Open Access Article
Open Access ArticleThe chemistry and chemical ecology of nudibranchs
Lewis J.
Dean
and
Michèle R.
Prinsep
 *
*
School of Science, University of Waikato, Hamilton 3240, New Zealand. E-mail: Michele@waikato.ac.nz
First published on 14th November 2017
Abstract
Covering: up to the end of February 2017
Nudibranchs have attracted the attention of natural product researchers due to the potential for discovery of bioactive metabolites, in conjunction with the interesting predator-prey chemical ecological interactions that are present. This review covers the literature published on natural products isolated from nudibranchs up to February 2017 with species arranged taxonomically. Selected examples of metabolites obtained from nudibranchs across the full range of taxa are discussed, including their origins (dietary or biosynthetic) if known and biological activity.
1 Introduction
Nudibranchs, often called sea slugs or more poetically, “butterflies of the sea”,1 are a diverse group of marine gastropod molluscs, representing over 4700 known species.2 After shedding their shells as larvae, nudibranchs thereafter remain shell-less; indeed the name nudibranch literally translates to “naked gill”, a reference to the exposed cerata on the backs of many species.3 Whilst essentially blind, nudibranchs perceive their environment through chemosensory interactions with two specialised rhinophores on their heads.3,4 These carnivores are important consumers in benthic communities, feeding mostly upon sessile organisms, (sponges, cnidarians, tunicates and bryozoans), although some species hunt other nudibranchs. Both specialist and generalist feeders are known and can be found in practically all oceans.3Having lost the physical protection of a shell, nudibranchs often utilise chemical defences to deter predators.3 Given that their typical prey species are some of the most prolific producers of natural products,5 it is perhaps unsurprising to find that many nudibranchs are known to sequester these defences.6 The vibrant colours associated with many nudibranch species are often correlated with those of their prey,6 however, de novo production of defences is also known.7 Nudibranchs have thus attracted many natural product researchers due to the potential for discovery of bioactive metabolites, in conjunction with the chemical ecological interactions between predator and prey that exist for many species.
2 Taxonomy
Broadly speaking, nudibranchs can be separated into two distinct groups based on their general morphology (Fig. 1)3 and digestive glands; dorids and aeolids.7 Dorid nudibranchs (clade: Euctenidiacea) have an intact digestive gland7 and are distinguished by a feather-like plume of gills on their dorsal side, circling the anus.3 They also commonly feature discrete pockets, bumps and other distortions, on their skin known as mantle dermal formations (MDFs), in which bioactive defence chemicals are typically stored.3,8 Aeolid nudibranchs (clade: Cladobranchia) have a branched digestive gland7 and lack gills.3 They are characterised by the presence of dorsal projections known as cerata, which function in place of gills by facilitating gas exchange through the epidermis.3 In many aeolid species, the digestive tract also extends into the cerata and the tips often contain cnidosacs; stinging cells absorbed from prey species that are used for the nudibranch's own defence.3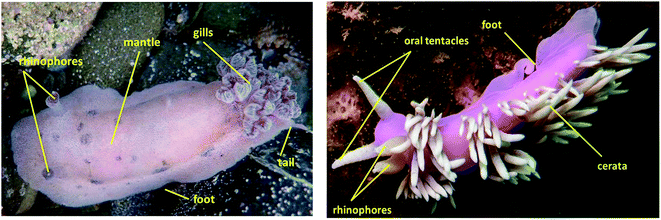 | ||
| Fig. 1 General morphology of a dorid nudibranch (Alloiodoris lanuginata) (left), highlighting the dorsal gill plumage and an aeolid nudibranch (Jason mirabilis) with characteristic cerata (right). Note the presence of rhinophores on both. Photographs courtesy of Tracey Bates. (After Picton and Morrow, 2006).3 | ||
Strictly speaking, a branched digestive gland is indicative of a non-dorid nudibranch, known as a cladobranch7 and true aeolids are only those belonging to the parvorder Aeolidida (Fig. 2), the only nudibranchs to possess cnidosacs.7,9 However, many of the non-dorid nudibranchs are said to be aeolid-like as they possess cerata, or cerata-like projections, instead of gills.3 Others do possess gills, often tucked between the mantle and foot, but they bear little resemblance to the dorsal plumes of dorids.3
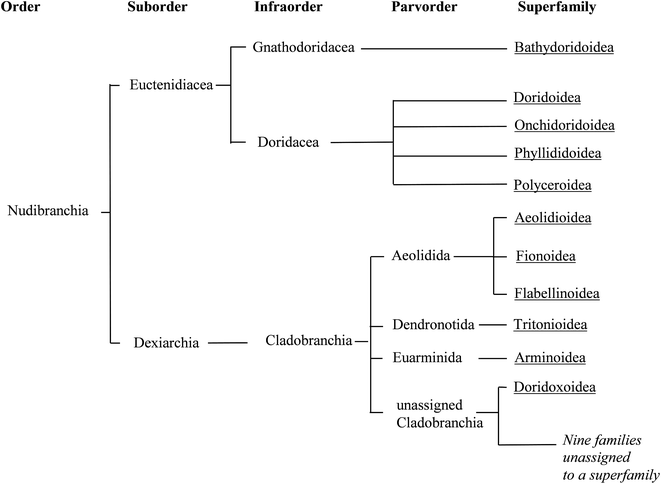 | ||
| Fig. 2 Phylogenetic relationships between the taxonomic divisions currently recognised for Nudibranchia, and that are used in this review. Branches are terminated with superfamilies (underlined), whilst the roots represent taxonomic divisions (as used in WoRMS13). | ||
The classification of nudibranchs is far from settled. There have been a number of taxonomic revisions in the past,10–12 the one presented here being that currently used as in WoRMs.13 A clear distinction between dorid and non-dorid nudibranchs is well established, although a number of cladobranch families have yet to be assigned to an appropriate superfamily.13
Molecular studies are offering new insights into phylogenetic relationships. A relatively recent report analysing RNA sequence data, attempted to reconcile the groupings within Cladobranchia.14 Whilst many of the relationships highlighted in past classifications12 are retained, some new clades have been suggested (although unnamed), incorporating some of the previously unassigned families. However, this analysis is by no means conclusive since only 10 of the 32 families currently recognised were investigated.14 A similar study analysed the mitochondrial and nuclear genes of the Doridoxa genus (the only genus in the current superfamily Doridoxoidea).15
The changes proposed by the above studies serve to highlight the ever evolving nature of taxonomy. This can lead to confusion for anyone attempting to navigate the literature covering nudibranch natural products, as invariably one will come across outdated phylogenetic relationships as well as old or repurposed taxonomic names. As an example, Doridacea is now recognised as an Infraorder within Euctenidiacea representing a subdivision of dorid nudibranchs,13 whereas historically it was used as a suborder covering all dorids.3,7 Some species may also have been renamed and others folded into one another. This confusion has in some, thankfully rare, cases extended to a non-nudibranch sea slug (Pleurobranchaea meckeli) being reported as such.16
3 The origin of nudibranch natural products
Given that both sequestration and de novo synthesis of natural products are known in nudibranchs, it is rarely immediately clear if an isolated metabolite is of dietary origin. Feeding experiments with isotopically labelled precursors, are the only accepted procedures to prove biosynthesis of a metabolite.17 Unfortunately, the low uptake of feedstock by many marine organisms often forces the use of radioactive isotopes.17 Advances in nuclear magnetic resonance (NMR) spectroscopy have improved sensitivity and thus stable isotope experiments are increasingly becoming viable options, yet the cost of precursors, radioactive or stable, combined with the costs of very sensitive NMR instruments has inevitably restricted the use of feeding experiments in nudibranch studies.There are many cases in the literature where de novo synthesis has been assumed, despite no feeding experiments being conducted. As the digestive glands of all nudibranchs are the source of nutrient distribution throughout the organism, it stands to reason that if a natural product is found therein, it is likely of dietary origin.7 Conversely if a metabolite is only present in the skin or mantle, and not in the viscera of a nudibranch, then it is probable that the compound is either biosynthesised in full or secondarily modified.7 Likewise, the absence or presence of the same, or structurally related, metabolite(s) in possible food species are also used to argue for biosynthesis or sequestration respectively.18 It has been further reasoned that any species of nudibranch that synthesises its own metabolites should consistently possess all metabolites, across all geographic collections.18 Species known to show variation in their natural products, especially from differing locations, may be indicative of metabolite sequestration.18 In addition, as one of the evolutionary advantages of de novo synthesis is to liberate a nudibranch from a singular food source, specialist feeders may be more likely to sequester metabolites.7,18
4 Scope of review
There have been a number of excellent reviews of nudibranch chemistry over the years, including reviews on chemical ecology,19–21 chemical defence7,22,23 and bioactive metabolites.24,25 Many focus on specific groups of metabolites such as diterpenes,26 terpenoids,27 cyanide and isothiocyanates28 and isocyanides29 (but all also include other marine invertebrates). A number of these reviews are regional in their focus, dealing with nudibranchs and other molluscs found in New Zealand,30,31 Australia,31,32 North America,33 Japan,34 Africa,35 South America35 and Antarctica.35The annual review of marine natural products that appears in Natural Product Reports has a section on molluscs36a-f but given the wide scope of this review, there is no capacity to discuss nudibranchs in any detail or in a comprehensive manner over a considerable time period. There have only been three relatively recent reviews that deal with nudibranch chemistry, but two of these are not specific to nudibranchs (focus respectively on the terpene chemistry of marine molluscs37 and marine molluscs as a source of antiviral drugs38) and the third is limited in scope (deals with lipids and fatty acids of nudibranchs but only eight species were examined).39
This current review focusses on the chemistry and chemical ecology of nudibranchs. It is arranged taxonomically and species are ordered as per their superfamily in the World Register of Marine Species (WoRMS)13 to highlight both the different and similar defensive strategies adopted by the various clades. Species are referred to and ordered by the name used in the original publication but where a taxonomic revision has taken place, the correct name is given in Table 1 and noted in brackets in the text. In each section, a selection of representative, significant or otherwise interesting, nudibranch natural products are discussed, including their origins (dietary or biosynthetic) if known and biological activity.
| Reported | Reclassification or (correction) | ||||
|---|---|---|---|---|---|
| Superfamily | Family | Subfamily | Genus | Species | |
| Dorids | |||||
| Bathydoridoidea | Bathydorididae | Bathydoris | hodgsoni | ||
| Doridoidea | Actinocyclidae | Actinocyclus | papillatus | ||
| Cadlinidae | Aldisa | andersoni | |||
| sanguinea cooperi | Aldisa cooperi 359 | ||||
| smaragdina | |||||
| Cadlina | laevis | ||||
| luteomarginata | |||||
| pellucida | |||||
| Chromodorididae | Ardeadoris | egretta | |||
| Casella | atromarginata | Doriprismatica atromarginata 360 | |||
| Ceratosoma | amoena | Ceratosoma amoenum 361 | |||
| brevicaudatum | |||||
| gracillimum | |||||
| trilobatum | |||||
| Chromodoris | albonotata | ||||
| albopunctata | Goniobranchus albopunctatus 362 | ||||
| annulata | Goniobranchus annulatus 363 | ||||
| cavae | Goniobranchus cavae 364 | ||||
| elisabethina | |||||
| epicura | |||||
| funerea | Chromodoris lineolata 365 | ||||
| geminus | Goniobranchus geminus 366 | ||||
| geometrica | Goniobranchus geometricus 367 | ||||
| gleniei | Goniobranchus gleniei 368 | ||||
| hamiltoni | |||||
| inopinata | |||||
| inornata | Chromodoris aspersa 369 | ||||
| kuniei | Goniobranchus kuniei 370 | ||||
| lochi | |||||
| luteorosea | Felimida luteorosea 371 | ||||
| macfarlandi | Felimida macfarlandi 372 | ||||
| maridadilus | Hypselodoris maridadilus 373 | ||||
| marislae | Felimida marislae 374 | ||||
| michaeli | |||||
| norrisi | Felimida norrisi 375 | ||||
| obsoleta | Goniobranchus obsoletus 376 | ||||
| petechialis | Goniobranchus petechialis 377 | ||||
| quadricolor | |||||
| reticulata | Goniobranchus reticulatus 378 | ||||
| sedna | Doriprismatica sedna 379 | ||||
| sinensis | Goniobranchus sinensis 380 | ||||
| tinctoria | Goniobranchus tinctorius 381 | ||||
| willani | |||||
| youngbleuthi | Glossodoris rufomarginata 382 | ||||
| Felimida | grahami | (Felimare grahami)383 | |||
| Glossodoris | atromarginata | Doriprismatica atromarginata 360 | |||
| averni | Ardeadoris averni 384 | ||||
| cincta | |||||
| dalli | Felimida dalli 385 | ||||
| hikuerensis | |||||
| pallida | |||||
| quadricolor | Chromodoris quadricolor 386 | ||||
| rufomarginata | |||||
| sedna | Doroprismatica sedna 379 | ||||
| tricolor | Felimare tricolor 387 | ||||
| valenciennesi | Felimare picta 388 | ||||
| vespa | |||||
| Goniobranchus | albonares | ||||
| collingwoodi | |||||
| daphne | |||||
| splendidus | |||||
| verrieri | |||||
| Hypselodoris | agassizii | Felimare agassizii 389 | |||
| californiensis | Felimare californiensis 390 | ||||
| cantabrica | Felimare cantabrica 391 | ||||
| daniellae | Thorunna daniellae 392 | ||||
| fontandraui | Felimare fontandraui 393 | ||||
| ghiselini | Felimare ghiselini 394 | ||||
| godeffroyana | Risbecia godeffroyana 395 | ||||
| infucata | |||||
| jacksoni | |||||
| lajensis | Felimare lajensis 396 | ||||
| orsini(i) | Felimare orsinii 397 | ||||
| porterae | Felimare porterae 398 | ||||
| tricolor | Felimare tricolor 387 | ||||
| villafranca | Felimare villafranca 399 | ||||
| webbi | Felimare picta 400 | ||||
| zebra | Felimare zebra 401 | ||||
| Miamira | magnifica | ||||
| miamirana | |||||
| Risbecia | tryoni | Hypselodoris tryoni 402 | |||
| Tyrinna | nobilis | Tyrinna delicata 403 | |||
| Discodorididae | Anisodoris | nobilis | Peltodoris nobilis 404 | ||
| fontainei | Doris fontainii 405 | ||||
| Asteronotus | cespitosus | ||||
| Diaulula | sandiegensis | ||||
| Halgerda | aurantiomaculata | ||||
| gunnessi | |||||
| rubicunda | Sclerodoris rubicunda 406 | ||||
| stricklandi | |||||
| theobroma | |||||
| willeyi | |||||
| Jorunna | funebris | ||||
| Peltodoris | atromaculata | ||||
| Platydoris | sp. | ||||
| Sclerodoris | tanya | ||||
| Dorididae | Archidoris | carvi | Doris fontainii 405 | ||
| Neodoris | carvi | Doris fontainii 405 | |||
| Archidoris | montereyensis | Doris montereyensis 407 | |||
| odhneri | Doris odhneri 408 | ||||
| pseudoargus | Doris pseudoargus 409 | ||||
| tuberculata | Doris pseudoargus 409 | ||||
| Austrodoris | kerguelenensis | Doris kerguelenensis 410 | |||
| Doris | verrucosa | ||||
| Onchidoridoidea | Goniodorididae | Hopkinsia | rosacea | Okenia rosacea 411 | |
| Okenia | zoobotryon | ||||
| Onchidorididae | Acanthodoris | nanaimoensis | |||
| Adalaria | loveni | ||||
| Phyllidioidea | Dendrodoridae | Dendrodoris | arborescens | ||
| carbunculosa | |||||
| denisoni | |||||
| fulva | |||||
| grandiflora | |||||
| krebsii | |||||
| limbata | |||||
| nigra | |||||
| tuberculosa | |||||
| Doriopsilla | albopunctata | ||||
| areolata | |||||
| janaina | |||||
| pelseneeri | |||||
| Phyllidiidae | Phyllidia | bourgini | Phyllidiella rosans 412 | ||
| coelestis | |||||
| ocellata | |||||
| pulitzeri | |||||
| pustulosa | Phyllidiella pustulosa 413 | ||||
| varicosa | |||||
| Phyllidiella | rosans | ||||
| Phyllidiopsis | krempfi | ||||
| Reticulidia | fungia | ||||
| Polyceroidea | Aegiridae | Notodoris | citrina | ||
| gardineri | |||||
| Hexabranchidae | Hexabranchus | sanguineus | |||
| Polyceridae | Nembrothinae | Nembrotha | kubaryana | ||
| Roboastra | tigris | ||||
| Tambja | abdere | ||||
| ceutae | |||||
| eliora | |||||
| stegosauriformis | |||||
| Polycerinae | Polycera | tricolor | |||
| Triophinae | Limacia | clavigera | |||
| Triopha | carpenteri | Triopha catalinae 414 | |||
| catalinae | |||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Cladobranchs | |||||
| Aeolidioidea | Aeolidiidae | Aeolidia | papillosa | ||
| Aeolidiella | stephanieae | Berghia stephanieae 415 | |||
| Spurilla | sp. | ||||
| Facelinidae | Cratena | peregrina | |||
| Hermissenda | crassicornis | ||||
| Phidiana | militaris | ||||
| Phyllodesmium | guamensis | (Phyllodesmium guamense)416 | |||
| lizardensis | (Phyllodesmium lizardense)417 | ||||
| longicirra | (Phyllodesmium longicirrum)418 | ||||
| magnum | |||||
| Pteraeolidia | |||||
| Arminoidea | Arminidae | Armina | maculata | ||
| Dermatobranchus | ornatus | ||||
| Doridomorphidae | Doridomorpha | gardineri | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Doridoxoidea | |||||
| Fionoidea | Fionidae | Phestilla | melanobrachia | Tenellia melanobrachia 419 | |
| sibogae | Tenellia sibogae 420 | ||||
| Flabellinoidea | Flabellinidae | Flabellina | affinis | ||
| Flabellinopsis | iodinea | Flabellina iodinea 421 | |||
| Tritonioidea | Tethydidae | Melibe | leonina | ||
| Tethys | fimbria | ||||
| Tritoniidae | Marionia | limceana | |||
| Tritonia | diomedea | Tritonia tetraquetra 422 | |||
| hamnerorum | |||||
| Tritoniidae | Tritoniella | belli | |||
| Tritoniopsis | elegans | ||||
| Tochuina | tetraquetra | Tochuina gigantea 423 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Unassigned | |||||
| Families | Charcotiidae | Charcotia | granulosa | ||
| Leminda | millecra | ||||
| Dotidae | Doto | antarctica | |||
| carinova | |||||
| pinnatifida | |||||
| Pinufidae | Pinufius | rebus | |||
| Proctonotidae | Janolus | cristatus | |||
| novozelandicus | |||||
5 Dorid nudibranchs
5.1 Bathydoridoidea
5.2 Doridoidea
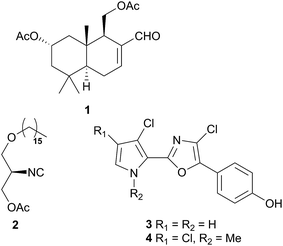
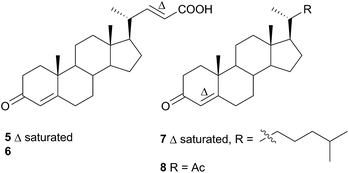
Comprising five families, Doridoidea is by far the most studied nudibranch superfamily in terms of species, genera and metabolites reported. Searching within the MarinLit database42 for natural products investigations into nudibranchs (Taxonomy filter: Phylum-Mollusca; Article free text filter: nudibranch) reveals 152 records (as of 28 February 2017). Of these, 80 concern species of Doridoidea (56%).Two steroidal acids, 3-oxo-chol-4-ene-24-oic acid (5) and its unsaturated analogue (6), were isolated from Aldisa sanguinea cooperi46 (Aldisa cooperi359). Cholestenone (7), an alkyl derivative of 5 and 6 was also present in the extract. A. cooperi was consistently found feeding upon the sponge Anthoarcuata graceae, which, whilst lacking 5 and 6, was found to contain 7 as one of its major metabolites. Cholestenone, 7, proved to be inactive in feeding experiments whereas 5 was an effective inhibitor of feeding behaviour (common goldfish). This indicated that A. cooperi was obtaining an inactive metabolite from its diet and modifying it to provide a defence against predators.46 It has been suggested that another steroidal metabolite, 24-norchol-4-ene-3,22-dione (8), isolated from the related nudibranch A. smaragdina may also be secondarily modified from 7 (ref. 47) or alternatively, result from the β-oxidation and subsequent decarboxylation of 5. However, these suggestions were tentative since no steroidal precursors were noted in the observed prey species Phorbas fictitius.47
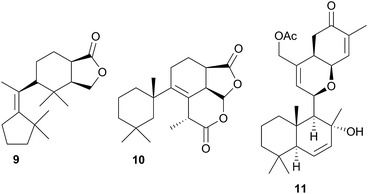
A nudibranch that has attracted significant interest is Cadlina luteomarginata, as it is one of only two species of nudibranchs known to both sequester prey metabolites and biosynthesise its own natural products48 (the other, Dendrodoris grandioflora, will be discussed below). To date, 38 terpenoid metabolites, with 22 carbon skeletons, representing monoterpenes,49 sesquiterpenes,49–51 diterpenes,52,53 sesterterpenes54 and degraded sesterterpenoids51,55 and diterpenoids53,56 have been isolated from C. luteomarginata.57 Examples of the sequestered metabolites are glaciolide (9)56 and cadlinolide A (10)53 from the sponge Aplysilla glacialis, and ansellone A (11)54 from a prey sponge of the Phorbas genus. Their ecological roles are not well defined,57 but 11 has been shown to be moderately activating towards cellular processes that use the cyclic adenosine monophosphate (cAMP) signal pathway.54
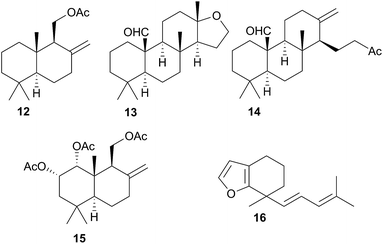
Terpenoid biosynthesis in C. luteomarginata was suspected when all previously analysed individuals from a British Columbian (Canadian) study consistently possessed the same three terpenoids.58 The de novo production of these terpenes; albicanyl acetate (12), cadlinaldehyde (13) and luteone (14) was subsequently proven by stable isotope labelling experiments.58 Antifeeding activity of 12 was detected, and it was found at high concentration in the mantle and mucus, indicating its role as a deterrent.51 A derivative of 12, 1α,2α-diacetoxyalbicanyl acetate (15), was isolated from the egg masses of C. luteomarginata but not detected in the nudibranch itself.57 Whilst no feeding assays of 15 were conducted, its similarity to 12 indicated that it may act to protect the egg masses.58 Likewise de novo synthesis may be expected for 15, despite it not being detected to date in C. luteomarginata nudibranchs.
