Open Access Article
Open Access ArticleInfluence of the [4Fe–4S] cluster coordinating cysteines on active site maturation and catalytic properties of C. reinhardtii [FeFe]-hydrogenase†
Leonie
Kertess
 a,
Agnieszka
Adamska-Venkatesh
b,
Patricia
Rodríguez-Maciá
b,
Olaf
Rüdiger
b,
Wolfgang
Lubitz
b and
Thomas
Happe
a,
Agnieszka
Adamska-Venkatesh
b,
Patricia
Rodríguez-Maciá
b,
Olaf
Rüdiger
b,
Wolfgang
Lubitz
b and
Thomas
Happe
 *a
*a
aAG Photobiotechnologie, Lehrstuhl für Biochemie der Pflanzen, Ruhr Universität Bochum, Universitätsstr. 150, 44801 Bochum, Germany. E-mail: thomas.happe@rub.de
bMax Planck Institute for Chemical Energy Conversion, Stiftstrasse 34-36, 45470 Mülheim an der Ruhr, Germany
First published on 9th October 2017
Abstract
[FeFe]-Hydrogenases catalyze the evolution and oxidation of hydrogen using a characteristic cofactor, termed the H-cluster. This comprises an all cysteine coordinated [4Fe–4S] cluster and a unique [2Fe] moiety, coupled together via a single cysteine. The coordination of the [4Fe–4S] cluster in HydA1 from Chlamydomonas reinhardtii was altered by single exchange of each cysteine (C115, C170, C362, and C366) with alanine, aspartate, or serine using site-directed mutagenesis. In contrast to cysteine 115, the other three cysteines were found to be dispensable for stable [4Fe–4S] cluster incorporation based on iron determination, UV/vis spectroscopy and electron paramagnetic resonance. However, the presence of a preformed [4Fe–4S] cluster alone does not guarantee stable incorporation of the [2Fe] cluster. Only variants C170D, C170S, C362D, and C362S showed characteristic signals for an inserted [2Fe] cluster in Fourier-transform infrared spectroscopy. Hydrogen evolution and oxidation were observed for these variants in solution based assays and protein-film electrochemistry. Catalytic activity was lowered for all variants and the ability to operate in either direction was also influenced.
Introduction
[FeFe]-Hydrogenase HydA1, from the unicellular green algae Chlamydomonas reinhardtii (C. reinhardtii), is able to reversibly catalyze the evolution and uptake of molecular hydrogen, however the former is generally favored.1,2 Although the enzyme is oxygen sensitive, its structural simplicity as a monomeric hydrogenase makes it an excellent candidate for determining structure–function relationships at the H-cluster, the site where catalytic turnover takes place.3 This cofactor (Fig. 1) comprises a [4Fe–4S] cluster linked to a unique [2Fe] cluster via the bridging cysteine 366 and is embedded in a hydrophobic pocket of the enzyme. The [4Fe–4S] cluster is coordinated by four highly conserved cysteine residues (C115, C170, C362 and C366).4,5 The presence of the [4Fe–4S] cluster in the active site in its oxidized state is suggested to be a prerequisite for the stable incorporation of the [2Fe] cluster via artificial maturation.6–9 This [2Fe] site consists of two iron atoms (Fep, proximal and Fed, distal relative to the [4Fe–4S] moiety) connected via an azadithiolate (adt) bridge, which is indispensable for efficient proton transfer from and to the open coordination site at Fed where the catalytic turnover of hydrogen takes place.8–13 Additionally, the Fep and Fed are each coordinated by one CO and one CN− ligand, with a third CO ligand bridging the two iron atoms. A native-like protein (holoHydA1), with respect to structural and catalytic features, has been obtained by maturating the apo form of HydA1 (apoHydA1) only harboring the [4Fe–4S] cluster in vitro with a chemically synthesized analogue of the [2Fe] cluster ([2Fe]MIM).8,11 This method allows for discrete modifications of the constitution of the [4Fe–4S] and [2Fe] cluster within the H-cluster.8,11,14,15 Besides targeted modifications of the bioinorganic cofactor, the fundamentals of the interplay between the H-cluster and the protein can also be assayed by site-directed mutagenesis of the surrounding amino acids. In a previous extensive study, selected amino acids involved in the hydrogen bonding network between the protein and the [2Fe] site were mutated leading to impaired in vivo maturation and catalytic activity.16 So far, the constitution of the first coordination sphere of the [4Fe–4S] cluster in HydA1 has not been analyzed with respect to its structural and functional integrity. The redox active [4Fe–4S] cluster is the supposed entry, exit and storage site for electrons and is essential for catalytic turnover via redox coupling to the [2Fe] site.17,18 It is known that the accessory [4Fe–4S] clusters found in other hydrogenases contain glutamine, asparagine (HupS from Nostoc punctiforme)19 and histidine ([NiFe]-hydrogenase from Desulfovibrio fructosovorans and [FeFe]-hydrogenase form Clostridium pasteurianum) in their first coordination sphere.4,20 This alternative coordination of a [4Fe–4S] cluster has also been observed for a wide range of [Fe–S] cluster containing proteins in nature (see review articles ref. 21–23). However, natural [4Fe–4S] cluster coordination by serine is unknown to date, despite being the most structurally homologous to cysteine. This might be due to a the possible occurrence of redox-dependent ligand protonation and subsequent discoordination providing an unnecessary complication for [4Fe–4S] cluster stability.24 Mutational studies of amino acids around the accessory [Fe–S] clusters were performed in [NiFe]-hydrogenases, a further class of hydrogenases harboring a [NiFe] core as the site for hydrogen turnover.2,25,26 Findings from this class of systems allowed for a more in depth understanding of intra- and intermolecular electron transfer,20 cluster stability towards oxygen27,28 and even the transformation of an uptake hydrogenase into a hydrogen evolving enzyme in vivo was achieved.19 Therefore, amino acid exchanges within the first coordination sphere of the [4Fe–4S] cluster of HydA1 have the potential to reveal important clues for H-cluster maturation and catalytic properties. In this study the [4Fe–4S] cluster coordinating cysteines in HydA1 were individually substituted with alanine, aspartate or serine creating a pool of 12 new variants. In contrast to alanine, serine29–31 and aspartate32,33 may serve as direct oxygenic ligands in the form of serinate or aspartate.23 Detailed analysis of the obtained variants by UV/vis, electron paramagnetic resonance (EPR), Fourier-transform infrared (FTIR) spectroscopy, solution based activity assays, and protein-film electrochemistry offered insights into the importance of the cysteines coordinating the [4Fe–4S] cluster. Notably, C115 was found to be responsible for stable [4Fe–4S] cluster integration. Variants in position C170, C362 and C366 maintain [4Fe–4S] cluster formation while only C170D, C170S, C362D and C362S allow for [2Fe] cluster incorporation and subsequent enzymatic activity, which, however, appears lowered and biased compared to the wild-type (WT).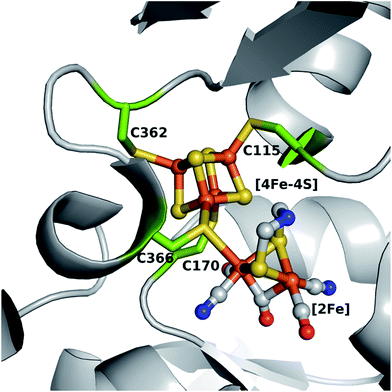 | ||
