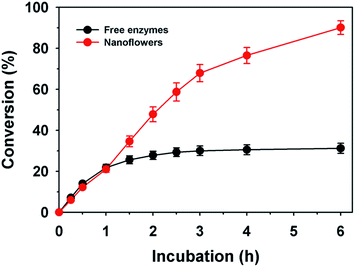
To improve L-xylulose production under co-factor recycling, free and immobilized enzymes of HjLAD and SpNox were compared (Fig. 7). These two enzymes have different optimum pH values 9.5 and 7.0, respectively. Thus, L-xylulose production was evaluated at pH 8.0.29 The free enzyme combination of HjLAD and SpNox resulted in the maximum conversion of L-arabinitol to L-xylulose with a yield of 31.2% after incubation for 6 h under optimum conditions. In contrast, immobilized HjLAD and SpNox enzymes nanoflowers showed 2.9-fold higher conversion with a maximum yield of 90.1%. This higher conversion may be associated with the higher stability of immobilized enzymes and co-factor regeneration as compared with that of the free forms of enzymes. In addition, significantly lower concentrations of NAD+ (2 mM) was required by nanoflowers to achieve a high conversion of L-arabinitol to L-xylulose compared to a previous study using 20 mM of NAD+ as a co-factor without a regeneration system.34 This is the first report on L-xylulose production under co-factor regeneration by protein–inorganic hybrid nanoflowers. Advantages of protein–inorganic hybrid biocatalysts for conversion of carbohydrates are mild synthesis conditions and high selectivity for desired products.46,47 However, the requirement of pure enzymes for the synthesis of hybrid biocatalysts is a disadvantage.474. Conclusions
In summary, we report the synthesis of his-tagged enzymes embedded in metal–organic hybrid nanoflower at 4 °C containing 2 mM of CuSO4 in phosphate-buffered saline (pH 7.4). This synthesis method appears to be very efficient for less stable enzymes such as the dehydrogenases HjLAD and SpNox. The stability of both enzymes was significantly improved after incubation at 25 °C and storage at 4 °C. The kinetic parameters of immobilized enzymes were significantly improved compared to those of the free forms of the enzymes. In addition, immobilized enzymes as nanoflowers showed high reusability. These significant improvements in the properties of immobilized his-tagged recombinant enzymes suggest that this process is very effective for thermally unstable enzymes.Acknowledgements
This work was supported by the Energy Efficiency & Resources Core Technology Program of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resource from the Ministry of Trade, Industry & Energy, Republic of Korea (20153030091450). This research was also supported by a grant from the Intelligent Synthetic Biology Center of Global Frontier Project (2013M3A6A8073184) funded by the Ministry of Science, ICT and Future Planning, Republic of Korea. This research was supported by the 2015 KU Brain Pool of Konkuk University.References
- J. Ge, J. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–532 CrossRef CAS PubMed.
- Z. Lin, Y. Xiao, Y. Yin, W. Hu, W. Liu and H. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 10775–10782 CAS.
- F. Lopez-Gallego and L. Yate, Chem. Commun., 2015, 51, 8753–8756 RSC.
- X. Wu, M. Hou and J. Ge, Catal. Sci. Technol., 2015, 5, 5077–5085 CAS.
- C. Ke, Y. Fan, Y. Chen, L. Xu and Y. Yan, RSC Adv., 2016, 6, 19413–19416 RSC.
- B. Zhang, P. Li, H. Zhang, L. Fan, H. Wang, X. Li, L. Tian, N. Ali, Z. Ali and Q. Zhang, RSC Adv., 2016, 6, 46702–46710 RSC.
- X. Zhang, Q. Liang, Q. Han, W. Wan and M. Ding, Analyst, 2016, 141, 4219–4226 RSC.
- J.-C. Zhao, Q.-Y. Zhu, L.-Y. Zhao, H.-Z. Lian and H.-Y. Chen, Analyst, 2016, 141, 4961–4967 RSC.
- T.-S. Kim, S. K. S. Patel, C. Selvaraj, W.-S. Jung, C.-H. Pan, Y. C. Kang and J.-K. Lee, Sci. Rep., 2016, 6, 33438 CrossRef CAS PubMed.
- R. K. Singh, M. K. Tiwari, R. Singh, J. R. Haw and J. K. Lee, Appl. Microbiol. Biotechnol., 2014, 98, 1095–1104 CrossRef CAS PubMed.
- S. K. S. Patel, V. C. Kalia, J. H. Choi, J. R. Haw, I. W. Kim and J. K. Lee, J. Microbiol. Biotechnol., 2014, 24, 639–647 CrossRef CAS PubMed.
- S. K. S. Patel, S. H. Choi, Y. C. Kang and J. K. Lee, Nanoscale, 2016, 8, 6728–6738 RSC.
- F. Lyu, Y. Zhang, R. N. Zare, J. Ge and Z. Liu, Nano Lett., 2014, 14, 5761–5765 CrossRef CAS PubMed.
- J. Sun, J. Ge, W. Liu, M. Lan, H. Zhang, P. Wang, Y. Wang and Z. Niu, Nanoscale, 2014, 6, 255–262 RSC.
- G. He, W. Hu and C. M. Li, Colloids Surf., B, 2015, 135, 613–618 CrossRef CAS PubMed.
- B. Somturk, M. Hancer, I. Ocsoy and N. Ozdemir, Dalton Trans., 2015, 44, 13845–13852 RSC.
- Y. Yu, X. Fei, J. Tian, L. Xu, X. Wang and Y. Wang, Colloids Surf., B, 2015, 130, 299–304 CrossRef CAS PubMed.
- J. Shi, S. Zhang, X. Wang, C. Yang and Z. Jiang, J. Mater. Chem. B, 2014, 2, 4289–4296 RSC.