Initially, there was speculation that C. luteomarginata may be divided into subspecies based on geographic distribution, as a Californian population was originally thought not to contain any of the biosynthesised compounds 12–14.48,58 However, reexamination of this population utilising gas chromatography (GC) indicated that 12–14 were present but at low concentrations.48 It was found that broadly speaking, individuals of C. luteomarginata, from British Columbia, could be separated into two groups. The first group possessed 12–14 at high concentration, with other terpenoids that could be detected by GC at much lower concentration, whilst the second group contained 12–14 at concentrations below those of other GC detectable terpenoids. This variance both between and within geographic populations suggested that C. luteomarginata was able to downregulate biosynthesis if sufficient dietary terpenoids could be sequestered.48
A number of terpenoids were obtained from C. pellucida and C. laevis, including the furanosesquiterpene laevidiene (16) from the latter. The structures indicated a dietary origin from sponge prey, although only C. pellucida was observed feeding upon a sponge (Spongia agaricina).59
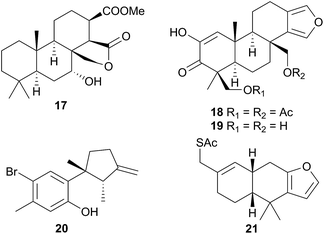
Six furanoditerpenoids were obtained from Casella atromarginata (Doriprismatica atromarginata360) from Sri Lanka.61 Of these, two were known metabolites of an Australian Spongia sp.62 and two more were minor structural variants but two (18 and 19) were novel metabolites, with a more highly oxidised A-ring, containing a monoenolised α-diketone function.61 It seems likely that C. atromarginata obtained these metabolites from its sponge prey.61
Ceratosoma amoena (Ceratosoma amoenum361) from New Zealand was observed on the red alga Hymenea variolosa.63 The red algal metabolite allolaurinterol64 (20) was isolated from both the nudibranch and alga, a surprising result since, as noted by the researchers, the nudibranch dietary system is not capable of breaking down algal tissue. As an alternative explanation for this result, it was suggested that C. amoenum could be preying on an organism (or more likely its egg masses) that does sequester this compound from the alga.63
Nine new spongiane diterpenes were isolated from a South Australian nudibranch tentatively identified as Ceratosoma brevicaudatum65 but later identified as Chromodoris epicura66 so perhaps it is unsurprising that these do not seem to be typical of the Cerastoma genus. A thiosesquiterpene (21) was isolated from the Australian nudibranch C. brevicaudatum but believed to be sequestered from a Dysidea sponge.67 The ecological role of 21 has not been studied, but it represents a relatively rare example of a sulphur containing sesquiterpene from the marine environment.67 Four “typical” sponge furanosesquiterpenoids, pallescensin-B,68 (−)-furodysinin,69 dehydroherbadysidolide70 and herbadysidolide71 were isolated from two species of Ceratosoma; C. trilobatum and C. gracillimum.66 As previously reported,72 (−)-furodysinin, exhibited feeding deterrent and ichthyotoxic properties, and additionally, concentration of the compound in the dorsal horn glands of the animals, supports the proposed defensive role of the dorsal horns.66
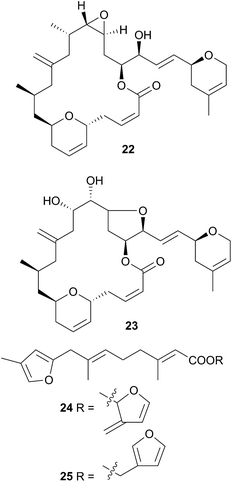
Chromodoris is the most examined of all nudibranch genera, with many reported studies,8,39,73–106 although many of the species in these studies originally classified as Chromodoris have been reclassified as Doriprismatica,75Felimida,73,76,79,80,85,87,90,93Glossodoris79,101Goniobranchus39,86,88,89,95,98,102–104 or Hypselodoris.74,106Chromodoris species feed exclusively on sponges and sequester a large number of terpenoid metabolites from their prey including sesquiterpenes,73,74,78,82,106 norditerpenes,80 diterpenes8,76,81,85–90,93,95,96,98,99,102–104 and sesterterpenes,79,84,91,92,92,97,100 although macrolides77,82,83,105 and bromophenols78 have also been reported. In some instances, metabolites have been simultaneously isolated from sponge and nudibranch, highlighting that they are of dietary origin. For example, the macrolides laulimalide (22)107,108 and isolaulimalide (23)109 were isolated from sponges of the Hyattella genus and from their nudibranch predator C. lochi.83
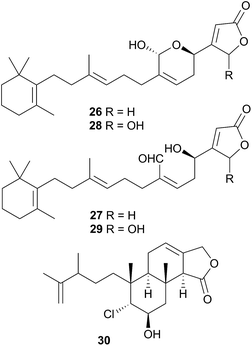
Latrunculin A, a sponge metabolite originally obtained from Latrunculia magnifica110 has been isolated from several Chromodoris species,77,82,94,105 and has been shown to be selectively stored in the mantle rim for protective purposes.105 In other cases, structural analogues of nudibranch metabolites have been detected in or isolated from, the sponge prey, indicating the likelihood of secondary modification of sponge metabolites. For example, the sesquiterpene marislin (24), obtained from C. marislae (Felimida marislae374), is related to the spongian metabolite pleraplysillin-2 (25), from Pleraplysilla spinifera, by a [3,3] sigmatropic rearrangement.73 Similarly, deoxymanoalide (26) and deoxysecomanoalide (27), isolated from C. willani, are modified from manoalide (28) and secomanoalide (29) respectively by loss of an hydroxyl group. Both 28 and 29 were identified in the sponge prey of C. willani.100 Some chlorinated homoditerpenes including hamiltonin A (30)94 were isolated from C. hamiltoni along with latrunculins and a sesterterpene but in this case, the origin of the chlorinated compounds was not clear.94 Where they have been assessed, virtually all of the metabolites obtained from Chromodoris species have displayed a moderate degree of bioactivity, indicating their role in limiting predation. Despite evidence suggesting secondary modification occurs, there have been no reports of de novo synthesis within Chromodoris to date.
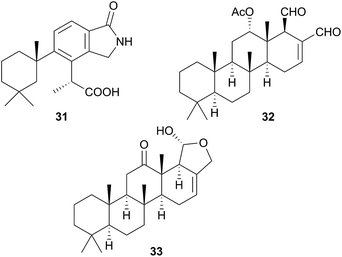
A species, denoted by researchers as Felimida grahami (Felimare grahami,383) sequestered rearranged terpenoids from its sponge prey Darwinella cf. oxeata.111 These included oxeatamide I (31), one of three new oxeatamides H–J111 found in the sponge to add to those already known112 and membranolide,113 previously isolated from the sponge Dendrilla membranosa113 and from nudibranchs Goniobranchus reticulatus102 and G. splendidus.114
As for Chromodoris, many of the species originally reported as Glossodoris have been reclassified, in this case as Ardeadoris,115Chromodoris,116Doriprismatica,39,117–120Felimare121 and Felimida.120 Although latrunculin B has been isolated from G. tricolor, this is now classified as Felimare387 and all other metabolites isolated from this genus are terpenoids and predominantly sponge derived.115,117–126 Spongian diterpenes have been obtained,117,119 but most of the metabolites isolated are based on the sponge-derived scalarane sesterterpene, scalaradial (32),115,119,121–125 including norscalaranes119 and homoscalaranes.120 A number of the scalarane metabolites obtained were 12-keto derivatives such as 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 124 and since in every case, they were only obtained from the nudibranch and not from the sponge prey, it is considered highly likely that biotransformation of the sponge scalarane occurs in the nudibranch,124 most likely through enzymatic oxidation.115
124 and since in every case, they were only obtained from the nudibranch and not from the sponge prey, it is considered highly likely that biotransformation of the sponge scalarane occurs in the nudibranch,124 most likely through enzymatic oxidation.115
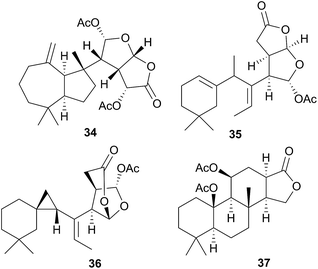
As noted above, taxonomic revisions have led many species that were formerly Chromodoris to be reclassified as Goniobranchus. From species originally reported as Goniobranchus, a range of diterpenes and norditerpenes have been obtained and all studies thus far have been of Australian specimens.60,114,126–128 In most cases, known sponge metabolites were co-isolated with the novel metabolites reported and compounds were sometimes isolated from both the nudibranch and its prey sponge. 12-Acetoxy dendrillolide (34) for example was obtained from both G. albonares and the crimson sponge on which it was feeding.126 The activity of isolated metabolites has been quite varied. For example, of the six new oxygenated norditerpene/bisnorditerpene metabolites isolated from G. splendidus, gracilins M–Q displayed significant potency against the HeLa S3 cell line, aplytandiene-3 was essentially inactive and the rearranged diterpene daphnelactone (35) reported from G. daphne in the same study was not tested.114 Another study of G. splendidus yielded some oxygenated diterpenes but these were not tested for activity.60G. verrieri yielded seven new norditerpenes and diterpenes, all with highly rearranged carbon skeletons, such as the cyclopropyl-containing verrielactone (36) and a biosynthetic pathway to these metabolites from spongialactone was proposed.127 All six new spongian-16-one diterpenes found localised in the mantle of G. collingwoodi (an example being 37) were inactive to a range of HTCLs but preliminary testing of whole body extracts indicated some antifeedant activity against Palaemon serenus (rock pool shrimp).128
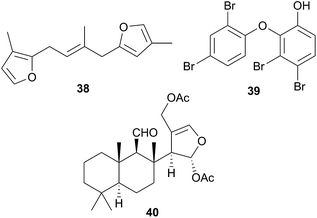
The majority of species originally reported as Hypselodoris have been reclassified as Felimare8,72,129–138 or in one case, Risbecia106 but the genus sequesters terpenes,8,72,98,106,129–139 predominantly furanosesquiterpenes,8,72,98,106,134–139 from various species of sponge, of which longifolin138 (38) is typical. Longifolin (38) is sequestered from Dysidea sponges106,130,136,137 and it and other antifeedant compounds are present in the MDFs of many Hypselodoris species8,72,131 and released when the nudibranch is molested. There is one reported isolation of a sesterterpene from this genus.133H. orsini(i) (Felimare orsinii)397 feeds on the sponge Cacospongia mollior but contains different metabolites to its prey. It is thought that the nudibranch converts the sponge sesterterpenoid scalaradial, into deoxoscalarin, then, a second chemical transformation produces the sesterterpenoid, 6-keto-deoxoscalarin which is also stored in MDFs.133
A number of oxy-polybrominated diphenyl ethers (O–PBDEs) have been isolated from the nudibranchs Miamira magnifica and M. miamirana from Australia, of which 39 from M. magnifica is a typical example.140 These compounds are sequestered from sponges and although such compounds have been isolated from nudibranchs previously,78,141 they were found in the gut tissue, whereas in the Miamira specimens, they were additionally located in the mantle and dorsal horn tissues, so likely play a role in chemical defence.140
As part of a study of Chromodorid species,8Risbecia tryoni (Hypselodoris tryoni402) yielded the sponge metabolite (−)-furodysinin142 and the lipid and fatty acid content has also been examined.39 There has only been one chemical study of the Tyrinna genus. Tyrinnal (40), a seco-11,12-spongiane was obtained from the mantle and MDFs of Patagonian Tyrinna nobilis (Tyrinna delicata403) along with three known furanosesquiterpenes.143
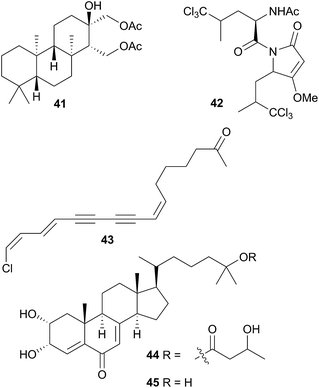
A number of diterpenoid diacylglycerols have been isolated from this family, including the anisodorins 1–5, which were obtained from the skin of the Patagonian species Anisodoris fontainei (Doris fontainii405) along with two further analogues.146 These last two compounds were previously reported from the same species147 (but erroneously classified as Archidoirs carvi146) and are diastereoisomers of two further diacylglycerols obtained from Archidoris tuberculata (Doris pseudoargus409), collected along the northern Spanish coast.147 Surprisingly, the isomers from each source display opposite absolute stereochemistry in their terpenoid moiety. The absolute stereochemistry of anisodorin 5, 41 was established through synthesis of its enantiomer148 and 41 itself has also been synthesised.149Anisodoris nobilis (Peltodoris nobilis404) was the source of doridosine,144 (1-methylguanosine), a hypotensive compound150 that was also obtained from the Australian sponge Tedania digitata.151 A degraded sesquiterpenoid was obtained from A. nobilis as the odoriferous principle.152
Asteronotus cespitosus specimens collected from Australia and the Philippines contained a range of halogenated metabolites and two sesquiterpenes.141 One of these was a novel chlorinated pyrrolidone (42), while all of the other metabolites had been previously isolated from Dysidea herbacea.141 Pyrrolidone (42) has a similar structure to dysideapyrrolidone,153 a sponge metabolite.