| Fig. 1 Model of the H-cluster within HydA1 from Chlamydomonas reinhardtii. Cysteine residues coordinating the [4Fe–4S] cluster are shown as green sticks (numbering is according to the expressed amino acid sequence). Protein Data Bank structures 3C8Y and 3LX4 were used for generating this model in PyMOL. Color coding is as follows: orange, iron; yellow, sulfur; gray, carbon; blue, nitrogen; red, oxygen. | ||
Results
Amino acid exchanges of the [4Fe–4S] cluster coordinating cysteines
The four coordinating cysteines (C115, C170, C362 and C366) were individually targeted by single amino acid exchanges. In total, 12 new variants of the [FeFe]-hydrogenase were successfully established by site-directed mutagenesis (see Table S1† for mutagenic primers): C115A, C115D, C115S, C170A, C170D, C170S, C362A, C362D, C362S, C366A, C366D, and C366S. Protein yields for the variants were decreased down to 15% compared to the WT. Furthermore, the C115 variants appeared less deep in the characteristic brown color of purified [FeFe]-hydrogenases. This color difference provided visual evidence that the site of replacement matters when looking at the effects of the individual replacement of the four cysteines.UV/vis and EPR spectroscopy to elucidate the presence of the [4Fe–4S] cluster in apoHydA1
The constitution of the [4Fe–4S] cluster in the apoHydA1 variants was determined by UV/vis and by EPR spectroscopy. Characteristic WT-like absorbance features around 320 and 420 nm, representing the oxidized [Fe–S] site within the [FeFe]-hydrogenases were observed by UV/vis spectroscopy for the C170, C362 and C366 variants (Fig. 2A), with C362D having an overall decrease in absorbance (Fig. S1A, bottom†). All C115 variants showed strongly reduced absorbances as expected from the pale color of the purified protein. Exposure to oxygen for 12 h led to an almost complete loss of signals for all variants in the characteristic region of 320 to 420 nm, pointing towards a retained oxygen sensitivity of the [4Fe–4S] clusters.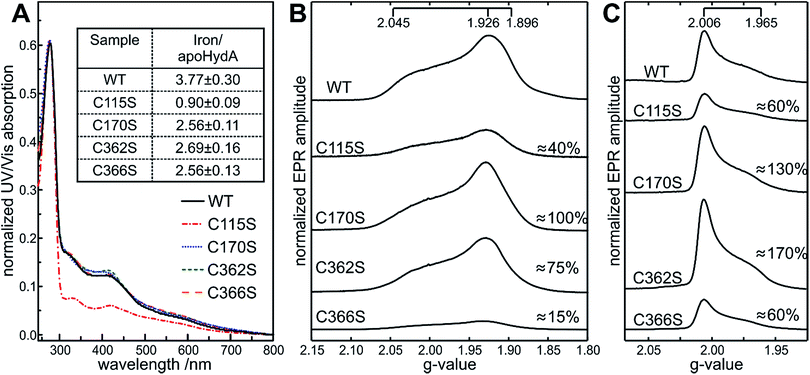 | ||
| Fig. 2 Spectroscopic characterization of apoHydA1 variants. UV/vis absorbance spectra of apoHydA1 WT and the apoHydA1 C115S (red), C170S (blue), C362S (green), and C366S (orange) variants in their oxidized form. UV/vis spectra were measured at 25 °C in 100 mM Tris–HCl buffer (pH 8). Spectra were normalized to the protein absorbance peak at 280 nm. The inset shows the iron content of apoHydA1 variants based on the method of Fish52 in [mol mol−1] determined in triplicate (A). Q-band FID detected EPR spectra obtained from the reduced (B) and oxidized (C) form of the 600 μM apoHydA1 variants measured at 10 K and 20 K respectively. The ratio of each variant to the WT integral value is given in [%] with 10–20% accuracy. Please note the WT sample was found to be composed of 90% [4Fe–4S] and 10% [3Fe–4S] clusters in total. | ||
EPR spectroscopy was used to unambiguously identify the nature of the [Fe–S] clusters formed. For the WT and all variants, the spectra of oxidized samples (“as purified”) and reduced samples (excess of sodium dithionite (NaDT), 10 mM) were investigated. In the reduced apoHydA1 samples, a [4Fe–4S]+ cluster is expected with a characteristic EPR signal, whereas in the oxidized samples, the [4Fe–4S]2+ cluster is EPR silent. Additional signals could result from oxidized [3Fe–4S] or reduced [2Fe–2S] clusters, indicating incomplete [4Fe–4S] cluster formation and/or oxygen damage.
In all oxidized samples, the presence of a narrow axial EPR spectrum with g = [2.006, 1.965] values is observed (Fig. 2C) that is typical for a S = 1/2[3Fe–4S]+ cluster.34 In the reduced samples, these [3Fe–4S] clusters were no longer detectable indicating that they are reduced. Instead, nearly axial EPR signals were detected, characteristic for a low-spin S = 1/2[4Fe–4S]+ cluster with g = [2.045, 1.926, 1.896] (Fig. 2B) as observed previously for apoHydA1.35 [2Fe–2S] clusters often exhibit g-values similar to [4Fe–4S] clusters, therefore the relaxation properties of the signals in the reduced samples were additionally checked and thus the presence of the [2Fe–2S] clusters could be excluded in all cases. The [3Fe–4S]+ cluster signals showed exactly the same g-values for all variants within the experimental accuracy. This was in contrast to the EPR spectra of the [4Fe–4S]+ clusters, where small shifts of the broad lines were detected. Most affected was the g1 value, with a maximum shift of 0.015, whereas g2 and g3 only varied up to 0.005, likely a result of missing or altered coordination of the [4Fe–4S] cluster. EPR signal integration of the cluster signals indicated differences in the distribution of [4Fe–4S] and [3Fe–4S] clusters between the different apoHydA1 variants compared to apoHydA1 WT. The WT sample was found to be composed of protein containing 90% [4Fe–4S] and 10% [3Fe–4S] clusters. As already seen from the UV/vis spectra, the occupancy within the C115 variants was strongly reduced to 40–60% (depending on the mutation) for the [4Fe–4S]+ (except for the C115A variant) and 20–60% for the [3Fe–4S]+ cluster signal (Fig. 2B and S1B†), which was additionally confirmed by the low molar iron/HydA1 ratio (Fig. 2A and S1A†). For the C170 and C362 variants the [4Fe–4S]+ cluster occupancy appeared native-like, while the [3Fe–4S]+ cluster content appeared 1.5 fold higher than observed for apoHydA1 WT. The only exceptions were the C170A variant with a decreased [4Fe–4S] cluster content (60%) and C362D where variation to aspartate led to a drastic decrease of [4Fe–4S]+ and [3Fe–4S]+ cluster content to roughly 20% and 60%, respectively (Fig. 2B and S1B†). EPR data of the C366 variants showed only 10% and 20–60% occupancy for [4Fe–4S]+ and [3Fe–4S]+ clusters respectively, despite a characteristic absorbance band at 420 nm and an average iron content of 2.5 iron/HydA1. These results show that the integrity of the [4Fe–4S] cluster depends on both the site of replacement and the choice of the amino acid used for exchange. In many cases the lack of a proper coordination for the [4Fe–4S] led to the absence of this cluster rather than its incomplete insertion.