- Y. Yin, Y. Xiao, G. Lin, Q. Xiao, Z. Lin and Z. Cai, J. Mater. Chem. B, 2015, 3, 2295–2300 RSC.
- Z. Lin, Y. Xiao, L. Wang, Y. Yin, J. Zheng, H. Yang and G. Chen, RSC Adv., 2014, 4, 13888–13891 RSC.
- C. Altinkaynak, I. Yilmaz, Z. Koksal, H. Ozdemir, I. Ocsoy and N. Ozdemir, Int. J. Biol. Macromol., 2016, 84, 402–409 CrossRef CAS PubMed.
- R. Wang, Y. Zhang, D. Lu, J. Ge, Z. Liu and R. N. Zare, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 320–328 CrossRef CAS PubMed.
- B. Zhang, P. Li, H. Zhang, X. Li, L. Tian, H. Wang, X. Chen, N. Ali, Z. Ali and Q. Zhang, Appl. Surf. Sci., 2016, 366, 328–338 CrossRef CAS.
- T. Zhang, Y. Zhou, Y. Wang, L. Zhang, H. Wang and X. Wu, Mater. Lett., 2014, 128, 227–230 CrossRef CAS.
- N. G. Balabushevich, A. V. L. de Guerenu, N. A. Feoktistova and D. Volodkin, Phys. Chem. Chem. Phys., 2015, 17, 2523–2530 RSC.
- P. Kittitheeranun, W. Sajomsang, S. Phanpee, A. Treetong, T. Wutikhun, K. Suktham, S. Puttipipatkhachorn and U. R. Ruktanonchai, Int. J. Pharm., 2015, 492, 92–102 CrossRef CAS PubMed.
- S. Donatan, A. Yashchenok, N. Khan, B. Parakhonskiy, M. Cocquyt, B.-E. Pinchasik, D. Khalenkow, H. Mohwald, M. Konrad and A. Skirtach, ACS Appl. Mater. Interfaces, 2016, 8, 14284–14292 CAS.
- Z. Li, Y. Zhang, Y. Su, P. Ouyang, J. Ge and Z. Liu, Chem. Commun., 2014, 50, 12465–12468 RSC.
- H. Gao, T.-S. Kim, P. Mardina, P. Zhou, F. Wen and J.-K. Lee, RSC Adv., 2016, 6, 66609–66616 RSC.
- H. Gao, I.-W. Kim, J.-H. Choi, E. Khera, F. Wen and J.-K. Lee, Biochem. Eng. J., 2015, 96, 23–28 CrossRef CAS.
- C. Roca, V. D. Alves, F. Freitas and M. A. M. Reis, Front. Microbiol., 2015, 6, 288 Search PubMed.
- K. Beerens, T. Desmet and W. Soetaert, J. Ind. Microbiol. Biotechnol., 2012, 39, 823–834 CrossRef CAS PubMed.
- M. K. Tiwari, R. K. Singh, R. Singh, M. Jeya, H. Zhao and J. K. Lee, J. Biol. Chem., 2012, 287, 19429–19439 CrossRef CAS PubMed.
- M. K. Tiwari, R. K. Singh, H. Gao, T. Kim, S. Chang, H. S. Kim and J. K. Lee, Bioorg. Med. Chem. Lett., 2014, 24, 173–176 CrossRef CAS PubMed.
- H. Gao, M. K. Tiwari, Y. C. Kang and J. K. Lee, Bioorg. Med. Chem. Lett., 2012, 22, 1931–1935 CrossRef CAS PubMed.
- S. K. S. Patel, C. Selvaraj, P. Mardina, J.-H. Jeong, V. C. Kalia, Y. C. Kang and J.-K. Lee, Appl. Energy, 2016, 171, 383–391 CrossRef CAS.
- S. K. S. Patel, P. Mardina, D. Kim, S.-Y. Kim, V. C. Kalia, I.-W. Kim and J.-K. Lee, Bioresour. Technol., 2016, 218, 202–208 CrossRef CAS PubMed.
- P. Ramachandran, S. S. Jagtap, S. K. S. Patel, J. Li, Y. C. Kang and J.-K. Lee, RSC Adv., 2016, 6, 48137–48144 RSC.
- S. V. Otari, S. K. S. Patel, J.-H. Jeong, J. H. Lee and J.-K. Lee, RSC Adv., 2016, 6, 86808–86816 RSC.
- R. Huang, M. Wu, M. J. Goldman and Z. Li, Biotechnol. Bioeng., 2015, 112, 1092–1101 CrossRef CAS PubMed.
- G. Begum, W. B. Goodwin, B. M. deGlee, K. H. Sandhage and N. Kroger, J. Mater. Chem. B, 2015, 3, 5232–5240 RSC.
- B. S. Batule, K. S. Park, M. Kim II and H. G. Park, Int. J. Nanomed., 2015, 10, 137–142 CAS.
- Y. Huang, X. Ran, Y. Lin, J. Ren and X. Qu, Chem. Commun., 2015, 51, 4386–4389 RSC.
- L. Liang, X. Fei, Y. Li, J. Tian, L. Xu, X. Wang and Y. Wang, RSC Adv., 2015, 5, 96997–97002 RSC.
- P. Han, X. Song, H. Wu, Z. Jiang, J. Shi, X. Wang, W. Zhang and Q. Ai, Ind. Eng. Chem. Res., 2015, 54, 597–604 CrossRef CAS.
- C. Chatterjee, F. Pong and A. Sen, Green Chem., 2015, 17, 40–71 RSC.
- Z. Xu, R. Wang, C. Liu, B. Chi, J. Gao, B. Chen and H. Xu, RSC Adv., 2016, 6, 30791–30794 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra24404a |
| This journal is © The Royal Society of Chemistry 2017 |