Isoguanosine, a compound previously isolated as a constituent of the croton bean154 was obtained from Diaulula sandiegensis.155 A Californian population of D. sandiegensis contained a series of nine chlorinated acetylenes, of which 43 is a typical example.156 Over twenty years later, six of the metabolites were isolated from the sponge Haliclona lunisimilis which was collected in the same area as the nudibranchs were originally.157 The two major metabolites found in D. sandiegensis (both ketones) were not present in the sponge, although the corresponding alcohols were, suggesting modification of the alcohols by the nudibranch.157 Steroidal metabolites, diaulusterols A (44) and B (45), have been isolated from a British Columbian collection of D. sandiegensis.158 Stable isotope experiments indicated that the polyketide chain of 44 was biosynthesised by the nudibranch itself, whereas there was no evidence of uptake in the steroidal nucleus.159 Radiolabelled incorporation also failed to prove biosynthesis of the steroid nucleus.159 This suggested that as for Aldisa cooperi,46 a precursor steroid was sequestered and subsequently modified, although a prey species has yet to be identified.158,159
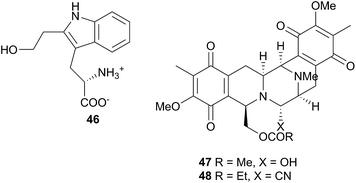
Five species of the Halgerda genus collected from various locations around Australia and from Okinawa, Japan were examined. Of these, H. aurantiomaculata yielded the new tryptophan derivative halgerdamine (46), in addition to four known nitrogenous compounds, while H. gunnessi yielded mixtures of acylated tetrasaccharides.145 No secondary metabolites were detected in H. theobroma, H. rubicunda (Sclerodoris rubicunda406) or H. willeyi, leading the authors to suggest that these nudibranchs may have evolved to exploit a wider range of food including nontoxic sponges rather than sequestering or synthesising protective compounds.145 Organic extracts of an Indian specimen of H. stricklandi displayed modest activity to Staphylococcus aureus and no activity against a range of other bacterial and fungal species.160
Jorunna funebris is another nudibranch to yield non terpenoid natural products. Studies of the nudibranch and its prey, sponges of the genus Xestospongia, indicate that J. funebris sequesters a range of isoquinolinequinones160–163 and bistetrahydroisoquinolines162,164,165 from the sponge. Some of the isoquinolinequinones have been synthesised166 and the metabolites possess a range of biological activities, including antibacterial activity,161,164 cytotoxicity to HTCLs,162,164,165 and NF–κB inhibitory activity.162 Jorumycin (47) was not found in the sponge but was isolated from the mucus and mantle of an Indian collection of J. funebris. It exhibited very potent cytotoxicity towards various tumour types but was only available in small quantities due to instability of the carbinolamine moiety.164 Pretreatment of the nudibranch tissues with potassium cyanide, resulted in isolation of jorunnamycins A–C (C pictured as 48), more stable compounds which retained high cytotoxicity to human tumour cell lines.165 Zalypsis®, a synthetic compound derived from jorumycin,167 is currently in Phase II clinical trials for treatment of endometrial and cervical cancer.168
Several studies have chemically linked the nudibranch Peltodoris atromaculata to its prey sponges Petrosia ficiformis169,170 and Haliclona fulva,171 indicating sequestration of sponge metabolites by P. atromaculata, including sterols,169 the polyacetylene alcohols, petroformynes170 and fulvinol-related polyacetylenes.170 One of these last compounds was moderately cytotoxic to the SKMEL-28 melanoma cell line.171 The lipid and fatty acid content of the related Platydoris sp. were examined as part of a study of eight nudibranch species.39
The sesquiterpene glyceride esters, tanyolides A and B were isolated from Sclerodoris tanya from Southern California. Located mainly in the mantle of the nudibranch, both deterred feeding by fish predators and were also synthesised.172 Feeding experiments utilising [2-13C]mevalonolactone indicated that Sclerodoris tanya is capable of de novo biosynthesis of the terpenoid fragment of tanyolide B.173
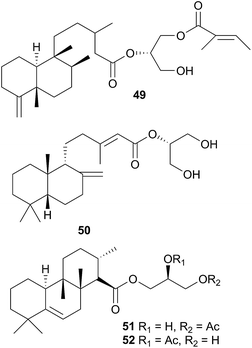
The Antarctic nudibranch Austrodoris kerguelenensis (Doris kerguelenensis410) has yielded a number of diterpene glycerides with a variety of skeletons, even within one population,181 including ent-labdane,182,183 labdane,184 halimane,184,185 clerodane184,186,187 and isocopalane188 frameworks.186,189 Some norsesquiterpenes190 have also been isolated from this well-studied species. Of the compounds isolated, some of the diterpene diacylglycerides had feeding deterrent activity against the predatory seastar Odontaster validus,191 some of the clerodane (palmadorins A and B) and labdane (palmadorins M–O) diterpenes were growth inhibitory to human erythroleukamia (HEL) cells and palmadorin M (50) was shown to be apoptotic to these cells.184 Not surprisingly, as for Archidoris, it is thought likely that the above compounds are biosynthesised de novo by the nudibranch.191,192
The verrucosins are also diterpenoid acid glycerides obtained from Doris verrucosa,193,194 and in addition to icthyotoxicity, verrucosins A and B (51 and 52) were shown to be potent activators of protein kinase C and to promote tentacle regeneration in the freshwater hydrozoan Hydra vulgaris.195 Biosynthetic feeding experiments with 14C labelled mevalonic acid and glycerol resulted in poor incorporation into the verrucosins so de novo biosynthesis was not proven but good incorporation into sterols did establish that the mevalonate pathway was operative in the nudibranch.196 A xylosyl thioether, 9-[5′-deoxy-5′(methylthio)-β-D-xylofuranosyl]adenine was also obtained from D. verrucosa197–199 and biosynthetic studies indicated that it arises in D. verrucosa from isomerisation of endogenous 5′-deoxy-5′-methylthioadenosine (MTA).200
5.3 Onchidoridoidea
The Onchidoridoidea consists of five families but only representatives of the Onchidorididae and the Goniodorididiae have been examined for natural products.5.4 Phyllidioidea
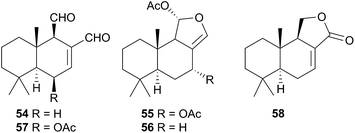
The Phyllidiodea superfamily comprises three families (Dendrodoridae, Mandeliidae and Phyllidiidae) and is very well studied, although no representatives of the Mandeliidae have been studied to date. All metabolites isolated thus far from this superfamily have been terpenes.Biosynthesis of 54 and related compounds has been proven in Dendrodoris grandiflora through radiolabelling experiments.212,213 Whilst 54 was extracted from the mantle, the fatty acid esterified sesquiterpenoids were found in the digestive glands along with a series of known spongian terpenes.212 Other studies have reported drimane sesquiterpenes from Dendrodoris carbunculosa,214D. nigra, D. tuberculosa and D. krebsii.215,216 The anatomical distribution and role that such terpenes may play has been investigated in Mediterranean Dendrodoris nudibranchs.217 The presence of sequestered sponge metabolites might indicate a dual defence for Dendrodoris species but they were completely absent from the mantle, where one would expect to find defence allomones. Unlike Cadlina luteomarginata, in which it appears de novo synthesis is downregulated when sequestered metabolites are available,48D. grandiflora still utilises its own natural products for defence.
Drimane biosynthesis has also been reported in Dendrodoris arborescens, in the form of 56, following radiolabelling studies and another drimane, 6β-acetoxypolygodial (57), was also isolated from the mantle of D. arborescens.213 Feeding experiments with 14C- and 13C-labelled glucose on D. limbata and D. grandiflora indicated that glucose was incorporated into the terpenoid portion of drimanes via the standard mevalonate pathway.218 A more recent study identified the presence of symbiotic bacteria in the outer tissues of D. nigra and it was suggested that they could be implicated in defence from predators.219
In a New Zealand study, the sesquiterpene cinnamolide (58) was detected in all specimens of D. denisoni examined, regardless of geographic location or age, potentially indicating that it was biosynthesised by the nudibranch.61 Cinnamolide (58) has previously only been obtained from terrestrial plants.220
Studies of species of the related Doriopsilla genus including D. albopunctata215,216D. janaina,215D. pelseneeri221 and D. areolata211,222–224 indicated that they also contained drimane sesquiterpenoids, as well as enantiomerically related sesquiterpenoids of the ent-pallescensin A skeleton.221–224 Biosynthetic investigations of D. areolata222,223 and D. pelseneeri225 indicated that they produce both drimane sesquiterpenoids and sesquiterpenoids with the ent-pallescensin A skeleton de novo and their presence in the mantle of D. areolata indicated that they too were involved in the chemical defence of the nudibranch.222,223
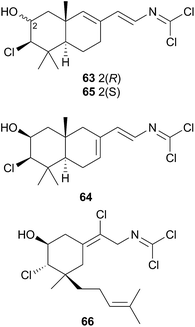
A series of cytotoxic, antifouling sesquiterpenes (63–66) was isolated from Reticulidia fungia247 which possess an unique carbonimidic dichloride functionality. Two of the metabolites, reticulidins A (63) and B (64) are novel to the nudibranch and were moderately cytotoxic to two HTCLs,247 whilst the other two metabolites (65 and 66) had previously been isolated from the sponge Pseudaxinyssa pitys.248,249 The structural similarities to 65, and the rarity of the carbonimidic dichloride functionality, suggests that 63 and 64 are also likely sequestered from an, as yet unidentified, Pseudaxinyssa sponge.
5.5 Polyceroidea
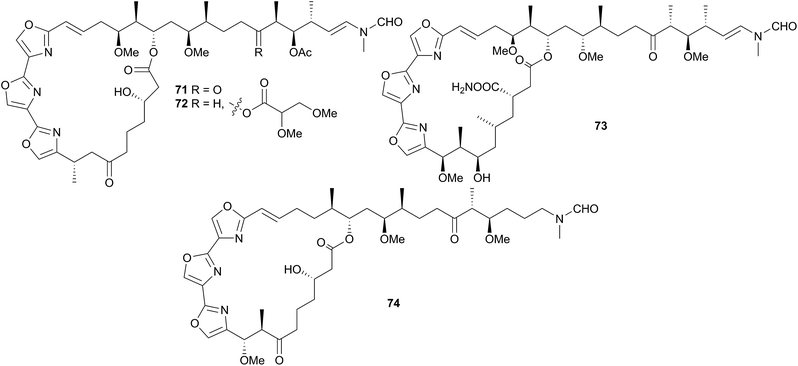
Although only three of the five families in the Polyceroidea have been chemically examined, the superfamily represents a remarkably diverse range of dorids that have attracted considerable attention from researchers.Other macrolides have also been isolated from H. sanguineus and its egg masses from various locations. These include kabiramide C (73)256–258 and related compounds256,258–260 and halichondramide derivatives258,260,261 such as dihydrohalichondramide (74). Ulapualides, kabiramides and halichondramide derivatives possessed a range of bioactivities, including cytotoxicity254,255,260,261 and antifungal properties254,256,259,261 and the latter two macrolide families have been shown to deter feeding in both fish (Thalassoma lunare) and crabs (Dardunus megistos).258 Macrolides have been found at higher concentrations in egg masses than in the adult nudibranchs,254,258 suggesting that their main function is to protect these vulnerable egg masses, although it has been postulated that the macrolides are also secreted in the mucus of H. sanguineus thus conferring protection on the adult as well.258 One investigation has highlighted that the bioactivity of 71 (and hence presumably that of related compounds), is derived from potent depolymerisation of the actin cytoskeleton within cells.262
Most of these macrolides are strongly suspected of being of dietary origin as they were found in both the mantle and digestive tracts of the nudibranch.258 Furthermore, halichondramide261 and derivatives have been isolated from Halichondria sponges and these sponges are known prey species of H. sanguineus. Halichondramide itself, is absent in all specimens of H. sanguineus, despite being the major metabolite of the sponge,258,261 so it has been suggested that the nudibranch is either secondarily converting halichondramide into compounds such as 74, or selectively excluding it.258 Since halichondramide is very cytotoxic (LDLO in mice when injected subcutaneously is 1.4 mg kg−1),261 it may be too potent for use by H. sanguineus.
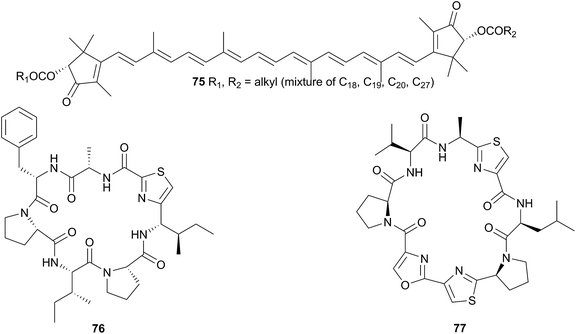
Hurghadin (75), the red pigment responsible for the colouration of H. sanguineus, has been isolated from an Egyptian specimen263 and as it is a carotenoid, it is almost certainly sequestered. The occurrence of 75 in the mantle follows an inverse relationship with the known macrolides, indicating that it may provide an alternative chemical defence, in addition to acting as a visual warning to potential predators,263 although with no bioactivity studies conducted on 75, this is only conjecture. The apocarotenoid apoastacenal was isolated from H. sanguineus egg masses from Japan264 as a new natural product (but known synthetic compound).265
H. sanguineus is a generalist feeder on many species of sponge. An adult specimen from the South China Sea was found to contain a mixture of nitrogenous sesquiterpenoids rather than macrolides,266 two of which were known metabolites from Axinella, Acanthella and Dysidea sponges, and two were novel.266 However, as both novel compounds were found in the digestive tract of the nudibranch, this suggests that they too are of dietary origin.
A Fijian specimen of H. sanguineus analysed by very sensitive NMR spectroscopy (600 MHz spectrometer with a 1 mm high-temperature superconducting cryoprobe), yielded, in addition to a number of related macrolides, two thiazole cyclic peptides, sanguinamide A (76) and sanguinamide B (77).259 Both 76 and 77 were present at extremely low concentrations across the whole individual (0.0023% and 0.011% dry weight respectively) suggesting that the compounds were not major components in H. sanguineus defence and likely represented metabolites of an as yet unidentified prey species.259 Although no dietary source has been identified for the sanguinamides, the structures suggest that cyanobacteria or tunicates may be their original source.259 Both 76 (ref. 267) and 77 (ref. 68) have been synthesised; synthesis of the former revised the configuration to (76). Sanguinamide B analogues were active against Pseudomonas aeruginosa, reducing twitching motility.268,269
The implication of these studies, is that H. sanguineus can sequester a variety of metabolites to utilise in defence and can seemingly do so from a range of prey species.259,266
Tambjamines have also been noted to deter feeding in fish, and thus likely act as a defence against both generalist and specialist feeders.271 Conversely, the Tambja nudibranchs and R. tigris have developed the ability to track their respective prey through chemoreception of these same compounds.271 In the slime trails of T. abdere, 78–81 were present at low concentrations, as well as being released in a mucus at relatively high concentrations when T. abdere was molested.270,271R. tigris was noted to track the slime trail of T. abdere, but would sometimes break off attack if the mucus was released,270 indicating that the antifeeding properties of the tambjamines still affected R. tigris if their concentration was sufficient.271 In the event that T. eliora was attacked, it would attempt to flee rather than secrete a defence, suggesting that the lower concentration of tambjamines in T. eliora prevents release of a mucus.271 Similarly, T. eliora preferentially selected low concentrations of tambjamines (consistent with natural concentrations in S. translucens) in a Y maze, but was deterred if concentrations were high.271 Thus the tambjamines, in addition to being an antifeedant, in Tambja species, act as a tracking pheromone at low concentrations and an alarm pheromone at higher concentrations.271
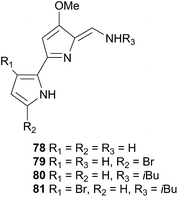
Tambjamines, including 78–81 and new variants, have since been reisolated from T. eliora272 and from a bryozoan.273 A related yellow pigment was obtained from a bacterium274 and a related blue tetrapyrrole pigment was isolated independently from Nembrotha kubaryana,98 a compound ascidian,275 a bryozoan276 and from the predator-prey pairing Nembrotha spp. and the ascidian Atapozoa sp.277 and was identical to that produced by a mutant bacterium,278 indicating that it is likely of bacterial origin in all cases. Tambjamines have also been isolated from several predator-prey pairings; Tambja ceutae and the bryozoan Bugula dentata,279Tambja stegosauriformis and B. dentata137 and from Nembrotha spp. and the ascidian Atapozoa sp.277,280 Some tambjamines, especially 81, have been investigated for pharmaceutical uses as they have been shown to be cytotoxic against a number of tumour cell lines.272,275 Proposed mechanisms for the cytotoxicity include intercalation into DNA as well as the promotion of single-strand DNA oxidative cleavage, although a lack of selectivity for tumour cells over healthy cells has been noted as a prominent obstacle to drug development.281
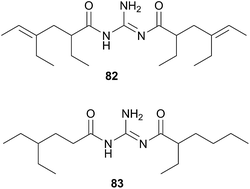
The Triophinae subfamily consists of nine genera of which only two (Triopha and Limacia) have been chemically examined. Two temperate nudibranchs from this subfamily, Triopha catalinae and Limacia clavigera also feed exclusively on bryozoans.282,283 The structurally related triophamine (82)282 and limaciamine (83)283 have been isolated from T. catalinae and L. clavigera respectively. Unlike the tambjamines, triophamine 82 has been shown to be de novo synthesised by T. catalinae284,285 and has also been isolated from a nudibranch in the closely related Polycerinae subfamily, Polycera tricolor.152 The geometry of the double bonds has been determined by total synthesis.286
The structural similarities between triophamine 82 and limaciamine 83 has led to speculation that 81 is also biosynthesised,283 but this has yet to be proven. As no bioactivity studies were conducted on these metabolites, their biological roles can only be speculated upon. The carotenoid triophaxanthin was first isolated as the main pigment of Triopha carpenteri287 (T. catalinae414) but subsequent isolations from a tunicate288 and a cuttlefish289 and its presence in the gut of T. carpenteri,287 indicate that it is dietary in nature.