FTIR spectroscopy for determination of [2Fe] cluster integration
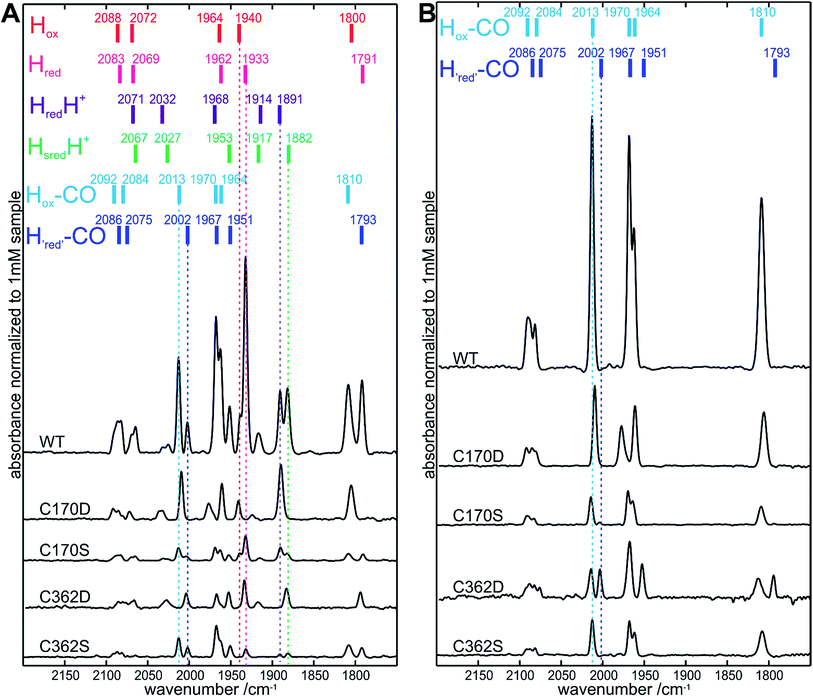
We further investigated if a preformed [4Fe–4S] cluster, as observed in the C170 and C362 apoHydA1 variants, was sufficient for in vitro [2Fe]MIM cluster insertion. This was assayed by iron content determination (Table S2†) and in more detail by FTIR spectroscopy. FTIR spectroscopy is an excellent method to check for successful [2Fe] cluster integration as its CO and CN− ligands give rise to characteristic absorption bands which appear sharp only after stable integration of the [2Fe] cluster. In all FTIR spectra obtained from variants with changes at position C115 or C366, no sharp peaks were observed in the FTIR spectra, showing that no cluster insertion took place. Interestingly, FTIR signals were also not detected for C170A and C362A, despite the presence of a preformed [4Fe–4S] cluster. On the other hand, for C170D, C170S, C362D, and C36S2 variants, characteristic absorption bands were observed (Fig. 3A, S3 and summary of the relevant redox states Table S3†) indicating the stable incorporation of the [2Fe]MIM site for these four variants only. However, maturation of these variants appeared only partial and did not directly correspond to the [4Fe–4S] cluster occupancies, except for C362D. Based on a comparison of the integrated area under the FTIR signals, the most highly maturated variant was C170D (≈25 ± 3%) followed by C362D (≈20 ± 3%), C170S (≈10 ± 3%) and C362S (≈10 ± 3%) (Fig. 3B). This indicated that the C170D and C362D variants were more likely to be maturated than the C170S and C362S variants, even though a higher occupancy of [4Fe–4S] for the C170S and C362S in comparison to C362D would suggest a lower [2Fe]MIM cluster loading for C362D. Additionally, longer maturation of the C170S and C362S variants followed by FTIR revealed that it is necessary for these variants to increase the time of maturation from 1 h to 30 h (Fig. S4 and S5†).Similar to the “as purified” WT FTIR spectrum, the “as purified” spectra of C170D, C170S, C362D, and C362S exhibited a mixture of active and CO inhibited states after in vitro maturation. Interestingly, for the C170D variant bands assigned to redox states with an oxidized [4Fe–4S]2+ cluster were predominant (HredH+, Hox, Hox–CO), while the C362D variant mainly exhibited bands assigned to redox states with a reduced [4Fe–4S]+ cluster (Hred, HredH+, Hred–CO).36Fig. 3 shows the indicative band positions for “as purified” samples, as well as for samples flushed with CO (15 minutes) to enrich the non-catalytic CO inhibited states (exact band positions are annotated in Fig. S3†). This is an easy method to obtain pure well-characterized states that allow for the direct comparison between FTIR spectra of all four variants. Small shifts of the band positions (up to 3 cm−1) were observed for all variants, except for bands assigned to the Hox–CO state of the C170D variant. Notably, the band assigned to its bridging CO was shifted by 5 cm−1, while the bands at 2010 and 1961 cm−1 appeared shifted by only 3 cm−1 to lower frequencies. The biggest overall shift to higher frequencies was detected for its band at 1977 cm−1 (7 cm−1). Taken together, the data demonstrate for the C170 and C362 variants that a preformed [4Fe–4S] cluster alone is not sufficient for [2Fe]MIM cluster insertion.
Solution based assays and protein-film electrochemistry for investigation of altered enzymatic activity
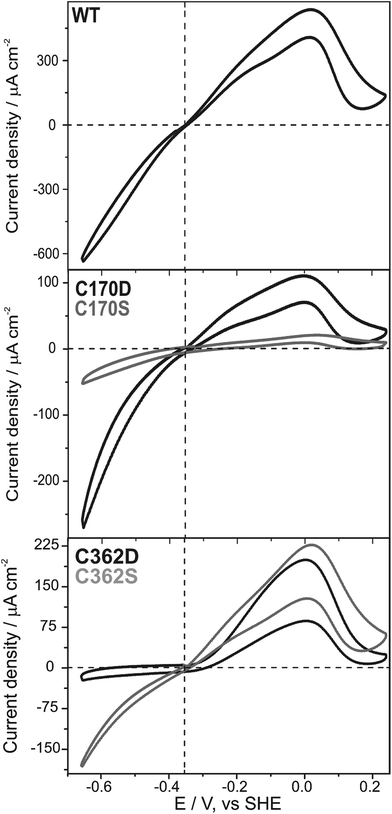
Subsequently, we questioned how the catalytic properties of the variants were influenced by the introduced amino acid exchanges. No hydrogen evolution above the detection limit of 1 μmol H2 per minute per mg HydA1 could be detected in a solution based assay using NaDT as electron donor and methyl viologen (MV) as electron mediator for all C115 and C366 variants as well as for C170A and C362A. In contrast, hydrogen evolution and uptake by the C170D, C170S, C362D, and C362S variants was clearly measurable (Table 1). As expected, only variants for which successful insertion of the [2Fe]MIM site was detected in FTIR spectroscopy showed catalytic turnover. Overall lower rates for hydrogen evolution and uptake compared to WT were detected ranging from about 0.2–8.0% and 0.5–32.1%, respectively. In line with the results from FTIR spectroscopy the C170S and C362S variants showed roughly a 3-fold increase in their catalytic rates once the time of maturation was prolonged from 1 to 30 h. The ratios of hydrogen uptake to hydrogen evolution rates revealed that the C362 variants had a tendency towards hydrogen uptake, while for the C170 variants a tendency towards hydrogen evolution was observed (Table 1). These tendencies are supported by findings from library screening plate tests where single colonies of Escherichia coli expressing the holoHydA1 variants were tested for their hydrogen oxidation ability. Herein, a MV suspension turning from colorless to blue once reduced (by enzymatic hydrogen uptake) was used as a readout system (for details see ref. 37). For the C362 variants, a blue coloring was detected, while the C170 remained clear (Fig. S2†). In an additional solution assay the natural electron mediator PetF, a [2Fe–2S]-ferredoxin, was used to determine hydrogen production leading to rates that followed the trends already observed for the MV based assay. Furthermore, enzymatic activity was further confirmed by protein-film electrochemistry for the four active variants (C170D, C170S, C362D, and C262S) adsorbed on a rotating disc pyrolytic graphite electrode and measured at pH 6 under 100% H2. Fig. 4 shows that all variants displayed a (partly) reversible oxidative inactivation. This oxidation leads to an inactive state at high potentials (E > +50 mV), which is characteristic for C. reinhardtii HydA1 and can be reactivated upon scanning in the negative direction.38 The ratios of currents for proton reduction and hydrogen oxidation showed that the C170 variants favored proton reduction, while the C362 variants favored hydrogen oxidation. As the most extreme case, the C362D variant barely reduced protons at pH 5 to 10 (Fig. S6†). Therefore, modifications of the cysteines coordinating the [4Fe–4S] site of the H-cluster truly influence the catalytic turnover at the [2Fe] site.| Sample | 1 h maturation | 30 h maturation | |||||
|---|---|---|---|---|---|---|---|
| PetF dependent H2 production | MV dependent H2 production | MV dependent H2 uptake | Ratio | MV dependent H2 production | MV dependent H2 uptake | Ratio | |
| a n.d.: not determined as FTIR spectroscopy did not show [2Fe]MIM cluster integration and H2 production yielded no significant activity. b Based on the used detection method for H2 production values below 1 μmol H2 per min per mg HydA1 cannot be determined. | |||||||
| WT | 410.6 ± 39.6 | 728.0 ± 5.0 | 271.0 ± 3.0 | 37.2 | n.d. | n.d. | |
| C115A | n.d.a | <1b | n.d. | n.d. | n.d. | ||
| C115D | n.d. | <1 | n.d. | n.d. | n.d. | ||
| C115S | n.d. | <1 | n.d. | n.d. | n.d. | ||
| C170A | n.d. | <1 | n.d. | <1 | <1 | ||
| C170D | 33.2 ± 2.0 | 101.1 ± 1.1 | 2.6 ± 1.3 | 2.6 | 141.2 ± 25.6 | 11.1 ± 3.0 | 7.9 |
| C170S | 11.7 ± 0.9 | 64.5 ± 2.1 | 11.8 ± 1.5 | 18.3 | 231.4 ± 58.2 | 29.6 ± 8.6 | 12.8 |
| C362A | n.d. | <1 | n.d. | <1 | <1 | ||
| C362D | 8.5 ± 0.9 | 5.7 ± 0.2 | 2.1 ± 0.3 | 36.8 | 6.4 ± 0.5 | 2.8 ± 0.2 | 43.8 |
| C362S | 9.4 ± 2.6 | 1.6 ± 0.1 | 1.4 ± 0.3 | 87.5 | 4.4 ± 1.4 | 6.9 ± 0.5 | 156.8 |
| C366A | n.d. | <1 | n.d. | n.d. | n.d. | ||
| C366D | n.d. | <1 | n.d. | n.d. | n.d. | ||
| C366S | n.d. | <1 | n.d. | n.d. | n.d. | ||
Discussion
Despite general observations that single exchanges of the cysteines coordinating the [4Fe–4S] cluster may lead to impairments in protein folding, stability, expression and purification, we were able to successfully generate such variants in their apoforms.23 So far only a few cases of cysteine to serine exchanges with retained [4Fe–4S] cluster integrity and enzymatic activity have been reported,29,39 since this exchange often leads to loss of the cluster, cluster interconversions and/or structural rearrangements.23,31,40,41 Exchanges of the [4Fe–4S] cluster coordinating cysteines to aspartate have been shown before to be functionally tolerated in the PsaC subunit of photosystem I, as well as in the small subunit of a [NiFe]-hydrogenase.32,33 By following the occupancies of the [4Fe–4S] and [2Fe] cluster of the catalytically relevant H-cluster, we have been able to gain new insights into the importance of each individual cysteine coordinating the [4Fe–4S] cluster.Variants at the C115 position showed that only a very small amount of the preformed [4Fe–4S] cluster was present in the purified proteins, with protein yields notably decreased. This finding was particularly surprising as the sequence alignment of putative hydrogenases suggests that serine is present in this position for about 20% of the analyzed sequences suggesting serine as an alternative ligand.3 As C115 is located in the core of the protein, structural flexibility at this position might be too limited to allow for alternative coordination by aspartate or serine. In the case of variation to alanine the neighboring cysteine 114 could act as an alternative ligand to the [4Fe–4S] cluster explaining the higher [4Fe–4S] cluster occupancy of C115A compared to the other C115 variants. Since a preformed [4Fe–4S] cluster is required for [2Fe] cluster integration in vitro,6,42 the observed deficiency of a [2Fe] cluster within the C115 variants is attributed to the low [4Fe–4S] cluster occupancy, which suggests that C115 is vital for the stability of the [4Fe–4S] cluster.
Variants in position C366 also showed no [2Fe]MIM cluster insertion, although they most likely do allow for [4Fe–4S] cluster formation according to WT-like UV/Vis spectra and findings from iron determination. However, the EPR spectroscopic signals for the [4Fe–4S]+ cluster appeared drastically decreased for the C366 variants compared to the WT (to 10–15 and 20–60%, respectively). One explanation is that the majority of the clusters were turned into an EPR silent state by the selected amino acid exchanges, an effect already observed for a [4Fe–4S] cluster within the PsaC subunit of photosystem I (exchange of cysteine to aspartate).33 Furthermore, in the native protein, the [4Fe–4S] cluster coordinating cysteine 366 is exposed to the surface of the maturation channel, providing the site of covalent attachment of the [2Fe] site.42 It was shown that upon variation of this bridging position the covalent linkage of the [2Fe] cluster to the [4Fe–4S] cluster, as well as to the protein, is no longer feasible.3 Subsequently, stable [2Fe] cluster insertion fails for the C366 variants herein.