6 Cladobranchs
6.1 Aeolidioidea
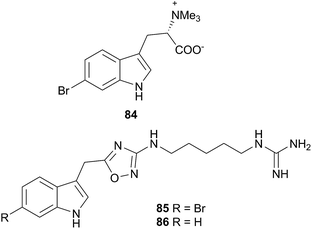
The superfamily Aeolidioidea, comprises seven families but only two (Aeolidiidae and Facelinidae) have been studied. Despite this, the superfamily has attracted much attention from researchers, largely due to the genus Phyllodesmium, the only aeolid genus known not to sequester cnidocysts or nematocysts.290L-6-Bromohypaphorine (84) was isolated from Hermissenda crassicornis and found to be an agonist of human α7 nicotinic acetylcholine receptor.297 A dietary origin of 84 is likely, as the compound has been found in the sponges Pachymatisma johnstoni298 and Aplysina sp.,299 as well as the tunicate Aplidium conicum.300 Whilst H. crassicornis is largely considered a generalist feeder, a recent report noted a preference for colonial tunicates in its diet, especially Aplidium solidum.301 It would therefore seem likely that the source of 84 in the nudibranch is from either A. conicum itself or another Aplidium species.
Two indole alkaloids, phidianidines A (85) and B (86), were isolated from Phidiana militaris and were cytotoxic against a number of cell lines, including C6 and HeLa tumour cells.302 They remain to date, the only known marine natural products with the 1,2,4-oxadiazole core. The rarity of this backbone, coupled with the total lack of any precursor metabolite in a known prey species, precludes the possibility of P. militaris sequestering 85 and 86. Whilst de novo synthesis might therefore be suspected, isotope studies are needed for confirmation. If isotope studies fail to show evidence of biosynthesis, then an unidentified prey species must possess the metabolites, or precursors thereof, or the source could potentially be a symbiotic organism. The biological activity of 85 and 86 coupled with their structural rarity, have led to syntheses of the natural products303–306 and analogues307–309 for exploration of their neuroprotective effects.303–309
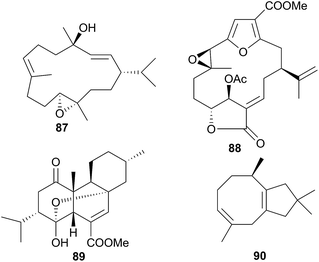
The genus Phyllodesmium is the only aeolid genus known not to sequester cnidocysts or nematocysts.290 Without the physical defence utilised by many aeolids, it might be expected that Phyllodesmium sp. would obtain a chemical defence in its place and indeed, this has been shown to be the case.290,310–312 The cytotoxic diterpene trocheliophorol (87) was isolated from Phyllodesmium longicirra (P. longicirrum418) and the soft coral Sarcophyton trocheliophorum, upon which it was found.310 The cerata of the nudibranch were concentrated with 87 and were noted to detach when molested, suggesting that P. longicirra was sequestering a defensive metabolite.310 Similarly, Phyllodesmium guamensis (P. guamense416) was also found grazing upon soft corals; Sinularia maxima, S. polydactyla and an unidentified Sinularia species.311 The diterpene 11β-acetoxypukalide (88) was sequestered from these corals by P. guamense and concentrated in its cerata. Diterpene 88 deterred feeding by the predatory pufferfish Canthigaster solandri at concentrations lower than those found in the cerata.311 Four polycyclic diterpenes were isolated from a single specimen of Phyllodesmium longicirrum from the Great Barrier Reef, Australia, of which 4-oxochatancin (89) deterred feeding by Canthigaster solandri.313 Although the food source of the nudibranch was not collected, it was noted that P. longicirrum usually feeds on soft coral of the Sarcophyton genus.313 This, coupled with 89 possessing a planar structure the same as that of isosarcophytin from Indian Ocean S. elegans314 but stereochemistry the same as that of chatacin315 from an Okinawan Sarcophyton sp., means that 89 is very likely a sequestered metabolite.
Sesquiterpenes are also sequestered by Phyllodesmium aeolids. Those obtained from P. lizardensis (P. lizardense417), were also found at an elevated concentration in the cerata290 and their dietary source was found to be a coral of the Heteroxenia genus. Although these sesquiterpenes were inactive in antifungal, antibacterial or antialgal assays and were not tested for feeding deterrence, a related compound, (+)-6-hydroxy-α-muurolene, has been found to be lethal to brine shrimp.312 A mixture of eight diverse sesquiterpenes was also isolated from P. magnum, including a novel asteriscane terpene (90).316 Of the remaining seven metabolites, four were known from corals of the Sinularia genus and three were known from the brown alga Dictyopteris divaricata,316 suggesting sequestration and that P. magnum may graze upon corals as well as brown algae. Whilst 90 was a novel natural product, a more recent study of Sinularia capillosa highlighted compounds with similar backbones,317 indicating that 90 may be a metabolite of an unidentified Sinularia sp., or be otherwise secondarily modified, as opposed to being biosynthesised de novo.
6.2 Arminoidea
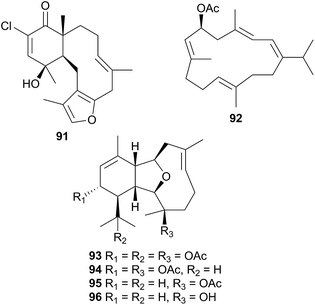
Consisting of the two families, Arminidae (seven genera) and Doridomorphidae (one genus), this superfamily has received minimal attention from natural products researchers, with only two species of the Arminidae (Armina and Dermatobranchus) being investigated to date. A series of seven briarane diterpenes, including the chlorinated example 91, was isolated from the nudibranch Armina maculata and its octocoral prey Veretillum cynomorium318–320 as was a cembranoid, preverecynarmin (92),320 highlighting that all were of dietary origin for the nudibranch. Similarly, four related diterpenoids, ophirin (93), calicophirin B (94), 13-deacetoxyl calicophirin B (95) and 13-deacetoxy-3-deacetyl calicophirin B (96), were isolated from the nudibranch Dermatobranchus ornatus.321 Of these, 94 and 95 were subsequently found within a prey species, a gorgonian of the Muricella genus.321 This indicated that these compounds may be of dietary origin, with 93 and 96 being sourced from an unidentified prey species or otherwise secondarily modified.6.3 Doridoxoidea
Of the unassigned Cladobranchs, there has been one superfamily designated, the Doridoxoidea which in turn consists of one family (the Doridoxidae) containing one genus (Doridoxa). Under former classification systems,12 Doridoxoidea was not strictly a cladobranch, but evidence emerged to suggest that this superfamily should be included under Cladobranchia,15 and it is now classified as such.13 There have been no natural product studies of Doridoxoidea species, possibly as they, like the Bathydoridoidea, are polar, deep water nudibranchs and are thus considerably more difficult to collect.126.4 Fionoidea
This superfamily contains the families Fionidae and Pseudovermidae but has barely been examined. A study of Phestilla melanobranchia (Tenellia melanobranchia419) (Fionidae) which feeds on the coral Tubastrea coccinea, yielded the known sponge alkaloids, 4′-de-N-methylaplysinopsin (97) and 6-bromo-4′-de-N-methylaplysinopsin (98), which were also obtained from the prey coral.322 A study of Phestilla sibogae (Tenellia sibogae420) highlighted that the nudibranch can track its coral prey through chemoreception using its rhinophores4 but the study did not involve isolating any metabolites that could potentially be implicated. The researchers noted that the rhinophores responded to various amino acids, with the most intense responses to aspartic and glutamic acids,4 so suggested that P. sibogae tracked corals through amino acids.4 However, as no natural product investigations have been conducted, the possibility remains that the nudibranch may detect and respond to secondary metabolites in corals that it wishes to sequester. After all, tracking of metabolites is not unprecedented in nudibranchs, as the response of Roboastra tigris to the tambjamines highlights (see Section 5.5.3 above).2706.5 Flabellinoidea
The Flabellinoidea is divided into two families, the Flabellinidae (eight genera including Flabellina and Flabellinopsis) and the Notaeolidiidae (one genus) but there has been very little chemical research into these nudibranchs. As noted above, chitin was found in skin epidermal cells and the epithelial cells of the stomach of Flabellina affinis as intracellular granules.296F. affinis was also noted to feed and lay its eggs upon several species of hydroid of the Eudendrium genus and to contain the same polyhydroxylated sterols as the hydroid but only the major component of this sterol mix was isolated and only from the hydroid.323 The ubiquitous carotenoid astaxanthin324 was found in Flabellinopsis iodina and its sponge diet.2876.6 Tritonioidea
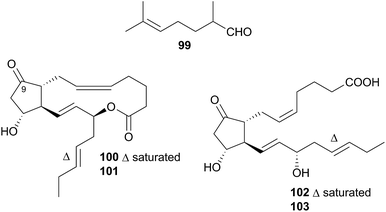
Nudibranchs of this superfamily, sometimes referred to as dendronotoids, are further separated into nine families, of which only two, the Tethydidae (two genera) and the Tritoniidae (nine genera) have received any attention from natural product chemists.Prostaglandin (PG) lactones have been isolated from the related species Tethys fimbria326,331 which is known to synthesise some of these de novo.326,332 These are synthesised from, and act as protected forms of, the free PG acids,326 compounds known to promote a range of hormonal responses in many organisms. PG molecules are classified by the structure of their central ring system, donated by a letter, which indicates the membrane protein that the PG will interact with.333 A subscript indicates the number of double bonds present in the aliphatic side chains. T. fimbria synthesises and stores PG lactones of the E class, PGE2-1,15-lactone (100) and PGE3-1,15-lactone (101), in its cerata.326 When molested, T. fimbria emits a mucus from its body and cerata and if the cerata become detached, they can continue to release mucus for a number of hours. When molested, the nudibranch converted 100 and 101 back to the free acid forms PGE2 (102) and PGE3 (103) respectively,326 suggesting that the free acid forms 102 and 103 were being utilised in the nudibranch's defence. PG-lactones of the F series have also been detected in T. fimbria (not pictured, but 9(S)-hydroxy derivatives of 100 and 101)332 but are only present in the ovotestis (hermaphroditic gland of the nudibranch) and not the cerata, indicating that they may play a role in reproduction as opposed to defence.332
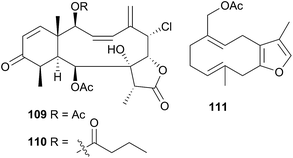
A series of six terpenoid metabolites has been isolated from two nudibranch populations of Tochuina tetraquetra337 (T. gigantea423). Specimens from Port Hardy, British Columbia were noted to contain tochuinyl acetate (105), dihydrotochuinyl acetate (106), rubifolide (107) and pukalide (108). Analysis of a prey species, the soft coral Gersemia rubiformis, present at Port Hardy, indicated that 105 and 106 were minor metabolites of this species. Rubifolide 107 had been previously detected in G. rubiformis.338 Whilst a known source of 108 was not found at Port Hardy, it has been previously reported from tropical corals, Sinularia,339 and gorgonians of the genus Lophogorgia.337 A second collection of T. tetraquetra, from Bamfield, British Columbia, revealed the diterpenoid ptilosarcenone (109) and its butanoate analogue (110).337 The sea pen Ptilosarcus gurneyi, a prey species of T. tetraquetra in Bamfield waters, is known to produce 109.340 As it appears that the nudibranch sequesters natural products from various prey species,337 it is therefore not surprising that the geographically separated populations possess different compositions of natural products.
Tritonia hamnerorum was observed on its prey, the sea fan Gorgonia ventalina at very high densities, in the Florida Keys, U.S.A., resulting in the isolation of the furano-germacrene julieannafuran (111) from both sea fan and nudibranch. T. hamnerorum concentrated 111 relative to other sea fan metabolites and 111 either significantly reduced or entirely prevented reef fish feeding on the nudibranch in both natural and laboratory environments.341
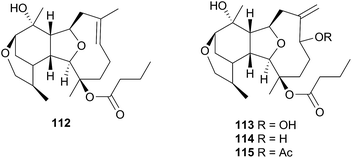
1-O-Hexadecyl glycerol (chimyl alcohol) was detected in the Antarctic nudibranch Tritoniella belli and its usual prey, the stoloniferan coral Clavularia frankliniana. The common predatory Antarctic sea star Odontaster validus, showed feeding deterrence to both T. belli mantle tissue and to chimyl alcohol.342 A series of sesquiterpenes, tritoniopsins A–D (112–115), were isolated from the nudibranch Tritoniopsis elegans and its soft coral prey Cladiella krempfi.334 The major metabolites in both species were found to be 112 and 113 but the relative ratio of these was inverted in the predator-prey pair, such that 112 dominated the T. elegans extract and 113 dominated the extract of C. krempfi. Tritoniopsin A 112 was concentrated within the mantle tissue of T. elegans, to a greater degree than were 113–115, suggesting that 112 plays a defensive role for the nudibranch and that it was preferentially sequestered from the coral. No bioassays were conducted for 112, but weak to moderate cytotoxicity was observed for tritoniopsin B 113 against a selection of rat cell lines.334
6.7 Unassigned families
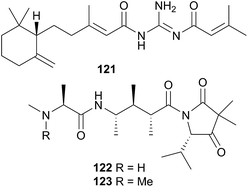
L. millecra is known to sequester sesquiterpenes.345,346 Millecrones A (117) and B (118), as well as millecrols A (119) and B (120), were isolated from a South African population of L. millecra.345 Analysis of the nudibranch's stomach contents revealed spicules from the soft corals Alcyonium foliatum, A. valdiviae and Capnella thyrsoidea.345 Limited antimicrobial activity was noted for 117, 119 and 120. A separate analysis of another South African population of L. millecra and its local prey, identified the dietary origin of 117 and 118.346 GC analysis indicated that 117 originated from the soft coral Alcyonium fauri, whilst 118 was from the gorgonian Leptogorgia palma.346 At least eleven other sesquiterpenes were detected (structures not shown) in the latter study by GC. These were also expected to be of dietary origin, from various octocoral prey species, due to the backbone common with typical octocoral metabolites.346 Some of the triprenylated toluquinones and toluhydroquinones obtained from Leminda millecra induced apoptosis in oesophageal cancer cell lines.347
 | ||
| Fig. 3 Janolus cristatus feeding on Bugulina flabellata in the Shetland Islands. Photograph courtesy of Joanne Porter. | ||
A subsequent investigation into a different New Zealand population of B. flabellata found evidence of both 122 and 123 within the bryozoan.355 Small scale extraction of New Zealand collections of Janolus novozealandicus and Bugulina flabellata and screening of the extracts by tandem liquid chromatography mass spectrometry (LCMS) indicated the presence of both 122 and 123 and suggested the presence of a series of at least six related tripeptides. The relative abundance of 122 and 123 is inverted within J. novozealandicus (122 dominant) with respect to B. flabellata (123 dominant) suggesting preferential uptake of 122. An egg mass of J. novozealandicus was also extracted and screened and of the two tripeptides, only 122 was found (as the main metabolite), highlighting its potential role as a defensive compound,355 although further studies are needed to confirm this.
7 Nematocysts and zooxanthellae
Sequestration of whole organisms (zooxanthellae) or nematocysts by some aeolid nudibranchs has been given its own section as, strictly speaking, it is not natural products chemistry. Nevertheless these processes deserve a brief mention. As has already been noted, virtually all aeolid species except those of the Phyllodesmium genus are known to sequester the stinging cells from their cnidarian prey.9,290 The nematocysts are taken up by the aeolids and transported to the tips of cerata where multiple cells are incorporated into a single cnidosac.9 The use of a cnidosac is almost certainly in defence, as aeolids which have had their cerata removed are considerably more likely to be consumed. In most species, the cerata are also known to regenerate, indicating that they serve an important function to the aeolids.9 The exact mechanisms by which the nudibranch is able to protect itself from the sting of these cells are not fully understood.9,356 The presence of epithelial linings within the viscera has been observed and noted to absorb the damage from a discharged nematocyst, thus protecting the nudibranch.9 Application of aeolid mucus to the sea anemone Anthopleura elegantissima lowered the number of nematocyst discharges, with respect to mucus of other nudibranchs.9 This highlighted that aeolids used multiple strategies to cope with the stinging cells. A relatively recent study tested the hypothesis that aeolids transfer immature nematocysts to the cnidosacs and then allow them to mature, thus avoiding the potential damage caused by transporting a charged nematocyst.356 Utilising a fluorescent dye, researchers were able to show that there was a change in pH as the nematocysts were incorporated into the cnidosacs. As an accumulation of protons is necessary for nematocysts to mature, this supported the hypothesis that aeolids themselves were maturing the nematocysts.356Whilst sequestration of nematocysts is common in all aeolids except those of Phyllodesmium, an interesting symbiotic relationship has been observed in a number of cladobranchs. Zooxanthellae, photosynthetic dinoflagellates, are frequently found within the cells of cladobranchs' branched digestive gland.357 This symbiosis has been observed in a number of Phyllodesmium species and may highlight why this genus does not sequester nematocysts.357,358 Aeolids of the Phestilla (Tenellia419,420) (Fionidae family) and Pteraeolidia (Facelinidae family) have also been noted to possess zooxanthellae symbionts. A number of non-aeolid cladobranchs also obtain these symbiotic organisms, namely the dendronotoids of Melibe and Doto genera as well as Doridomorpha gardineri (Doridomorphidae family) and Pinufius rebus (unassigned Cladobranchia, Superfamily Arminoidea, Family Pinufidae).357,358 The advantages offered to these cladobranchs are obvious, in that the transfer of (primary) metabolites can enhance survivability if food is scarce.357 Interestingly, there have been no reported cases of zooxanthellae symbiosis within any of the dorid nudibranchs. This is presumably because their unbranched digestive gland and lack of cerata (or similar projections) limits the available surface area for zooxanthellae photosynthesis.
8 Conclusions
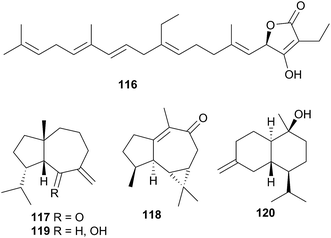
The natural products chemistry of nudibranchs is extensive and well documented. Terpenes and sesquiterpenes in particular, dominate those metabolites that have been isolated, although virtually all classes of natural products are represented. The sequestering of these metabolites from prey species is seemingly the most common source of nudibranch natural products. Indeed, metabolites have been sequestered from sponges, corals, tunicates, gorgonians and bryozoans. Whilst de novo synthesis is rarer than sequestration of natural products, it can still be found throughout all of Nudibranchia. Secondary modification is also suspected in a number of cases. Unsurprisingly, terpenes still dominate the types of structures observed.It is certainly true that dorids have received considerably more research attention than their cladobranch counterparts. This is perhaps due to a perceived notion that cladobranchs, especially aeolids, utilise a physical defence (cnidosacs) over a chemical one. Furthermore, many dorids are known to feed exclusively on sponges, and sponges themselves are known to be the most prolific producers of marine natural products,36c so the natural products chemistry of those dorids that feed on sponges might also be expected to be rich in metabolites. However, as this review has highlighted, natural products are found within all branches of Cladobranchia (that have been studied). De novo synthesis is known within the dendronotoids and suspected in some aeolids. There have even been reports of unprecedented novelty, including the first examples of marine metabolites with a 1,2,4-oxadiazole core (85 and 86)302 as well as the first known linear homosesterterpene (116).343 These examples challenge the historical notion that the dorids represent the greatest source of nudibranch natural products. In light of this, more investigations into cladobranchs should be conducted moving forwards.