Unlike cysteines C115 and C366, cysteine 170 is located in a flexible loop and appears to have enough structural flexibility to maintain WT-like [4Fe–4S] cluster occupancy for the C170D and C170S, variants as well as roughly 60% occupancy for the C170A variant. Overall, more [3Fe–4S] cluster compared to WT was present for variants in position C170, which is a clear indication, that [4Fe–4S] cluster integrity was affected by altering the fourth coordination site.23 The general observation in the EPR spectroscopy that [3Fe–4S] clusters were present for the HydA1 variants, as well as for the HydA1 WT, in EPR spectroscopy might be a consequence of insufficient [4Fe–4S] cluster maturation due to the heterologous overexpression, lack of the fourth coordinating ligand, or oxidative damage during sample preparation.6,15,28 EPR signals corresponding to the [4Fe–4S] clusters only displayed small shifts and changes in the line broadening for all variants, which was probably caused by a change in cluster coordination. It should be noted that while exchange of cysteinate to serinate retains monodentate ligation of the iron, aspartate may act as a mono- or bidentate ligand. The possibility of coordination by solvent molecules (OH− or H2O) may explain the stability of the cluster in the case of alanine substitutions. The nature of the actual coordination however remains unclear, since NMR or X-ray crystallographic studies are required for definitive conclusions.24
Despite the presence of a preformed [4Fe–4S] cluster for all of the C170 variants, only C170D and C170S showed [2Fe]MIM cluster insertion, which however was only partial. Prolongation of the in vitro maturation improved the final [2Fe] cluster occupancy notably for the C170S variants, indicating a slowed insertion of the subcluster as seen for the [FeFe]-hydrogenase HydAB from Desulfovibrio desulfuricans.43 Interestingly, for the C170A variant, any stable [2Fe]MIM cluster incorporation could not be detected even after 30 h of in vitro maturation. This clearly shows that a preformed [4Fe–4S] cluster alone does not guarantee for stable [2Fe]MIM incorporation and additional factors must certainly contribute to the maturation process. Very recent studies suggest a sequence of events in which the [2Fe]MIM cluster first attaches tightly to the apoprotein forming a stable intermediate.7 During this initial step, the [2Fe]MIM cluster passes through a maturation channel lined with positively charged amino acids where a lysine residue at the end of the maturation channel plays a crucial role in orientation and anchoring.16,42 The preformed [4Fe–4S] cluster is part of this channel being located at its base. Its redox state appears crucial for covalent attachment of the [2Fe]MIM cluster as under strongly reducing conditions the activation of the protein fails, indicating that the [4Fe–4S] cluster needs to be oxidized to form a bond with the [2Fe]MIM cluster.7 As a last step structural rearrangements and CO release take place.
In all variants for which successful [2Fe]MIM cluster incorporation occurred, the vibrational features of the CO and CN− ligands could be assessed by FTIR spectroscopy. No significant change in the overall spectra for the catalytically relevant states was observed for any of the variants. A similar observation was made recently when even for a chalcogenide exchange within the [4Fe–4S] cluster from sulfur to selenium, which did not lead to any altered FTIR patterns.14 Slight shifts in the spectra were observed for the CO inhibited form of the C170D variant. In particular, a shift of 7 cm−1 towards higher frequencies was detected for the band at 1977 cm−1, containing major contributions from the equatorial CO ligand of the proximal iron (COp).44 The close proximity of the COp ligand to C170 (C170 Sγ atom to COp 4.0 Å and C170 Cβ atom to COp 3.1 Å, based on an overlay structure of PDB ID: 3LX4 and 3C8Y, Fig. 1) might be the reason for this effect. Aspartate is slightly bigger and has a different charge distribution than cysteine therefore influencing the structural and electronic environment of the COp. Despite the FTIR data showing a higher [2Fe] occupancy for other variants, the C170S variant appears to be the most active in hydrogen evolution and uptake in solution based activity assays, pointing towards a high tolerance for amino acid exchanges at this position. The ratios of the obtained evolution and uptake rates of the functional variants in this study allowed for some quantification of the catalytic bias under the given conditions.15,45 Compared to the WT, the C170 variants were biased towards hydrogen evolution in these assays in agreement with protein-film electrochemistry experiments.
EPR and FTIR spectroscopy provided a similar picture for the C362 and the C170 variants, except for the C362D variant which showed only 20% [4Fe–4S] cluster occupancy. Despite its location in a flexible loop and findings for the variants at other positions, the slightly larger size and different charge distribution of aspartate compared to cysteine appears to destabilize the [4Fe–4S] cluster at this position. However, the obtained absolute numbers suggest that for the C362D variant, all protein containing a [4Fe–4S] cluster can be maturated whereas the C362S variant is only partially maturated. As seen for C170S, prolongation of the in vitro maturation led to increased [2Fe] cluster occupancy for the C362S variant.
The importance of the C362 position for catalytic turnover was highlighted by the fact that the C362D and C362S variants displayed enzymatic activities of less than 2.5% compared to the WT in either direction, despite 20% and 10% [2Fe] cluster occupancy, respectively. The C362 variants are biased towards hydrogen uptake in solution assays using MV as an artificial electron mediator as well as in protein-film electrochemistry experiments, while the bias towards hydrogen oxidation for the C362D appeared stronger in the latter. It has been proposed that the catalytic bias is controlled by the potential of the electron relay center at which the electrons enter and/or exit the hydrogenase (Eox/red).17 The actual ability of the enzyme to either evolve or oxidize hydrogen subsequently originates from the separation of Eox/red and the equilibrium potential of hydrogen oxidation and proton reduction (E2H+/H2) under the given conditions. Following up this basic model leads to the assumption that if E2H+/H2 ≫ Eox/red hydrogen evolution is favored and in case of E2H+/H2 ≪ Eox/red hydrogen uptake is favored.17,38 This would lead to the speculative conclusion that the redox potential of electrochemical control center, in the case of HydA1 the potential of the [4Fe–4S] cluster, is shifted to higher potentials for C362 variants and lower potentials for C170 variants compared to the WT. C362D represents an extreme case, as it loses its reversibility, similarly to the situation in oxygen tolerant [NiFe]-hydrogenases.17,46 Another consequence of this potential shift to more positive values is the appearance of an overpotential for catalytic H2 oxidation. The active site needs to be completely oxidized in order to accept the electrons from hydrogen, since the transitions are shifted to more positive values in the C362D variant, hydrogen oxidation does not start until a potential more positive than −300 mV is applied, that is a 50 mV overpotential (Fig. 4). The same effect, but to a lesser extent, could be observed for the C170 variants in the H2 evolution direction. These shifts in catalytic bias were observed for a broad pH range (Fig. S6†), indicating that the shift in the cluster potential dominates over any changes in the pKa of the cluster. The characteristic catalytic bias of the C170 and C362 variants can be related to the findings from FTIR spectroscopy where a shift in the equilibrium redox properties of the H-cluster is observed. The large population of states with a reduced [4Fe–4S] cluster for the C362D variant indicates a more positive potential while the absence of reduced [4Fe–4S] cluster species for the C170D variant when measured under the same conditions, indicate a more negative potential for the cluster. Since the characteristic bias of the C170 and C362 variants was observed in all assays performed, it became, therefore, evident that alteration around the [4Fe–4S] cluster influences the bias of catalytic turnover at the remote Fed.
Conclusion
The importance of each coordinating cysteine around the [4Fe–4S] cluster of the H-cluster within [FeFe]-hydrogenase HydA1 has been analyzed in detail within this study. The results clearly showed that C115 is crucial for [4Fe–4S] cluster binding, C170 and C362 are replaceable by aspartate or serine while maintaining catalytic activity and C366 appears responsible for anchoring the [2Fe] site. Furthermore a preformed [4Fe–4S] cluster does not guarantee stable insertion of the [2Fe] cluster. It became evident herein that alterations around the [4Fe–4S] cluster influence the catalytic turnover at the remote Fed, demonstrating the synergy between the active site and the protein scaffold (see schematic summary S1†). Since the [4Fe–4S] cluster is part of the catalytically relevant H-cluster via redox coupling, it must be considered when thinking about reducing hydrogenases to their minimal functional units. This study therefore contributes to the understanding of the catalytic site of [FeFe]-hydrogenases and at the same time contributes to the future design of such engineered enzymes as it reveals important structural and electronical prerequisites for tuning enzymatic turnover.Experimental procedure
Generation of HydA1 variants
Site-directed mutagenesis at position C115, C170, C362 and C366 (numbering according to the expressed amino acid sequence) was performed by QuikChange using pET21(b)-hydA1 as the template plasmid47 and the mutagenic primers listed in Table S1.† Introduction of the desired mutations was verified by sequencing.Protein expression and purification
[FeFe]-Hydrogenase HydA1 from C. reinhardtii1 (48 kDa) with C-terminally fused Strep-tagII and its variants were heterologous expressed in Escherichia coli strain BL21(DE3)ΔiscR48 in the absence of specific maturases, therefore lacking the [2Fe] subcluster (apoHydA1).47 Strep-tactin affinity chromatography (iba, Göttingen, Germany) was used for protein purification under strictly anaerobic conditions using 100 mM Tris–HCl buffer (pH 8) containing 2 mM NaDT.49 Protein preparations used for EPR and UV/vis spectroscopy were purified without the reducing agent NaDT. Protein yields were about 6 to 9 mg for C115 variants, 13 to 19 mg for C170 variants, 5 to 12 mg for C362 variants, and 6 to 11 mg for C366 variants out of 1 L expression culture compared to up to 30 mg for the WT. Recombinant [2Fe–2S]-ferredoxin PetF from C. reinhardtii was prepared as described earlier.50 Protein concentrations were determined by the method of Bradford51 using bovine serum albumin as a standard and protein purity was assessed by SDS-PAGE. Bovine serum albumin was obtained commercially from Carl Roth, Karlsruhe, Germany.In vitro maturation
[Fe2[μ-(SCH2)2NH](CN)2(CO)4][Et4N]2 ([2Fe]MIM) was prepared as described before11 and dissolved in 100 mM K2HPO4/KH2PO4 buffer (pH 6.8) to concentrations of 21 to 55 mM. A 5-fold molar excess of [2Fe]MIM was added to 200 to 400 μM apoHydA1 in a final volume of up to 500 μL 100 mM K2HPO4/KH2PO4 buffer (pH 6.8) with 2 mM NaDT initiating the maturation process at 25 °C for 1 h or 30 h.8,15 Subsequently the excess of [2Fe]MIM was removed via size exclusion chromatography using a NAP-5 column (GE Healthcare, Chicago, USA) equilibrated with 100 mM Tris–HCl (pH 8) with 2 mM NaDT. Protein preparations were concentrated up to 2 mM using 30 kDa Amicon Ultra centrifugal filter units (Merck Millipore, Billerica, USA) and stored anaerobically at −80 °C.UV/vis spectroscopy
UV/vis spectra were recorded with a UV-2450 spectrometer (Shimadzu, Kyoto, Japan) at 25 °C using sealed 1 mL UV-cuvettes filled with 500 μL apoHydA1 solution at a concentration of 10 μM in 100 mM Tris–HCl buffer (pH 8). After the initial spectra was measured under anaerobic conditions, the seal was removed and apoHydA1 solution was exposed to air at room temperature for 12 h before a further spectra were recorded. Obtained spectra were normalized to the protein peak at 280 nm.Iron quantification
Iron content of samples prepared for EPR and FTIR analysis was quantified in triplicate using the method of Fish52 and protein concentrations were determined in parallel.EPR analysis
ApoHydA1 samples at a concentration of 600 μM in 100 mM Tris–HCl buffer (pH 8) were analyzed by EPR. A final concentration of 10 mM NaDT was used to obtain reduced samples. Q-band EPR spectra were recorded using free induction decay detected EPR with a microwave pulse length of 1 μs. All pulse experiments were performed on a Bruker ELEXYS E580 Q-band spectrometer with a SuperQ-FT microwave bridge and home built resonator described earlier.53 X-band continuous wave EPR measurements were performed on a Bruker ELEXYS E-580 X-band spectrometer with a SuperX-FT microwave bridge and Bruker ER EN4118X-MD5 dielectric resonator. Cryogenic temperatures (10–20 K) were obtained by an Oxford CF935 flow cryostat.FTIR analysis
FTIR spectra of as purified in vitro maturated HydA1 samples at concentrations of 0.9 to 2 mM in 100 mM Tris–HCl buffer (pH 8) with 2 mM NaDT were recorded. For selected variants protein samples were additionally flushed with CO gas for 15 min prior to recording CO inhibited spectra. All experiments were performed using a Bruker IFS 66v/s FTIR spectrometer equipped with a nitrogen cooled MCT detector. For each spectrum 1000 scans were accumulated in the double-sided, forward–backward mode at a resolution of 2 cm−1 at 15 °C. Data processing was facilitated by home written routines in the MATLAB™ programing environment.Hydrogen evolution assay