Even within the dorid nudibranchs, there has been an uneven allocation of research attention. Certainly the Doridoidea are well represented but many of these investigations are from species of a single genus, Chromodoris. Similarly, Hexabranchus sanguineus, has received considerable attention but no reports into other Hexabranchus species have been carried out. This preference of some superfamilies and some species over others means that there are still many nudibranchs to be properly studied. Perhaps the biggest message to be gained from this review, is that very few natural products have proven ecological roles. Indeed in some cases, only structures have been reported with absolutely no consideration for their bioactivity. Of all the metabolites presented within this review for which an ecological role is known or suspected, all but the PGF-lactones isolated from Tethys fimbria, are used in defence. Although the defensive capabilities of natural products are assumed in many cases, very few have been shown to actually deter predation. However, proving a chemical ecological role is not a simple task. Typically this requires dissection of the nudibranch into its mantle, viscera and other parts, instead of whole organism extraction, as well as conducting a number of antifeeding tests and bioassays. In many cases, natural product investigations are pharmaceutically driven and the chemical ecology is only of secondary interest. Thus, extensive ecological studies are relatively rare in the literature, given the time and amount of metabolite required to adequately perform these experiments.
9 Conflicts of interest
There are no conflicts to declare.10 Acknowledgements
We thank Tracey Bates, Rex Fairweather and Joanne Porter for the photographs provided for this review, Tracey Bates and Rex Fairweather for helpful discussions and the anonymous reviewers for their helpful suggestions which have improved this manuscript.11 References
- R. C. Anderson, Int. Zoo Yb., 1995, 34, 65–70, DOI:10.1111/j.1748-1090.1995.tb00659.x
.
-
S. Gofas, Nudibranchia In MolluscaBase, 2017, 15 May, 2017, accessed through: World Register of Marine Species (WoRMS) at, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=1762 Search PubMed
.
-
B. E. Picton and C. C. Morrow, Nudibranchs of the British Isles, 1996, http://www.seaslug.org.uk/nudibranchs/index.html, accessed 7 January, 2016 Search PubMed
.
- B. F. Murphy and M. G. Hadfield, Comp. Biochem. Physiol., 1997, 118A, 727–735, DOI:10.1016/S0300-9629(97)00014-5
.
- G.-P. Hu, J. Yuan, L. Sun, Z.-G. She, J.-H. Wu, X.-J. Lan, X. Zhu, Y.-C. Lin and S.-P. Chen, Mar. Drugs, 2011, 9, 514–525, DOI:10.3390/md9040514
.
- D. J. Faulkner and M. T. Ghiselin, Mar. Ecol.: Prog. Ser., 1983, 13, 295–301, DOI:10.3354/meps013295
.
- G. Cimino and M. T. Ghiselin, Chemoecology, 1999, 9, 187–207, DOI:10.1007/s000490050052
.
- M. Carbone, M. Gavagnin, M. Haber, Y.-W. Guo, A. Fontana, E. Manzo, G. Genta-Jouve, M. Tsoukatou, W. B. Rudman, G. Cimino, M. T. Ghiselin and E. Mollo, PLoS One, 2013, 8, e62075 CAS
.
- P. G. Greenwood, Toxicon, 2009, 54, 1065–1070, DOI:10.1016/j.toxicon.2009.02.029
.
- H. Wägele and R. C. Willan, Zool. J. Linn. Soc., 2000, 130, 83–181, DOI:10.1111/j.1096-3642.2000.tb02196.x
.
- M. Schrödl, H. Wägele and R. C. Willan, Zool. Anz., 2001, 240, 83–97, DOI:10.1078/0044-5231-00008
.
- P. Bouchet, J. Fryda, B. Hausdorf, W. Ponder, A. Valdés and A. Warén, Malacologia, 2005, 47, 261–263 Search PubMed
.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=1762, accessed 15 May, 2017.
- J. A. Goodheart, A. L. Bazinet, A. G. Collins and M. P. Cummings, R. Soc. Open Sci., 2015, 2, 150196, DOI:10.1098/rsos.150196
.
- J. Mahguib and A. Valdés, Polar Biol., 2015, 38, 1369–1377, DOI:10.1007/s00300-015-1700-5
.
- A. W. W. van Wyk and M. T. Davies-Coleman, Tetrahedron, 2007, 63, 12179–12184 CrossRef CAS
.
- M. J. Garson, Chem. Rev., 1993, 93, 1699–1733, DOI:10.1021/cr00021a003
.
- D. J. Faulkner, T. F. Molinski, R. J. Anderson, E. J. Dumdei and E. Dilip de Silva, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1990, 97, 233–240 Search PubMed
.
- P. Karuso, Bioorg. Mar. Chem., 1987, 1, 31–60, DOI:10.1007/978-3-642-72726-9_2
.
- P. Proksch, Biol. Unserer Zeit, 1991, 21, 26–30, DOI:10.1002/biuz.19910210110
.
- G. Cimino, A. Fontana and M. Gavagnin, Curr. Org. Chem., 1999, 3, 327–372 CAS
.
- P. Proksch, Toxicon, 1994, 32, 639–655, DOI:10.1016/0041-0101(94)90334-4
.
-
G. Cimino and G. Sodano, in Sponges Time Space, Proc. 4th Int. Porifera Congr, ed. R. W. M. Van Soest, T. M. G. Van Kempen and J.-C. Braekman, 1994, pp. 459–472 Search PubMed
.
-
M. Ishibashi, Y. Yamaguchi and Y. J. Hirano, in Biomaterials from Aquatic and Terrestrial Organisms, ed. M. Fingerman and R. Nagabhushanam, 2006, pp. 513–535 Search PubMed
.
-
G. Cimino, M. L. Ciavatta, A. Fontana and M. Gavagnin, in Bioactive Compounds from Natural Sources, ed. C. Tringali, 2001, pp. 577–637 Search PubMed
.
- M. Gavagnin and A. Fontana, Curr. Org. Chem., 2000, 4, 1201–1248, DOI:10.2174/1385272003375798
.
-
R. J. Andersen, E. D. De Silva, E. J. Dumdei, P. T. Northcote, C. Pathirana and M. Tischler, in Recent Advances in Phytochemistry, Biochem. Mevalonic Acid Pathway Terpenoids, 1990, vol. 24, pp. 265–282 Search PubMed
.
-
M. J. Garson, J. S. Simpson, A. E. Flowers and E. J. Dumdei, Bioactive Natural Products (Part B), in Studies in Natural Products Chemistry, 2000, vol. 21, pp. 329–372 Search PubMed
.
- M. J. Garson and J. S. Simpson, Nat. Prod. Rep., 2004, 21, 164–179, 10.1039/b302359c
.
- T. Grkovic, D. R. Appleton and B. R. Copp, Chem. N.Z., 2005, 69, 12–15 CAS
.
- M. J. Garson, Prog. Mol. Subcell. Biol., 2006, 43(Molluscs), 159–174 CrossRef CAS PubMed
.
- M. J. Garson, Chem. Aust., 2004, 71, 16–18 CAS
.
- R. J. Andersen, K. Desjardine and K. Woods, Prog. Mol. Subcell. Biol., 2006, 43(Molluscs), 277–301 CrossRef CAS PubMed
.
- T. Miyamoto, Prog. Mol. Subcell. Biol., 2006, 43(Molluscs), 199–214 CrossRef CAS PubMed
.
- M. T. Davies-Coleman, Prog. Mol. Subcell. Biol., 2006, 43(Molluscs), 133–157 CrossRef CAS PubMed
.
-
(a) J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2017, 34, 235–294, 10.1039/C6NP00124F
; (b) J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2016, 33, 382–431, 10.1039/C5NP00156K
; (c) J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2015, 32, 116–211, 10.1039/C4NP00144C
; (d) J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2014, 31, 160–258, 10.1039/c3np70117d
; (e) J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2013, 30, 237–323, 10.1039/C2NP20112G
; (f) J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2012, 29, 144–222, 10.1039/C2NP00090C
.
- I. W. Mudianta, A. M. White, Suciati, P. L. Katavic, R. R. Krishnaraj, A. E. Winters, E. Mollo, K. L. Cheney and M. J. Garson, Pure Appl. Chem., 2014, 86, 995–1002, DOI:10.1515/pac-2013-1111
.
- V. T. Dang, K. Benkendorff, T. Green and P. Speck, J. Virol., 2015, 89, 8114–8118, DOI:10.1128/JVI.00287-15
.
- N. V. Zhukova, Mar. Drugs, 2014, 12, 4578–4592, DOI:10.3390/md12084578
.
- K. Iken, C. Avila, M. L. Ciavatta, A. Fontana and G. Cimino, Tetrahedron Lett., 1998, 39, 5635–5638, DOI:10.1016/S0040-4039(98)01095-8
.
- C. Avila, K. Iken, A. Fontana and G. Cimino, J. Exp. Mar. Biol. Ecol., 2000, 252, 27–44, DOI:10.1016/S0022-0981(00)00227-6
.
- MarinLit, The Royal Society of Chemistry, http://pubs.rsc.org/marinlit/, accessed 29 June 2017.
- E. Manzo, M. Carbone, E. Mollo, C. Irace, A. Di Pascale, Y. Li, M. L. Ciavatta, G. Cimino, Y.-W. Guo and M. Gavagnin, Org. Lett., 2011, 13, 1897–1899, DOI:10.1021/ol200377w
.
- G. Nuzzo, M. L. Ciavatta, R. Kiss, V. Mathieu, H. Leclercqz, E. Manzo, G. Villani, E. Mollo, F. Lefranc, L. D'Souza, M. Gavagnin and G. Cimino, Mar. Drugs, 2012, 10, 1799–1811, DOI:10.3390/md10081799
.
- A. Rudi, Z. Stein, S. Green, I. Goldberg, Y. Kashman, Y. Benayahu and M. Schleyer, Tetrahedron Lett., 1994, 35, 2589–2592, DOI:10.1016/S0040-4039(00)77179-6
.
- S. W. Ayer and R. J. Andersen, Tetrahedron Lett., 1982, 23, 1039–1042, DOI:10.1016/S0040-4039(00)87016-1
.
- M. Gavagnin, N. Ungur, E. Mollo, J. Templado and G. Cimino, Eur. J. Org. Chem., 2002, 1500–1504, DOI:10.1002/1099-0690(200205)2002:9<1500::AID-EJOC1500>3.0.CO;2-D
.
- J. Kubanek, D. J. Faulkner and R. J. Anderson, J. Chem. Ecol., 2000, 26, 377–389, DOI:10.1023/A:1005405304862
.
- D. L. Burgoyne, E. J. Dumdei and R. J. Andersen, Tetrahedron, 1993, 49, 4503–4510, DOI:10.1016/S0040-4020(01)81280-1
.
- J. E. Thompson, R. P. Walker, S. J. Wratten and D. J. Faulkner, Tetrahedron, 1982, 38, 1865–1873, DOI:10.1016/0040-4020(82)80035-5
.
- J. Hellou, R. J. Anderson and J. E. Thompson, Tetrahedron, 1982, 38, 1875–1879, DOI:10.1016/0040-4020(82)80036-7
.
- K. Gustafson, R. J. Andersen, C. H. He and J. Clardy, Tetrahedron Lett., 1985, 26, 2521–2524, DOI:10.1016/S0040-4039(00)98826-9
.
- M. Tischler, R. J. Andersen, M. I. Choudhary and J. Clardy, J. Org. Chem., 1991, 56, 42–47, DOI:10.1021/jo00001a010
.
- J. Daoust, A. Fontana, C. E. Merchant, N. J. de Voogd, B. O. Patrick, T. J. Kieffer and R. J. Andersen, Org. Lett., 2010, 12, 3208–3211, DOI:10.1021/ol101151f
.
- J. Hellou, R. J. Andersen, S. Rafii, E. Arnold and J. Clardy, Tetrahedron Lett., 1981, 22, 4173–4176, DOI:10.1016/S0040-4039(01)82096-7
.
- M. Tischler and R. J. Andersen, Tetrahedron Lett., 1989, 30, 5717–5720, DOI:10.1016/S0040-4039(00)76179-X
.
- E. J. Dumdei, J. Kubanek, J. E. Coleman, J. Pika, R. J. Anderson, J. R. Steiner and J. Clardy, Can. J. Chem., 1997, 75, 773–789, DOI:10.1139/v97-094
.
- J. Kubanek, E. I. Graziani and R. J. Andersen, J. Org. Chem., 1997, 62, 7239–7246, DOI:10.1021/JO970695V
.
- A. Fontana, M. Gavagnin, E. Mollo, E. Trivellone, J. Ortea and G. Cimino, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1995, 111, 283–290, DOI:10.1016/0305-0491(94)00250-X
.
- A. M. White, A. S. Dewi, K. L. Cheney, A. E. Winters, J. T. Blanchfield and M. J. Garson, Nat. Prod. Commun., 2016, 11, 921–924 Search PubMed
.
- D. E. de Silva and P. J. Scheuer, Heterocycles, 1982, 17, 167–170, DOI:10.3987/S-1982-01-0167
.
- R. Kazlauskas, Rymantas, P. T. Murphy, R. J. Wells, K. Noack, W. E. Oberhaensli and P. Schoenholzer, Aust. J. Chem., 1979, 32, 867–880, DOI:org/10.1071/CH9790867
.
- T. Grkovic, D. R. Appleton and B. R. Copp, Chem. N. Z., 2005, 69, 12–15 CAS
.
- R. Kazlauskas, P. T. Murphy, R. J. Quinn and R. J. Wells, Aust. J. Chem., 1976, 29, 2533–2539, DOI:10.1071/CH9762533
.
- M. B. Ksebati and F. J. Schmitz, J. Org. Chem., 1987, 52, 3766–3773, DOI:10.1021/jo00226a008
.
- E. Mollo, M. Gavagnin, M. Carbone, Y.-W. Guo and G. Cimino, Chemoecology, 2005, 15, 31–36, DOI:10.1007/s00049-005-0289-5
.
- M. B. Ksebati and F. J. Schmitz, J. Nat. Prod., 1988, 51, 857–861, DOI:10.1021/np50059a007
.
- G. Cimino, S. De Stefano, A. Guerriero and L. Minale, Tetrahedron Lett., 1975, 1425–1428, DOI:10.1016/S0040-4039(00)72159-9
.
- R. Kazlauskas, P. T. Murphy, R. J. Wells, J. J. Daly and P. Schoenholzer, Tetrahedron Lett., 1978, 4951–4954, DOI:10.1016/S0040-4039(01)85779-8
.
- G. M. Cameron, B. L. Stapleton, S. M. Simonsen, D. J. Brecknell and M. J. Garson, Tetrahedron, 2000, 56, 5247–5252, DOI:10.1016/S0040-4020(00)00434-8
.
- C. Charles, J. C. Braekman, D. Daloze, B. Tursch, J. P. Declercq, G. Germain and M. Van Meerssche, Bull. Soc. Chim. Belg., 1978, 87, 481–486 CrossRef CAS
.
- A. Fontana, C. Avila, E. Martinez, J. Ortea, E. Trivellone and G. Cimino, J. Chem. Ecol., 1993, 19, 339–356, DOI:10.1007/BF00993700
.
- J. E. Hochlowski and D. J. Faulkner, Tetrahedron Lett., 1981, 22, 271–274, DOI:10.1016/0040-4039(81)80073-1
.
- G. R. Schulte and P. J. Scheuer, Tetrahedron, 1982, 38, 1857–1863, DOI:10.1016/0040-4020(82)80034-3
.
- J. E. Hochlowski, D. J. Faulkner, L. S. Bass and J. Clardy, J. Org. Chem., 1983, 48, 1738–1740, DOI:10.1021/jo00158a030
.
- J. E. Hochlowski, D. J. Faulkner, G. K. Matsumoto and J. Clardy, J. Org. Chem., 1983, 48, 1141–1142, DOI:10.1021/jo00155a055
.
- R. K. Okuda and P. J. Scheuer, Experientia, 1985, 41, 1355–1356 CrossRef CAS
.
- B. Carte, M. R. Kernan, E. B. Barrabee, D. J. Faulkner, G. K. Matsumoto and J. Clardy, J. Org. Chem., 1986, 51, 3528–3532, DOI:10.1021/jo00368a025
.
- B. Terem and P. J. Scheuer, Tetrahedron, 1986, 42, 4409–4412, DOI:10.1016/S0040-4020(01)87279-3
.
- T. F. Molinski and D. J. Faulkner, J. Org. Chem., 1986, 51, 2601–2603, DOI:10.1021/jo00363a040
.
- T. F. Molinski, D. J. Faulkner, C. H. He, G. D. Van Duyne and J. Clardy, J. Org. Chem., 1986, 51, 4564–4567, DOI:10.1021/jo00374a014
.
- Y. Kakou, P. Crews and G. J. Bakus, J. Nat. Prod., 1987, 50, 482–484, DOI:10.1021/np50051a023
.
- D. G. Corley, R. Herb, R. E. Moore, P. J. Scheuer and V. J. Paul, J. Org. Chem., 1988, 53, 3644–3646, DOI:10.1021/jo00250a053
.
- M. R. Kernan, E. B. Barrabee and D. J. Faulkner, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1988, 89B, 275–278, DOI:10.1016/0305-0491(88)90223-4
.
- S. C. Bobzin and D. J. Faulkner, J. Org. Chem., 1989, 54, 3902–3907, DOI:10.1021/jo00277a029
.
- E. J. Dumdei, E. D. De Silva, R. J. Andersen, M. I. Choudhary and J. Clardy, J. Am. Chem. Soc., 1989, 111, 2712–2713, DOI:10.1021/ja00189a055
.
- G. Cimino, A. Crispino, M. Gavagnin and G. Sodano, J. Nat. Prod., 1990, 53, 102–106, DOI:10.1021/np50067a013
.
- E. Dilip de Silva, S. A. Morris, S. Miao, E. Dumdei and R. J. Andersen, J. Nat. Prod., 1991, 54, 993–997, DOI:10.1021/np50076a011
.
- S. A. Morris, E. Dilip de Silva and R. J. Andersen, Can. J. Chem., 1991, 69, 768–771, DOI:10.1139/v91-114
.
- M. Gavagnin, R. R. Vardaro, C. Avila, G. Cimino and J. Ortea, J. Nat. Prod., 1992, 55, 368–371, DOI:10.1021/np50081a014
.
- K. Sakamoto, T. Miyamoto, H. Amano, R. Higuchi, T. Komori and T. Sasaki, Tennen Yuki Kagobutsu Toronkai Koen Yoshishu, 1992, 34, 455–462 Search PubMed
.
- T. Miyamoto, K. Sakamoto, H. Amano, R. Higuchi, T. Komori and T. Sasaki, Tetrahedron Lett., 1992, 33, 5811–5814, DOI:10.1016/0040-4039(92)89038-E
.
- R. Puliti, M. Gavagnin, G. Cimino, C. A. Mattia and L. Mazzarella, Acta Crystallogr., Sect. C: Struct. Chem., 1992, C48, 2145–2147, DOI:10.1107/S0108270192003196
.
- J. Pika and D. J. Faulkner, Tetrahedron, 1995, 51, 8189–8198, DOI:10.1016/0040-4020(95)00440-J
.
- T. Miyamoto, K. Sakamoto, K. Arao, T. Komori, R. Higuchi and T. Sasaki, Tetrahedron, 1996, 52, 8187–8198, DOI:10.1016/0040-4020(96)00388-2
.
- K. McPhail and M. T. Davies-Coleman, Tetrahedron, 1997, 53, 4655–4660, DOI:10.1016/S0040-4020(97)00198-1
.
- T. Miyamoto, K. Sakamoto, H. Amano, Y. Arakawa, Y. Nagarekawa, T. Komori, R. Higuchi and T. Sasaki, Tetrahedron, 1999, 55, 9133–9142, DOI:10.1016/S0040-4020(99)00477-9
.
- P. Karuso and P. J. Scheuer, Molecules, 2003, 8, 1–6, DOI:10.3390/70100001-rev
.
- K. W. Yong, A. A. Salim and M. J. Garson, Tetrahedron, 2008, 64, 6733–6738, DOI:10.1016/j.tet.2008.05.008
.
- M. H. Uddin, M. Otsuka, T. Muroi, A. Ono, N. Hanif, S. Matsuda, T. Higa and J. Tanaka, Chem. Pharm. Bull., 2009, 57, 885–887, DOI:10.1248/cpb.57.885
.
- M. Agena, C. Tanaka, N. Hanif, M. Yasumoto-Hirose and J. Tanaka, Tetrahedron, 2009, 65, 1495–1499, DOI:10.1016/j.tet.2008.11.101
.
- Suciati, L. K. Lambert and M. J. Garson, Aust. J. Chem., 2011, 64, 757–765, DOI:10.1071/CH11036
.
- P. L. Katavic, P. Jumaryatno, J. N. A. Hooper, J. T. Blanchfield and M. J. Garson, Aust. J. Chem., 2012, 65, 531–538, DOI:10.1071/CH12010
.
- P. L. Katavic, P. Jumaryatno, J. N. A. Hooper, J. T. Blanchfield and M. J. Garson, Aust. J. Chem., 2013, 66, 1461, DOI:10.1071/CH12010_NC
.
- K. L. Cheney, A. White, I. W. Mudianta, A. E. Winters, M. Quezada, R. J. Capon, E. Mollo and M. J. Garson, PLoS One, 2016, 11, e0145134, DOI:10.1371/journal.pone.0145134
.
- G. Schulte, P. J. Scheuer and O. J. McConnell, Helv. Chim. Acta, 1980, 63, 2159–2167, DOI:10.1002/hlca.19800630805
.
- C. W. Jefford, G. Bernardinelli, J. Tanaka and T. Higa, Tetrahedron Lett., 1996, 37, 159–162, DOI:10.1016/0040-4039(95)02113-2
.
- A. K. Ghosh and Y. Wang, J. Am. Chem. Soc., 2000, 122, 11027–11028, DOI:10.1021/ja0027416
.
- A. Gollner and J. Mulzer, Org. Lett., 2008, 10, 4701–4704, DOI:10.1021/ol802075v
.
- Y. Kashman, A. Groweiss, R. Lidor, D. Blasberger and S. Carmely, Tetrahedron, 1985, 41, 1905–1914, DOI:10.1016/S0040-4020(01)96553-6
.
- M. C. A. Ramirez, D. E. Williams, J. R. Gubiani, L. L. L. Parra, M. F. C. Santos, D. D. Ferreira, J. T. Mesquita, A. G. Tempone, A. G. Ferreira, V. Padula, E. Hajdu, R. J. Andersen and R. G. S. Berlinck, J. Nat. Prod., 2017, 80, 720–725, DOI:10.1021/acs.jnatprod.6b01160
.
- J. M. Wojnar, K. O. Dowle and P. T. Northcote, J. Nat. Prod., 2014, 77, 2288–2295, DOI:10.1021/np500549g
.
- T. F. Molinski and D. J. Faulkner, J. Org. Chem., 1987, 52, 296–298, DOI:10.1021/jo00378a031
.
- Y. Hirayama, P. L. Katavic, A. M. White, G. K. Pierens, L. K. Lambert, A. E. Winters, H. Kigoshi, M. Kita and M. J. Garson, Aust. J. Chem., 2016, 69, 136–144, DOI:10.1071/CH15203
.
- E. Manzo, M. Gavagnin, M. J. Somerville, S.-C. Mao, M. L. Ciavatta, E. Mollo, P. J. Schupp, M. J. Garson, Y.-W. Guo and G. Cimino, J. Chem. Ecol., 2007, 33, 2325–2336, DOI:10.1007/s10886-007-9387-x
.
- D. Mebs, J. Chem. Ecol., 1985, 11, 713–716, DOI:10.1007/BF00988300
.
- M. J. Somerville, E. Mollo, G. Cimino, W. Rungprom and M. J. Garson, J. Nat. Prod., 2006, 69, 1086–1088, DOI:10.1021/np060002i
.
- M. J. Somerville, E. Mollo, G. Cimino, W. Rungprom and M. J. Garson, J. Nat. Prod., 2007, 70, 1836, DOI:10.1021/np070553o
.
- K. W. L. Yong, I. W. Mudianta, K. L. Cheney, E. Mollo, J. Blanchfield and M. J. Garson, J. Nat. Prod., 2015, 78, 421–430, DOI:10.1021/np500797b
.
- A. Fontana, E. Mollo, J. Ortea, M. Gavagnin and G. Cimino, J. Nat. Prod., 2000, 63, 527–530, DOI:10.1021/np990506z
.
- G. Cimino, S. De Rosa, S. De Stefano and G. Sodano, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1982, 73B, 471–474, DOI:10.1016/0305-0491(82)90061-X
.
- S. D. Rogers and V. J. Paul, Mar. Ecol.: Prog. Ser., 1991, 77, 221–232, DOI:10.3354/meps077221
.
- C. Avila and V. J. Paul, Mar. Ecol.: Prog. Ser., 1997, 150, 171–180, DOI:10.3354/meps150171
.
- A. Fontana, P. Cavaliere, N. Ungur, L. D'Souza, P. S. Parameswaram and G. Cimino, J. Nat. Prod., 1999, 62, 1367–1370, DOI:10.1021/np9900932
.
- M. Gavagnin, E. Mollo, T. Docimo, Y.-W. Guo and G. Cimino, J. Nat. Prod., 2004, 67, 2104–2107, DOI:10.1021/np040087s
.
- I. W. Mudianta, A. M. White and M. J. Garson, Nat. Prod. Commun., 2015, 10, 865–868 Search PubMed
.
- A. M. White, G. K. Pierens, L. C. Forster, A. E. Winters, K. L. Cheney and M. J. Garson, J. Nat. Prod., 2016, 79, 477–483, DOI:10.1021/acs.jnatprod.5b00866
.
- L. C. Forster, A. E. Winters, K. L. Cheney, P. Dewapriya, R. J. Capon and M. J. Garson, J. Nat. Prod., 2017, 80, 670–675, DOI:10.1021/acs.jnatprod.6b00936
.
- J. E. Hochlowski, R. P. Walker, C. Ireland and D. J. Faulkner, J. Org. Chem., 1982, 47, 88–91, DOI:10.1021/jo00340a018
.
- S. H. Grode and J. H. Cardellina II, J. Nat. Prod., 1984, 47, 76–83, DOI:10.1021/np50031a009
.
- J. C. Garcia-Gomez, G. Cimino and A. Medina, Mar. Biol., 1990, 106, 245–250 CrossRef CAS
.
- C. Avila, G. Cimino, A. Fontana, M. Gavagnin, J. Ortea and E. Trivellone, J. Chem. Ecol., 1991, 17, 625–636, DOI:10.1007/BF00982131
.
- G. Cimino, A. Fontana, F. Gimenez, A. Marin, E. Mollo, E. Trivellone and E. Zubia, Experientia, 1993, 49, 582–586 CrossRef CAS
.
- A. Fontana, E. Trivellone, E. Mollo, G. Cimino, C. Avila, E. Martinez and J. Ortea, J. Nat. Prod., 1994, 57, 510–513, DOI:10.1021/np50106a011
.
- M. Haber, S. Cerfeda, M. Carbone, G. Calado, H. Gaspar, R. Neves, V. Maharajan, G. Cimino, M. Gavagnin, M. T. Ghiselin and E. Mollo, Biol. Bull., 2010, 218, 181–188, DOI:10.1086/BBLv218n2p181
.
- J. F. Cruz, H. Gaspar and G. Calado, Chemoecology, 2012, 22, 47–53, DOI:10.1007/s00049-011-0097-z
.
- F. R. Pereira, R. G. S. Berlinck, E. Rodrigues Filho, K. Veloso, A. G. Ferreira and V. Padula, Quim. Nova, 2012, 35, 2194–2201, DOI:10.1590/S0100-40422012001100018
.
- G. Cimino, S. De Stefano, L. Minale and E. Trivellone, Tetrahedron Lett., 1975, 3727–3730, DOI:10.1016/S0040-4039(00)91320-0
.
- I. W. Mudianta, V. L. Challinor, A. E. Winters, K. L. Cheney, J. J. De Voss and M. J. Garson, Beilstein J. Org. Chem., 2013, 9, 2925–2933, DOI:10.3762/bjoc.9.329
.
- A. S. Dewi, K. L. Cheney, H. H. Urquhart, J. T. Blanchfield and M. J. Garson, Mar. Drugs, 2016, 14, 198, DOI:10.3390/md14110198
.
- S. J. Fahey and M. J. Garson, J. Chem. Ecol., 2002, 28, 1773–1785 CrossRef CAS PubMed
.
- G. Guella, I. Mancini, A. Guerriero and F. Pietra, Helv. Chim. Acta, 1985, 68, 1276–1282, DOI:10.1002/hlca.19850680523
.
- A. Fontana, C. Muniain and G. Cimino, J. Nat. Prod., 1998, 61, 1027–1029, DOI:10.1021/NP980073K
.
- F. A. Fuhrman, G. J. Fuhrman, Y. H. Kim, L. A. Pavelka and H. S. Mosher, Science, 1980, 207, 193–195, DOI:10.1126/science.7350655
.
- S. J. Fahey and A. R. Carroll, J. Chem. Ecol., 2007, 33, 1226–1234, DOI:10.1007/s10886-007-9288-z
.
- M. Gavagnin, N. Ungur, F. Castelluccio, C. Muniain and G. Cimino, J. Nat. Prod., 1999, 62, 269–274, DOI:10.1021/NP980344R
.
- E. Zubia, M. Gavagnin, A. Crispino, E. Martinez, J. Ortea and G. Cimino, Experientia, 1993, 49, 268–271 CrossRef CAS
.
- N. Ungur, M. Gavagnin, E. Mollo and G. Cimino, Tetrahedron: Asymmetry, 1999, 10, 1635–1636, DOI:10.1016/S0957-4166(99)00160-3
.
- J. Ren, P. Zhao, X. Xiao, T. Chen and B.-B. Zeng, Synthesis, 2016, 48, 4161–4166, DOI:10.1055/s-0035-1561493
.
- Y. H. Kim, R. J. Nachman, L. Pavelka, H. S. Mosher, F. A. Fuhrman and G. J. Fuhrman, J. Nat. Prod., 1981, 44, 206–214, DOI:10.1021/np50014a011
.
- R. J. Quinn, R. P. Gregson, A. F. Cook and R. T. Bartlett, Tetrahedron Lett., 1980, 21, 567–568, DOI: org/10.1016/S0040-4039(01)85558-1
.
- K. Gustafson and R. J. Andersen, Tetrahedron, 1985, 41, 1101–1108, DOI:10.1016/S0040-4020(01)96478-6
.
- M. D. Unson, C. B. Rose, D. J. Faulkner, L. S. Brinen, J. R. Steiner and J. Clardy, J. Org. Chem., 1993, 58, 6336–6343, DOI:10.1021/jo00075a029
.
- E. Cherbuliez and K. Bernhard, Helv. Chim. Acta, 1932, 15, 978–980, DOI:10.1002/hlca.193201501106
.
- F. A. Fuhrman, G. J. Fuhrman, R. J. Nachman and H. S. Mosher, Science, 1981, 212, 557–558, DOI:10.1126/science.7209552
.
- R. P. Walker and D. J. Faulkner, J. Org. Chem., 1981, 46, 1475–1478, DOI:10.1021/jo00320a046
.
- R. P. de Jesus and D. J. Faulkner, J. Nat. Prod., 2003, 66, 671–674, DOI:10.1021/np020542p
.
- D. E. Williams, S. W. Ayer and R. J. Anderson, Can. J. Chem., 1986, 64, 1527–1529, DOI:10.1139/v86-250
.
- J. Kubanek and R. J. Anderson, J. Nat. Prod., 1999, 62, 777–779, DOI:10.1021/NP9804839
.
- K. V. Reddy, R. Mohanrajd, K. N. Murthy, C. Ramesh and P. Karthick, J. Coastal Life Med., 2015, 3, 582–584, DOI:10.12980/JCLM.3.201514J65
.
-
N. K. Gulavita, P. J. Scheuer and E. D. de Silva, Bioact. Compd. Mar. Org. Indo-U. S. Symp., ed. M. F. Thompson, R. Sarojini and R. Nagabhushanam, 1991, pp. 229–233 Search PubMed
.
- W.-F. He, Y. Li, M.-T. Feng, M. Gavagnin, E. Mollo, S.-H. Mao and Y.-W. Guo, Fitoterapia, 2014, 96, 109–114, DOI:10.1016/j.fitote.2014.04.011
.
- R.-Y. Huang, W.-T. Chen, T. Kurtan, A. Mandi, J. Ding, J. Li, X.-W. Li and Y.-W. Guo, Future Med. Chem., 2016, 8, 17–27, DOI:10.4155/fmc.15.169
.
- A. Fontana, P. Cavaliere, S. Wahidulla, C. G. Naik and G. Cimino, Tetrahedron, 2000, 56, 7305–7308, DOI:10.1016/S0040-4020(00)00629-3
.
- K. Charupant, K. Suwanborirux, S. Amnuoypol, E. Saito, A. Kubo and N. Saito, Chem. Pharm. Bull., 2007, 55, 81–86, DOI:10.1248/cpb.55.81
.
- A. Kubo, Y. Kitahara and S. Nakahara, Chem. Pharm. Bull., 1989, 37, 1384–1386, DOI:10.1248/cpb.37.1384
.
-
C. Cuevas, M. Perez, A. Francesch, C. Fernandez, J. L. Chicharro, P. Gallego, M. Zarzuelo, F. De la Calle and I. Manzanares, PCT Int. Appl., WO 2000069862 A2 20001123, 2000
.
- B. J. Petek and R. L. Jones, Molecules, 2014, 19, 12328–12335, DOI:10.3390/molecules190812328
.
- D. Castiello, G. Cimino, S. De Rosa, S. De Stefano, G. Izzo and G. Sodano, Colloq. Int. C. N. R. S., 1979, 291, 413–416 CAS
.
- D. Castiello, G. Cimino, S. De Rosa, S. De Stefano and G. Sodano, Tetrahedron Lett., 1980, 21, 5047–5050, DOI:10.1016/S0040-4039(00)71129-4
.
- M. L. Ciavatta, G. Nuzzo, K. Takada, V. Mathieu, R. Kiss, G. Villani and M. Gavagnin, J. Nat. Prod., 2014, 77, 1678–1684, DOI:10.1021/np500298h
.
- P. J. Krug, K. G. Boyd and D. J. Faulkner, Tetrahedron, 1995, 51, 11063–11074, DOI:10.1016/0040-4020(95)00679-3
.
- E. I. Graziani, R. J. Andersen, P. J. Krug and D. J. Faulkner, Tetrahedron, 1996, 52, 6869–6878, DOI:10.1016/0040-4020(96)00327-4
.
- N. Ungur, M. Gavagnin, A. Fontana and G. Cimino, Tetrahedron: Asymmetry, 1999, 10, 1263–1273, DOI:10.1016/S0957-4166(99)00119-6
.
- R. J. Andersen and F. W. Sum, Tetrahedron Lett., 1980, 21, 797–800, DOI:10.1016/S0040-4039(00)71508-5
.
- K. Gustafson, R. J. Andersen, M. H. M. Chen, J. Clardy and J. E. Hochlowski, Tetrahedron Lett., 1984, 25, 11–14, DOI:10.1016/S0040-4039(01)91135-9
.
- A. Soriente, G. Sodano, K. C. Reed and C. Todd, Nat. Prod. Lett., 1993, 3, 31–35, DOI:10.1080/10575639308043834
.
- G. Cimino, A. Crispino, M. Gavagnin, E. Trivellone, E. Zubia, E. Martinez and J. Ortea, J. Nat. Prod., 1993, 56, 1642–1646, DOI:10.1021/np50099a033
.