For determination of the MV dependent hydrogen evolution, a standard in vitro enzymatic activity assay54 was performed within 8 mL anaerobically sealed vials using 800 ng of the in vitro maturated HydA1 variants, 100 mM NaDT and 10 mM MV in 100 mM K2HPO4/KH2PO4 buffer (pH 6.8) in a final volume of 2 mL. Hydrogen formation was detected via gas chromatography after incubation at 37 °C for 30 min. For determination of the PetF dependent hydrogen evolution, an activity assay described in literature50 was performed within 2 mL anaerobically sealed vials using 160 ng of the in vitro maturated HydA1 variants, 25 mM NaDT and 80 μM PetF in 100 mM K2HPO4/KH2PO4 buffer (pH 6.8) in a final volume of 200 μL. Hydrogen formation was detected via gas chromatography after incubation at 37 °C for 20 min.Hydrogen uptake assay
Hydrogen uptake was assayed based on procedures described in the literature.15,47 A sealed 1 mL UV-cuvette was flushed with H2 before adding 955 μL 100 mM K2HPO4/KH2PO4 buffer (pH 6.8) containing 40 mM MV and 5 μL in vitro maturated HydA1 corresponding to 10 to 1250 ng of total protein via a syringe. Evolution of MV therefore its colorimetric change from clear to dark purple, was followed at 25 °C spectrophotometrically using a Beckman Coulter PARADIGM™ absorbance detection cartridge at 604 nm. Activity was estimated based on the initial slope of absorbance versus time determined using linear regression.Protein-film electrochemistry
A pyrolytic graphite rotating disk electrode (0.07 cm2 homemade pyrolytic graphite from Momentive Materials) was first polished with alumina MasterPrep Polishing Suspension (0.05 mm, Buehler, Esslingen, Germany) and sonicated for 5 min in Milli-Q water. Then, the electrode was transferred into the Glove-box (MBRAUN, Garching, Germany), filled with N2 where the surface-adsorbed oxygen was removed by cycling the potential 10 times from +250 mV to −650 mV (vs. SHE) in a buffer a mix at pH 6 containing mixture of 15 mM MES, HEPES, TAPS, CHES and sodium acetate and 0.1 M NaCl. The enzyme was then adsorbed following the protocol described previously and the measurements were done using the described set-up55 under the conditions: 25 °C, buffer mix pH 6, 20 mV s−1, 1 atm H2 (1 L min−1 total flow) and 2000 rpm electrode rotation rate to avoid mass-diffusion limitation. The potential was controlled by a PARSTAT MC-1000 multi-channel potentiostat (Princeton Applied Research, Oak Ridge, USA). All potentials are quoted versus the standard hydrogen electrode (SHE) using a conversion of +241 mV from SCE. The reference electrode potential was periodically controlled using (hydroxymethyl)ferrocene (+420 mV vs. SHE).56Conflicts of interest
There are no conflicts to declare.Abbreviations
| [2Fe] | [2Fe–2S] subcluster of the H-cluster |
| [2Fe]MIM | Chemically synthesized [2Fe] cluster analogue |
| [4Fe–4S] | [4Fe–4S] subcluster of the H-cluster |
| adt | Azadithiolate |
| apoHydA1 | Apoform of HydA1 only harboring the [4Fe–4S] subcluster |
| COp | Equatorial CO ligand of the proximal iron |
| C. reinhardtii | Chlamydomonas reinhardtii |
| EPR | Electron paramagnetic resonance |
| Fed | Distal iron atom of the [2Fe] cluster relative to the [4Fe–4S] moiety |
| Fep | Proximal iron atom of the [2Fe] cluster relative to the [4Fe–4S] moiety |
| FTIR | Fourier-transform infrared |
| holoHydA1 | Holoform of HydA1 harboring the [4Fe–4S] and [2Fe] subcluster |
| MV | Methyl viologen |
| NaDT | Sodium dithionite |
| SHE | Standard hydrogen electrode |
| WT | Wild-type |
Acknowledgements
We kindly thank Florian Wittkamp and Ulf-Peter Apfel for providing the [2Fe]MIM used for in vitro maturation of apoHydA1 in this study. We thank Birgit Nöring for her technical support in protein-film electrochemical experiments. This work was supported by the Max Planck Society. The authors are grateful to Volkswagen Foundation (LigH2t), the Cluster of Excellence RESOLV (EXC1069) and the DIP Programme (LU 315/17-1) both funded by the Deutsche Forschungsgemeinschaft (DFG). We thank the Fond der Chemischen Industrie for supporting LK by a Kekulé Mobility Fellowship.References
- T. Happe and J. D. Naber, Eur. J. Biochem., 1993, 214, 475–481 CrossRef CAS PubMed.
- W. Lubitz, H. Ogata, O. Rüdiger and E. Reijerse, Chem. Rev., 2014, 114, 4081–4148 CrossRef CAS PubMed.
- M. Winkler, J. Esselborn and T. Happe, Biochim. Biophys. Acta, Bioenerg., 2013, 1827, 974–985 CrossRef CAS PubMed.
- J. W. Peters, W. N. Lanzilotta, B. J. Lemon and L. C. Seefeldt, Science, 1998, 282, 1853–1858 CrossRef CAS PubMed.
- Y. Nicolet, C. Piras, P. Legrand, C. E. Hatchikian and J. C. Fontecilla-Camps, Structure, 1999, 7, 13–23 CrossRef CAS PubMed.
- D. W. Mulder, D. O. Ortillo, D. J. Gardenghi, A. V. Naumov, S. S. Ruebush, R. K. Szilagyi, B. Huynh, J. B. Broderick and J. W. Peters, Biochemistry, 2009, 48, 6240–6248 CrossRef CAS PubMed.
- C. F. Megarity, J. Esselborn, S. V. Hexter, F. Wittkamp, U.-P. Apfel, T. Happe and F. A. Armstrong, J. Am. Chem. Soc., 2016, 138, 15227–15233 CrossRef CAS PubMed.
- J. Esselborn, C. Lambertz, A. Adamska-Venkatesh, T. Simmons, G. Berggren, J. Noth, J. Siebel, A. Hemschemeier, V. Artero and E. Reijerse, et al. , Nat. Chem. Biol., 2013, 9, 607–609 CrossRef CAS PubMed.
- G. Berggren, A. Adamska, C. Lambertz, T. R. Simmons, J. Esselborn, M. Atta, S. Gambarelli, J.-M. Mouesca, E. Reijerse, W. Lubitz, T. Happe, V. Artero and M. Fontecave, Nature, 2013, 499, 66–69 CrossRef CAS PubMed.
- B. J. Lemon and J. W. Peters, Biochemistry, 1999, 38, 12969–12973 CrossRef CAS PubMed.
- J. Esselborn, N. Muraki, K. Klein, V. Engelbrecht, N. Metzler-Nolte, U.-P. Apfel, E. Hofmann, G. Kurisu and T. Happe, Chem. Sci., 2016, 7, 959–968 RSC.
- D. W. Mulder, Y. Guo, M. W. Ratzloff and P. W. King, J. Am. Chem. Soc., 2016, 139, 83–86 CrossRef PubMed.
- E. J. Reijerse, C. C. Pham, V. Pelmenschikov, R. Gilbert-Wilson, A. Adamska-Venkatesh, J. F. Siebel, L. B. Gee, Y. Yoda, K. Tamasaku, W. Lubitz, T. B. Rauchfuss and S. P. Cramer, J. Am. Chem. Soc., 2017, 139, 4206–4309 CrossRef PubMed.