- N. Ungur, M. Gavagnin and G. Cimino, Tetrahedron Lett., 1996, 37, 3549–3552, DOI:10.1016/0040-4039(96)00609-0
.
- R. M. Young and B. J. Baker, Planta Med., 2016, 81, S1–S381 Search PubMed
.
- A. Cutignano, W. Zhang, C. Avila, G. Cimino and A. Fontana, Eur. J. Org. Chem., 2011, 5383–5389, S5383, DOI:10.1002/ejoc.201100552
.
- M. T. Davies-Coleman and D. J. Faulkner, Tetrahedron, 1991, 47, 9743–9750, DOI:10.1016/S0040-4020(01)80714-6
.
- M. Gavagnin, A. De Napoli, G. Cimino, K. Iken, C. Avila and F. J. Garcia, Tetrahedron: Asymmetry, 1999, 10, 2647–2650, DOI:10.1016/S0957-4166(99)00273-6
.
- J. A. Maschek, E. Mevers, T. Diyabalanage, L. Chen, Y. Ren, J. B. McClintock, C. D. Amsler, J. Wu and B. J. Baker, Tetrahedron, 2012, 68, 9095–9104, DOI:10.1016/j.tet.2012.08.045
.
- M. Gavagnin, E. Trivellone, F. Castelluccio, G. Cimino and R. Cattaneo-Vietti, Tetrahedron Lett., 1995, 36, 7319–7322, DOI:10.1016/0040-4039(95)01476-X
.
- M. Gavagnin, M. Carbone, E. Mollo and G. Cimino, Tetrahedron, 2003, 59, 5579–5583, DOI:10.1016/S0040-4020(03)00775-0
.
- T. Diyabalanage, K. B. Iken, J. B. McClintock, C. D. Amsler and B. J. Baker, J. Nat. Prod., 2010, 73, 416–421, DOI:10.1021/np900617m
.
- M. Gavagnin, A. De Napoli, F. Castelluccio and G. Cimino, Tetrahedron Lett., 1999, 40, 8471–8475, DOI:10.1016/S0040-4039(99)01777-3
.
- L. Núñez-Pons and C. Avila, Nat. Prod. Rep., 2015, 32, 1114–1130, 10.1039/C4NP00150H
.
- M. Gavagnin, M. Carbone, E. Mollo and G. Cimino, Tetrahedron Lett., 2003, 44, 1495–1498, DOI:10.1016/S0040-4039(02)02849-6
.
- K. Iken, C. Avila, A. Fontana and M. Gavagnin, Mar. Biol., 2002, 141, 101–109, DOI:10.1007/s00227-002-0816-7
.
- M. Gavagnin, A. Fontana, M. L. Ciavatta and G. Cimino, Ital. J. Zool., 2000, 67(Suppl.), 101–109, DOI:10.1080/11250000009356363
.
- G. Cimino, M. Gavagnin, G. Sodano, R. Puliti, C. A. Mattia and L. Mazzarella, Tetrahedron, 1988, 44, 2301–2310, DOI:10.1016/S0040-4020(01)81739-7
.
- M. Gavagnin, N. Ungur, F. Castelluccio and G. Cimino, Tetrahedron, 1997, 53, 1491–1504, DOI:10.1016/S0040-4020(96)01083-6
.
- L. De Petrocellis, V. Di Marzo, B. Arca, M. Gavagnin, R. Minei and G. Cimino, Comp. Biochem. Physiol., C: Comp. Pharmacol., 1991, 100C, 603–607 CrossRef CAS
.
- C. Avila, M. Ballesteros, G. Cimino, A. Crispino, M. Gavagnin and G. Sodano, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1990, 97B, 363–368, DOI:10.1016/0305-0491(90)90294-4
.
- G. Cimino, A. Crispino, S. De Stefano, M. Gavagnin and G. Sodano, Experientia, 1986, 42, 1301–1302 CrossRef CAS
.
- M. Porcelli, G. Cacciapuoti, A. Oliva and V. Zappia, J. Chromatogr., 1988, 440, 151–155, DOI:10.1016/S0021-9673(00)94519-9
.
- A. C. Granato, R. G. S. Berlinck, A. Magalhaes, A. B. Schefer, A. G. Ferreira, B. De Sanctis, J. C. De Freitas, E. Hajdu and A. E. Migotto, Quim. Nova, 2000, 23, 594–599, DOI:10.1590/S0100-40422000000500005
.
- M. Porcelli, G. Cacciapuoti, G. Cimino, M. Gavagnin, G. Sodano and V. Zappia, Biochem. J., 1989, 263, 635–640, DOI:10.1042/bj2630635
.
- S. W. Ayer, J. Hellou, M. Tischler and R. J. Andersen, Tetrahedron Lett., 1984, 25, 141–144, DOI:10.1016/S0040-4039(00)99824-1
.
- S. W. Ayer, R. J. Andersen, C. H. He and J. Clardy, J. Org. Chem., 1984, 49, 2653–2654, DOI:10.1021/jo00188a036
.
- E. I. Graziani and R. J. Andersen, J. Am. Chem. Soc., 1996, 118, 4701–4702, DOI:10.1021/JA9539305
.
- E. I. Graziani, T. M. Allen and R. J. Andersen, Tetrahedron Lett., 1995, 36, 1763–1766, DOI:10.1016/0040-4039(95)00118-V
.
- H. H. Strain, Biol. Bull., 1949, 97, 206–209, DOI:10.2307/1538298
.
- J. W. McBeth, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1972, 41, 69–77, DOI:10.1016/0305-0491(72)90008-9
.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=851581, accessed 23 May 2017.
- G. Cimino, S. De Rosa, S. De Stefano, G. Sodano and G. Villani, Science, 1983, 219, 1237–1238, DOI:10.1126/science.219.4589.1237
.
- G. Cimino, S. De Rosa, S. De Stefano and G. Sodano, Experientia, 1985, 41, 1335–1336 CrossRef CAS
.
- G. Cimino, G. Sodano and A. Spinelli, J. Nat. Prod., 1988, 51, 1010–1011, DOI:10.1021/np50059a039
.
- G. Cimino, S. De Rosa, S. De Stefano and G. Sodano, Tetrahedron Lett., 1981, 22, 1271–1272, DOI:10.1016/S0040-4039(01)90293-X
.
- G. Cimino, S. De Rosa, S. De Stefano, R. Morrone and G. Sodano, Tetrahedron, 1985, 41, 1093–1100, DOI:10.1016/S0040-4020(01)96477-4
.
- A. Fontana, M. L. Ciavatta, T. Miyamoto, A. Spinella and G. Cimino, Tetrahedron, 1999, 55, 5937–5946, DOI:10.1016/S0040-4020(99)00256-2
.
- Y. Sakio, Y. J. Hirano, M. Hayashi, K. Komiyama and M. Ishibashi, J. Nat. Prod., 2001, 64, 726–731, DOI:10.1021/np000639g
.
- R. K. Okuda, P. J. Scheuer, J. E. Hochlowski, R. P. Walker and D. J. Faulkner, J. Org. Chem., 1983, 48, 1866–1869, DOI:10.1021/jo00159a014
.
- M. Gavagnin, E. Mollo, G. Calado, S. Fahey, M. Ghiselin, J. Ortea and G. Cimino, Chemoecology, 2001, 11, 131–136, DOI:10.1007/PL00001843
.
- C. Avila, G. Cimino, A. Crispino and A. Spinella, Experientia, 1991, 47, 306–310, DOI:10.1007/BF01958169
.
- A. Fontana, G. Villani and G. Cimino, Tetrahedron Lett., 2000, 41, 2429–2433, DOI:10.1016/S0040-4039(00)00135-0
.
- N. V. Zhukova and M. G. Eliseikina, Mar. Biol., 2012, 159, 1783–1794, DOI:10.1007/s00227-012-1969-7
.
- Y. Asakawa and T. Aratani, Bull. Soc. Chim. Fr., 1976, 9–10, 1469–1470 Search PubMed
.
- H. Gaspar, M. Gavagnin, G. Calado, F. Castelluccio, E. Mollo and G. Cimino, Tetrahedron, 2005, 61, 11032–11037, DOI:10.1016/j.tet.2005.08.096
.
- A. Fontana, A. Tramice, A. Cutignano, G. d'Ippolito, M. Gavagnin and G. Cimino, J. Org. Chem., 2003, 68, 2405–2409, DOI:10.1021/jo026131v
.
- M. Gavagnin, E. Mollo, F. Castelluccio, M. T. Ghiselin, G. Calado and G. Cimino, Tetrahedron, 2001, 57, 8913–8916, DOI:10.1016/S0040-4020(01)00876-6
.
- A. Spinella, L. A. Alvarez, C. Avila and G. Cimino, Tetrahedron Lett., 1994, 35, 8665–8668, DOI:10.1016/S0040-4039(00)78466-8
.
- H. Gaspar, A. Cutignano, T. Ferreira, G. Calado, G. Cimino and A. Fontana, J. Nat. Prod., 2008, 71, 2053–2056, DOI:10.1021/np8004387
.
- B. J. Burreson, P. J. Scheuer, J. Finer and J. Clardy, J. Am. Chem. Soc., 1975, 97, 4763–4764, DOI:10.1021/ja00849a053
.
- M. R. Hagadone, B. J. Burreson, P. J. Scheuer, J. S. Finer and J. Clardy, Helv. Chim. Acta, 1979, 62, 2484–2894, DOI:10.1002/hlca.19790620742
.
- N. Fusetani, H. J. Wolstenholme and S. Matsunaga, Tetrahedron Lett., 1990, 31, 5623–5624, DOI:10.1016/S0040-4039(00)97915-2
.
- R. A. Edrada, V. Wray and P. Proksch, J. Nat. Prod., 2003, 66, 1512–1514, DOI:10.1021/np030237j
.
- N. K. Gulavita, E. D. De Silva, M. R. Hagadone, P. Karuso, P. J. Scheuer, G. D. Van Duyne and J. Clardy, J. Org. Chem., 1986, 51, 5136–5139, DOI:10.1021/jo00376a015
.
- K. E. Kassühlke, B. C. M. Potts and D. J. Faulkner, J. Org. Chem., 1991, 56, 3747–3750, DOI:10.1021/jo00011a065
.
- T. Okino, E. Yoshimura, H. Hirota and N. Fusetani, Tetrahedron, 1996, 52, 9447–9454, DOI:10.1016/0040-4020(96)00481-4
.
- N. Fusetani, H. J. Wolstenholme, K. Shinoda, N. Asai and S. Matsunaga, Tetrahedron Lett., 1992, 33, 6823–6826 CrossRef CAS
.
- A. M. White, G. K. Pierens, T. Skinner-Adams, K. T. Andrews, P. V. Bernhardt, E. H. Krenske, E. Mollo and M. J. Garson, J. Nat. Prod., 2015, 78, 1422–1427, DOI:10.1021/acs.jnatprod.5b00354
.
- S. Jaisamut, S. Prabpai, C. Tancharoen, S. Yuenyongsawad, S. Hannongbua, P. Kongsaeree and A. Plubrukarn, J. Nat. Prod., 2013, 76, 2158–2161, DOI:10.1021/np4007074
.
- N. Fusetani, H. J. Wolstenholme and S. Matsunaga, Tetrahedron Lett., 1991, 32, 7291–7294, DOI:10.1016/0040-4039(91)80501-V
.
- H. Hirota, T. Okino, E. Yoshimura and N. Fusetani, Tetrahedron, 1998, 54, 13971–13980, DOI:10.1016/S0040–4020(98)00867-9
.
- E. J. Dumdei, A. E. Flowers, M. J. Garson and C. J. Moore, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 1997, 118A, 1385–1392, DOI:10.1016/S0300-9629(97)00051-0
.
- A. D. Wright, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2003, 134A, 307–313, DOI:10.1016/S1095-6433(02)00265-9
.
- E. Manzo, M. L. Ciavatta, M. Gavagnin, E. Mollo, Y.-W. Guo and G. Cimino, J. Nat. Prod., 2004, 67, 1701–1704, DOI:10.1021/np0400961
.
- E. G. Lyakhova, S. A. Kolesnikova, A. I. Kalinovskii and V. A. Stonik, Chem. Nat. Compd., 2010, 46, 534–538, DOI:10.1007/s10600-010-9670-x
.
- T. Jomoria, T. Shibutani, P. Ahmadi, T. Suzuka and J. Tanaka, Nat. Prod. Commun., 2015, 10, 1913–1914 Search PubMed
.
- N. Fusetani, H. Hiroto, T. Okino, Y. Tomono and E. Yoshimura, J. Nat. Toxins, 1996, 5, 249–259 CAS
.
- K. Nishikawa, H. Nakahara, Y. Shirokura, Y. Nogata, E. Yoshimura, T. Umezawa, T. Okino and F. Matsuda, J. Org. Chem., 2011, 76, 6558–6573, DOI:10.1021/jo2008109
.
- N. V. Zhukova, Lipids, 2007, 42, 1169–1175, DOI:10.1007/s11745-007-3123-8
.
- A. Alqudahi, S. B. Saad, D. Susanti and N. F. Hadry, Malays. Appl. Biol., 2016, 45, 23–28 Search PubMed
.
- J. Tanaka and T. Higa, J. Nat. Prod., 1999, 62, 1339–1340, DOI:10.1021/np990186j
.
- S. J. Wratten and D. J. Faulkner, Tetrahedron Lett., 1978, 16, 1395–1398, DOI:10.1016/S0040-4039(01)94554-X
.
- S. J. Wratten, D. J. Faulkner, D. Van Engen and J. Clardy, Tetrahedron Lett., 1978, 16, 1391–1394 CrossRef
.
- S. Carmely, M. Ilan and Y. Kashman, Tetrahedron, 1989, 45, 2193–2200 CrossRef CAS
.
- K. A. Alvi, P. Crews and D. G. Loughhead, J. Nat. Prod., 1991, 54, 1509–1515, DOI:10.1021/np50078a004
.
- K. A. Alvi, B. M. Peters, L. M. Hunter and P. Crews, Tetrahedron, 1993, 49, 329–336, DOI:10.1016/S0040-4020(01)80302-1
.
- A. R. Carroll, B. F. Bowden and J. C. Coll, Aust. J. Chem., 1993, 46, 1229–1234, DOI:10.1071/CH9931229
.
- J. A. Roesener and P. J. Scheuer, J. Am. Chem. Soc., 1986, 108, 846–847, DOI:10.1021/ja00264a052
.
- S. M. Parrish, W. Yoshida, B. Yang and P. G. Williams, J. Nat. Prod., 2017, 80, 726–730, DOI:10.1021/acs.jnatprod.6b00896
.
- S. Matsunaga, N. Fusetani, K. Hashimoto, K. Koseki and M. Noma, Tennen Yuki Kagobutsu Toronkai Koen Yoshishu, 1985, 27, 375–382 Search PubMed
.
- S. Matsunaga, N. Fusetani, K. Hashimoto, K. Koseki and M. Noma, J. Am. Chem. Soc., 1986, 108, 847–849, DOI:10.1021/ja00264a053
.
- J. R. Pawlik, M. R. Kernan, T. F. Molinski, M. K. Harper and D. J. Faulkner, J. Exp. Mar. Biol. Ecol., 1988, 119, 99–109, DOI:10.1016/0022-0981(88)90225-0
.
- D. S. Dalisay, E. W. Rogers, A. S. Edison and T. F. Molinski, J. Nat. Prod., 2009, 72, 732–738, DOI:10.1021/np8007649
.
- S. Matsunaga, N. Fusetani, K. Hashimoto, K. Koseki, M. Noma, H. Noguchi and U. Sankawa, J. Org. Chem., 1989, 54, 1360–1363, DOI:10.1021/jo00267a024
.
- M. R. Kernan, T. F. Molinski and D. J. Faulkner, J. Org.
Chem., 1988, 53, 5014–5020, DOI:10.1021/jo00256a021
.
- E. Vincent, J. Saxton, C. Baker-Glenn, I. Moal, J. D. Hirst, G. Pattenden and P. E. Shaw, Cell. Mol. Life Sci., 2007, 64, 487–497, DOI:10.1007/s00018-007-6427-1
.
- Y. Guo, M. Gavagnin, E. Mollo, E. Trivellone and G. Cimino, Tetrahedron Lett., 1998, 39, 2635–2638, DOI:10.1016/S0040-4039(98)00225-1
.
- Y. Tanaka, S. Yamada and M. Sameshima, Nippon Suisan Gakkaishi, 1992, 58, 1549, DOI:10.2331/suisan.58.1549
.
- R. E. Coman and B. C. L. Weedon, J. Chem. Soc., Perkin Trans. 1, 1975, 2529–2532 RSC
.
- W. Zhang, M. Gavagnin, Y.-W. Guo, E. Mollo, M. T. Ghiselin and G. Cimino, Tetrahedron, 2007, 63, 4725–4729, DOI:10.1016/j.tet.2007.03.082
.
- D. S. Nielsen, H. N. Hoang, R.-J. Lohman, F. Diness and D. P. Fairlie, Org. Lett., 2012, 14, 5720–5723, DOI:10.1021/ol3027347
.
- E. K. Singh, D. M. Ramsey and S. R. McAlpine, Org. Lett., 2012, 14, 1198–1201, DOI:10.1021/ol203290n
.
- H. Wahyudi, W. Tantisantisom, X. Liu, D. M. Ramsey, E. K. Singh and S. R. McAlpine, J. Org. Chem., 2012, 77, 10596–10616, DOI:10.1021/jo3017499
.
- B. Carté and D. J. Faulkner, J. Org. Chem., 1983, 48, 2314–2318 CrossRef
.
- B. Carté and D. J. Faulkner, J. Chem. Ecol., 1986, 12, 795–804 CrossRef PubMed
.
- A. C. Granato, J. H. H. L. de Oliveira, M. H. R. Seleghim, R. G. S. Berlinck, M. L. Macedo, A. G. Ferreira, R. M. da Rocha, E. Hajdu, S. Peixinho, C. O. Pessoa, M. O. Moraes and B. C. Cavalcanti, Quim. Nova, 2005, 28, 192–198, DOI:10.1590/S0100-40422005000200005
.
- A. J. Blackman and C. Li, Aust. J. Chem., 1994, 47, 1625–1629 CrossRef CAS
.
- A. Franks, P. Haywood, C. Holmstrom, S. Egan, S. Kjelleberg and N. Kumar, Molecules, 2005, 10, 1286–1291 CrossRef CAS PubMed
.
- R. Kazlauskas, J. F. Marwood, P. T. Murphy and R. J. Wells, Aust. J. Chem., 1982, 35, 215–217, DOI:10.1071/CH9820215
.
- S. Matsunaga, N. Fusetani and K. Hashimoto, Experientia, 1986, 42, 84 CrossRef CAS
.
- V. J. Paul, N. Lindquist and W. Fenical, Mar. Ecol.: Prog. Ser., 1990, 59, 109–118 CrossRef CAS
.
- H. H. Wasserman, D. J. Friedland and D. A. Morrison, Tetrahedron Lett., 1968, 641–644, DOI:10.1016/S0040-4039(00)75602-4
.
- M. Carbone, C. Irace, F. Costagilola, F. Castelluccio, G. Villani, G. Calado, V. Padula, G. Cimino, J. L. Cervera, R. Santamaria and M. Gavagnin, Bioorg. Med. Chem. Lett., 2010, 20, 2668–2670, DOI:10.1016/j.bmcl.2010.02.020
.
- N. Lindquist and W. Fenical, Experientia, 1991, 47, 504–506 CrossRef CAS
.
- B. C. Cavalcnti, H. V. N. Júnior, M. H. R. Seleghim, R. G. S. Berlinck, G. M. A. Cunha, M. O. Moraes and C. Pessoa, Chem.-Biol. Interact., 2008, 174, 155–162, DOI:10.1016/j.cbi.2008.05.029
.
- K. Gustafson and R. J. Anderson, J. Org. Chem., 1982, 47, 2167–2169, DOI:10.1021/jo00132a036
.
- E. I. Graziani and R. J. Anderson, J. Nat. Prod., 1998, 61, 285–286, DOI:10.1021/NP970397T
.
- E. I. Graziani and R. J. Andersen, Chem. Commun., 1996, 2377–2378, 10.1039/cc9960002377
.
- J. Kubanek and R. J. Andersen, Tetrahedron Lett., 1997, 38, 6327–6330, DOI:10.1016/S0040-4039(97)01455-X
.
- E. Piers, J. M. Chong, K. Gustafson and R. J. Andersen, Can. J. Chem., 1984, 62, 1–5, DOI:10.1139/v84-001
.
- J. W. McBeth, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1972, 41, 55–68, DOI:10.1016/0305-0491(72)90007-7
.
- T. Matsuno, M. Ookubo and T. Komori, J. Nat. Prod., 1985, 48, 606–613, DOI:10.1021/np50040a015
.
- B. Czeczuga, Hydrobiologia, 1976, 51, 71–75, DOI:10.1007/BF00007987
.
- S. Affeld, S. Kehraus, H. Wägele and G. M. König, J. Nat. Prod., 2009, 72, 298–300, DOI:10.1021/np800583e
.
- N. R. Howe and L. G. Harris, J. Chem. Ecol., 1978, 4, 551–561, DOI:10.1007/BF00988919
.
- M. C. Leal, C. Nunes, D. Alexandre, T. L. Silva, A. Reis, M. T. Dinis and R. Calado, Mar. Biol., 2012, 159, 1745–1751, DOI:10.1007/s00227-012-1962-1
.
- A. Garese, S. Garcia-Matucheski, F. H. Acuna and C. Muniain, Zool. Stud., 2012, 51, 905–912 CAS
.
- M. L. Ciavatta, E. Trivellone, G. Villani and G. Cimino, Gazz. Chim. Ital., 1996, 126, 707–710 CAS
.
- S. J. Rochfort and R. J. Capon, J. Nat. Prod., 1994, 57, 849–851, DOI:10.1021/np50108a029
.
- R. Martin, S. Hild, P. Walther, K. Ploss, W. Boland and K.-H. Tomaschko, Biol. Bull., 2007, 213, 307–315, DOI:10.2307/25066648
.
- I. E. Kasheverov, I. V. Shelukhina, D. S. Kudryavtsev, T. N. Makarieva, E. N. Spirova, A. G. Guzii, V. A. Stonik and V. I. Tsetlin, Mar. Drugs, 2015, 13, 1255–1266, DOI:10.3390/md13031255
.
- W. D. Raverty, R. H. Thomson and T. J. King, J. Chem. Soc., Perkin Trans. 1, 1977, 10, 1204–1211 RSC
.
- K. Kondo and J. Nishi, J. Nat. Prod., 1994, 57, 1008–1011, DOI:10.1021/np50109a023
.
- A. Aiello, F. Borrelli and R. Capasso, Bioorg. Med. Chem. Lett., 2003, 13, 4481–4483, DOI:10.1016/j.bmcl.2003.08.081
.
- C. Megina, T. Gosliner and J. L. Cervera, Cah. Biol. Mar., 2007, 48, 1–7 Search PubMed
.
- M. Carbone, Y. Li, C. Irace, E. Mollo, F. Castelluccio, A. Di Pascale, G. Cimino, R. Santamaria, Y.-W. Guo and M. Gavagnin, Org. Lett., 2011, 13, 2516–2519, DOI:10.1021/ol200234r
.
- J. T. Brogan, S. L. Stoops and C. W. Lindsley, ACS Chem. Neurosci., 2012, 3, 658–664, DOI:10.1021/cn300064r
.
- H.-Y. Lin and B. B. Snider, J. Org. Chem., 2012, 77, 4832–4836, DOI:10.1021/jo300449n
.
- E. Manzo, D. Pagano, M. Carbone, M. L. Ciavatta and M. Gavagnin, ARKIVOC, 2012, 220–228, DOI:10.3998/ark.5550190.0013.919
.
- J. C. Buchanan, B. P. Petersen and S. Chamberland, Tetrahedron Lett., 2013, 54, 6002–6004, DOI:10.1016/j.tetlet.2013.08.063
.
- C. V. Maftei, E. Fodor, P. G. Jones, M. H. Franz, G. Kelter, H. Fiebig and I. Neda, Beilstein J. Org. Chem., 2013, 9, 2202–2215, DOI:10.3762/bjoc.9.259
.
- C.-S. Jiang, Y. Fu, L. Zhang, J.-X. Gong, Z.-Z. Wang, W. Xiao, H.-Y. Zhang and Y.-W. Guo, Bioorg. Med. Chem. Lett., 2015, 25, 216–220, DOI:10.1016/j.bmcl.2014.11.068
.
- L. Zhang, C.-S. Jiang, L.-X. Gao, J.-X. Gong, Z.-H. Wang, J.-Y. Li, J. Li, X.-W. Li and Y.-W. Guo, Bioorg. Med. Chem. Lett., 2016, 26, 778–781, DOI:10.1016/j.bmcl.2015.12.097
.
- J. C. Coll, B. F. Bowden, D. M. Tapiolas, R. H. Willis, P. Djura, M. Streamer and L. Trott, Tetrahedron, 1985, 41, 1085–1092, DOI:10.1016/S0040-4020(01)96476-2
.
- M. Slattery, C. Avila, J. Starmer and V. J. Paul, J. Exp. Mar. Biol. Ecol., 1998, 226, 33–49, DOI:10.1016/S0022–0981(97)00240-2
.
- R. A. Edrada, V. Wray, L. Witte, L. van Ofwegen and P. Proksch, Z. Naturforsch., C: J. Biosci., 2000, 55, 82–86, DOI:10.1515/znc-2000-1-216
.
- A. Bogdanov, C. Hertzer, S. Kehraus, S. Nietzer, S. Rohde, P. J. Schupp, H. Waegele and G. M. König, J. Nat. Prod., 2016, 79, 611–615, DOI:10.1021/acs.jnatprod.5b00860
.
- A. S. R. Anjaneyulu, P. M. Gowri, M. J. R. V. Venugopal, P. Sarada, M. V. R. K. Murthy, G. V. Rao, P. S. N. Murthy, C. V. Rao and G. Kumar, J. Indian Chem. Soc., 1999, 76, 651–659 CAS
.
- M. Sugano, T. Shindo, A. Sato, Y. Iijima, T. Oshima, H. Kuwano and T. Hata, J. Org. Chem., 1990, 55, 5803–5804, DOI:10.1021/jo00310a001
.
- S.-C. Mao, M. Gavagnin, E. Mollo and Y.-W. Guo, Biochem. Syst. Ecol., 2011, 39, 408–411, DOI:10.1016/j.bse.2011.05.018
.
- D. Chen, W. Cheng, D. Liu, L. van Ofwegen, P. Proksch and W. Lin, Tetrahedron Lett., 2014, 55, 3077–3082, DOI:10.1016/j.tetlet.2014.03.132
.
- A. Guerriero, M. D'Ambrosio and F. Pietra, Helv. Chim. Acta, 1987, 70, 984–991, DOI:10.1002/hlca.19870700408
.
- A. Guerriero, M. D'Ambrosio and F. Pietra, Helv. Chim. Acta, 1988, 71, 472–485, DOI:10.1002/hlca.19880710221
.
- A. Guerriero, M. D'Ambrosio and F. Pietra, Helv. Chim. Acta, 1990, 73, 277–283, DOI:10.1002/hlca.19900730206
.
- W. Zhang, M. Gavagnin, Y.-W. Guo, E. Mollo and G. Cimino, Chin. J. Org. Chem., 2006, 26, 1667–1672 CAS
.
- R. K. Okuda, D. Klein, R. B. Kinnel, M. Li and P. J. Scheuer, Pure Appl. Chem., 1982, 54, 1907–1914, DOI:10.1351/pac198254101907
.
- G. Cimino, S. De Rosa, S. De Stefano and G. Sodano, Tetrahedron Lett., 1980, 21, 3303–3304, DOI:10.1016/S0040-4039(00)78673-4
.
- R. Kuhn, J. Stene and N. A. Sorensen, Ber. Dtsch. Chem. Ges. B, 1939, 72B, 1688–1701 CrossRef CAS
.
- T. Barsby, R. G. Linington and R. J. Anderson, Chemoecology, 2002, 12, 199–202, DOI:10.1007/PL00012669
.
- G. Cimino, A. Crispino, V. Di Marzo, G. Sodano, A. Spinella and G. Villani, Experientia, 1991, 47, 56–60 CrossRef CAS PubMed
.
- S. W. Ayer and R. J. Andersen, Experientia, 1983, 39, 255–256, DOI:10.1007/BF01955289
.
- D. Barnard and L. Bateman, J. Chem. Soc., 1950, 932–936, 10.1039/JR9500000932
.
- J. L. Sevigny, L. E. Kirouac, W. K. Thomas, J. S. Ramsdell, K. E. Lawlor, O. Sharifi, S. Grewal, C. Baysdorfer, K. Curr, A. A. Naimie, K. Okamoto, J. A. Murray and J. M. Newcomb, PLoS One, 2015, 10, e0127519, DOI:10.1371/journal.pone.0127519
.
- J. L. Sevigny, L. E. Kirouac, W. K. Thomas, J. S. Ramsdell, K. E. Lawlor, O. Sharifi, S. Grewal, C. Baysdorfer, K. Curr, A. A. Naimie, K. Okamoto, J. A. Murray and J. M. Newcomb, PLoS One, 2015, 10, e0132861, DOI:10.1371/journal.pone.0132861
.
- G. Cimino, A. Spinella and G. Sodano, Tetrahedron Lett., 1989, 30, 3589–3592, DOI:10.1016/S0040-4039(00)99449-8
.
- G. Cimino, A. Crispino, V. Di Marzo, A. Spinella and G. Sodano, J. Org. Chem., 1991, 56, 2907–2911, DOI:10.1021/jo00008a056
.
- K. Tsuboi, Y. Sugimoto and A. Ichikawa, Prostaglandins Other Lipid Mediators, 2002, 68–69, 535–556, DOI:10.1016/S0090-6980(02)00054-0
.
- M. L. Ciavatta, E. Manzo, E. Mollo, C. A. Mattia, C. Tedesco, C. Irace, Y.-W. Guo, X.-B. Li, G. Cimino and M. Gavagnin, J. Nat. Prod., 2011, 74, 1902–1907, DOI:10.1021/np200342k
.
- T. S. Sousa, G. Nuzzo, M. C. M. Torres, N. P. Lopes, A. Cutignano, P. C. Jimenez, E. A. Santos, B. A. Gomes, A. Sardo, O. D. L. Pessoa, L. V. Costa-Lotufo and A. Fontana, Rev. Bras. Farmacogn., 2015, 25, 600–604, DOI:10.1016/j.bjp.2015.08.010
.
- M. Ishibashi, M. Takahashi and J. Kobayashi, J. Org. Chem., 1995, 60, 6062–6066, DOI:10.1021/jo00124a015
.
- D. E. Williams and R. J. Anderson, Can. J. Chem., 1987, 65, 2244–2247, DOI:10.1139/v87-374
.
- D. E. Williams, R. J. Anderson, G. Van Duyne and J. Clardy, J. Org. Chem., 1987, 52, 332–335, DOI:10.1021/jo00379a002
.
- M. G. Missakian, B. J. Burreson and P. J. Scheuer, Tetrahedron, 1975, 31, 2513–2515, DOI:10.1016/0040-4020(75)80262-6
.
- S. J. Wratten, W. Fenical, D. J. Faulkner and J. C. Wekell, Tetrahedron Lett., 1977, 18, 1559–1562, DOI:10.1016/S0040-4039(01)93102-8
.
- G. Cronin, M. E. Hay, W. Fenical and N. Lindquist, Mar. Ecol.: Prog. Ser., 1995, 119, 177–189, DOI:10.3354/meps119177
.
- J. B. McClintock, B. J. Baker, M. Slattery, J. N. Heine, P. J. Bryan, W. Yoshida, M. T. Davies- Coleman and D. J. Faulkner, J. Chem. Ecol., 1994, 20, 3361–3372, DOI:10.1007/BF02033732
.
- A. Cutignano, J. Moles, C. Avila and A. Fontana, J. Nat. Prod., 2015, 78, 1761–1764, DOI:10.1021/acs.jnatprod.5b00378
.
- J. Moles, H. Wagele, A. Cutignano, A. Fontana and C. Avila, Mar. Biol., 2016, 163, 1–11, DOI:10.1007/s00227-016-2831-0
.
- J. Pika and D. J. Faulkner, Tetrahedron, 1994, 50, 3065–3070, DOI:10.1016/S0040-4020(01)81106-6
.
- K. L. McPhail, M. T. Davies-Coleman and J. Starmer, J. Nat. Prod., 2001, 64, 1183–1190, DOI:10.1021/np010085x
.
- C. E. Whibley, K. L. McPhail, R. A. Keyzers, M. F. Maritz, V. D. Leaner, M. J. Birrer, M. T. Davies-Coleman and D. T. Hendricks, Mol. Cancer Ther., 2007, 6, 2535–2543, DOI:10.1158/1535-7163.MCT-06-0760
.
- A. Putz, S. Kehraus, G. Diaz-Agras, H. Waegele and G. M. Koenig, Eur. J. Org. Chem., 2011, 3733–3737, S3733, DOI:10.1002/ejoc.201100347
.
- J. Moles, H. Waegele, M. Ballesteros, A. Pujals, G. Uhl and C. Avila, PLoS One, 2016, 11, e0157941, DOI:10.1371/journal.pone.0157941
.
- G. Sodano and A. Spinella, Tetrahedron Lett., 1986, 27, 2505–2508, DOI:10.1016/S0040-4039(00)84569-4
.
- J. Wang, M. R. Prinsep, D. P. Gordon, M. J. Page and B. R. Copp, J. Nat. Prod., 2015, 78, 530–533, DOI:10.1021/np500752y
.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=834002, accessed 12 June 2017.
-
G. Inglis, N. Gust, I. Fitridge, O. Floerl, C. Woods, B. Hayden and G. Fenwick, Port of Tauranga: Baseline survey for non-indigenous marine species, Ministry of Agriculture and Forestry Biosecurity, New Zealand: Wellington, NZ, 2005, p. 23 Search PubMed
.
- OPK, http://opistobranquis.info/en/guia/nudibranchia/dexiarchia/unassigned-cladobranchia/janolus-cristatus/#gsc.tab=0, accessed 29 July 2017.
-
L. Dean, M. Prinsep and R. Fairweather, Chemical Theft: The Sequestration of Natural Products by New Zealand Nudibranchs from their Bryozoan Prey, in NZIC-16, The New Zealand Institute of Chemistry Conference, Queenstown, New Zealand, August, 2016, pp. 21–24, http://www.nzic16.org/ Search PubMed
.
- D. Obermann, U. Bickmeyer and H. Wägele, Toxicon, 2012, 60, 1108–1116, DOI:10.1016/j.toxicon.2012.08.003
.
- H. Wägele and G. Johnsen, Org. Divers. Evol., 2001, 1, 193–210, DOI:10.1078/1439-6092-00016
.
- I. Burghardt, K. Stemmer and H. Wägele, Org. Divers. Evol., 2008, 8, 66–76, DOI:10.1016/j.ode.2007.01.001
.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=581809, accessed 19 May, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558499, accessed 11th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558620, accessed 5 October 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=565620, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597348, accessed 11th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597352, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558218, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597335, accessed 11th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597336, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597358, accessed 11th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558319, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597339, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597403, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597451, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=209601, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597452, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597406, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597365, accessed 14 June 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597367, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597341, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597380, accessed 11 May, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597343, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597374, accessed 14 June 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=559960, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=884441, accessed 28 July, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597315, accessed 11 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597398, accessed 11 May, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=139143, accessed 11 May, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597530, accessed 11 May, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597522, accessed 11 May, 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597514, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597520, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597543, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558680, accessed 21 September 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597820, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597823, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558649, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597827, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597533, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597534, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597536, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597522, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=597538, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558646, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=536910, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=594422, accessed 9th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=536827, accessed 9th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=872299, accessed 9th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=581816, accessed 9th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=581817, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=181228, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=411132, accessed 15 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=599485, accessed 23 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=536695, accessed 9th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=536574, accessed 9th May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=531260, accessed 30 May 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=730410, accessed 28 April 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=456805, accessed 28 April 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=457000, accessed 1 June 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=204424, accessed 28 April 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=890630, accessed 21 September 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=412595, accessed 1 June 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=558400, accessed 1 June 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=851422, accessed 28 July 2017.
- WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails%26id=572093, accessed 27 April 2017.
| This journal is © The Royal Society of Chemistry 2017 |