- J. Noth, J. Esselborn, J. Güldenhaupt, A. Brünje, A. Sawyer, U.-P. Apfel, K. Gerwert, E. Hofmann, M. Winkler and T. Happe, Angew. Chem., Int. Ed., 2016, 55, 8396–8400 CrossRef CAS PubMed.
- J. F. Siebel, A. Adamksa-Venkatesh, K. Weber, S. Rumpel, E. J. Reijerse and W. Lubitz, Biochemistry, 2015, 54, 1474–1483 CrossRef CAS PubMed.
- P. Knörzer, A. Silakov, C. E. Foster, F. A. Armstrong, W. Lubitz and T. Happe, J. Biol. Chem., 2012, 287, 1489–1499 CrossRef PubMed.
- S. V. Hexter, F. Grey, T. Happe, V. Climent and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11516–11521 CrossRef CAS PubMed.
- A. Adamska-Venkatesh, D. Krawietz, J. Siebel, K. Weber, T. Happe, E. Reijerse and W. Lubitz, J. Am. Chem. Soc., 2014, 136, 11339–11346 CrossRef CAS PubMed.
- P. Raleiras, N. Khanna, H. Miranda, L. S. Mészáros, H. Krassen, F. Ho, N. Battchikova, E.-M. Aro, A. Magnuson and P. Lindblad, et al. , Energy Environ. Sci., 2016, 9, 581–594 CAS.
- S. Dementin, V. Belle, P. Bertrand, B. Guigliarelli, G. Adryanczyk-Perrier, A. L. De Lacey, V. M. Fernandez, M. Rousset and C. Léger, J. Am. Chem. Soc., 2006, 128, 5209–5218 CrossRef CAS PubMed.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 CrossRef CAS PubMed.
- D. W. Bak and S. J. Elliott, Curr. Opin. Chem. Biol., 2014, 19, 50–58 CrossRef CAS PubMed.
- J.-M. Moulis, V. Davasse, M.-P. Golinelli, J. Meyer and I. Quinkal, J. Biol. Inorg Chem., 1996, 1, 2–14 CrossRef CAS.
- P. S. Brereton, R. E. Duderstadt, C. R. Staples, M. K. Johnson and M. W. W. Adams, Biochemistry, 1999, 38, 10594–10605 CrossRef CAS PubMed.
- P. M. Vignais and B. Billoud, Chem. Rev., 2007, 107, 4206–4272 CrossRef CAS PubMed.
- J. W. Peters, G. J. Schut, E. S. Boyd, D. W. Mulder, E. M. Shepard, J. B. Broderick, P. W. King and M. W. W. Adams, Biochim. Biophys. Acta, Mol. Cell Res., 2015, 1853, 1350–1369 CrossRef CAS PubMed.
- T. Goris, A. F. Wait, M. Saggu, J. Fritsch, N. Heidary, M. Stein, I. Zebger, F. Lendzian, F. A. Armstrong and B. Friedrich, et al. , Nat. Chem. Biol., 2011, 7, 310–318 CrossRef CAS PubMed.
- K. Karstens, S. Wahlefeld, M. Horch, M. Grunzel, L. Lauterbach, F. Lendzian, I. Zebger and O. Lenz, Biochemistry, 2015, 54, 389–403 CrossRef CAS PubMed.
- H. McTavish, L. A. Sayavedra-Soto and D. J. Arp, J. Bacteriol., 1995, 177, 3960–3964 CrossRef CAS PubMed.
- P. V. Warren, L. B. Smart, L. McIntosh and J. H. Golbeck, Biochemistry, 1993, 32, 4411–4419 CrossRef CAS PubMed.
- A. T. Kowal, M. T. Werth, A. Manodori, G. Cecchini, I. Schroeder, R. P. Gunsalus and M. K. Johnson, Biochemistry, 1995, 34, 12284–12293 CrossRef CAS PubMed.
- I. T. Yonemoto, B. R. Clarkson, H. O. Smith and P. D. Weyman, BMC Biochem., 2014, 15, 10 CrossRef PubMed.
- Y.-S. Jung, I. R. Vassiliev, J. Yu, L. McIntosh and J. H. Golbeck, J. Biol. Chem., 1997, 272, 8040–8049 CrossRef CAS PubMed.
- B. Guigliarelli and P. Bertrand, Adv. Inorg. Chem., 1999, 47, 421–497 CrossRef CAS.
- I. Czech, A. Silakov, W. Lubitz and T. Happe, FEBS Lett., 2010, 584, 638–642 CrossRef CAS PubMed.
- C. Sommer, A. Adamska-Venkatesh, K. Pawlak, J. A. Birrell, O. Rüdiger, E. J. Reijerse and W. Lubitz, J. Am. Chem. Soc., 2017, 139, 1440–1443 CrossRef CAS PubMed.
- S. Morra, A. Giraudo, G. D. Nardo, P. W. King, G. Gilardi and F. Valetti, PLoS One, 2012, 7, e48400 CAS.
- F. A. Armstrong, R. M. Evans, S. V. Hexter, B. J. Murphy, M. M. Roessler and P. Wulff, Acc. Chem. Res., 2016, 49, 884–892 CrossRef CAS PubMed.
- M. P. Golinelli, N. H. Chmiel and S. S. David, Biochemistry, 1999, 38, 6997–7007 CrossRef CAS PubMed.
- T. Mehari, F. Qiao, M. P. Scott, D. F. Nellis, J. Zhao and D. A. Bryant, et al. , J. Biol. Chem., 1995, 270, 28108–28117 CrossRef CAS PubMed.
- B. Shen, D. R. Jollie, T. C. Diller, C. D. Stout, P. J. Stephens and B. K. Burgess, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 10064–10068 CrossRef CAS.
- D. W. Mulder, E. S. Boyd, R. Sarma, R. K. Lange, J. A. Endrizzi, J. B. Broderick and J. W. Peters, Nature, 2010, 465, 248–251 CrossRef CAS PubMed.
- J. A. Birrell, K. Wrede, K. Pawlak, P. Rodriguez-Maciá, O. Rüdiger, E. J. Reijerse and W. Lubitz, Isr. J. Chem., 2016, 56, 852–863 CrossRef CAS.
- M. Senger, S. Mebs, J. Duan, F. Wittkamp, U.-P. Apfel, J. Heberle, M. Haumann and S. T. Stripp, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 8454–8459 CrossRef CAS PubMed.
- A. Abou Hamdan, S. Dementin, P.-P. Liebgott, O. Gutierrez-Sanz, P. Richaud, A. L. De Lacey, M. Rousset, P. Bertrand, L. Cournac and C. Léger, J. Am. Chem. Soc., 2012, 134, 8368–8371 CrossRef CAS PubMed.
- B. J. Murphy, F. Sargent and F. A. Armstrong, Energy Environ. Sci., 2014, 7, 1426–1433 CAS.
- J. M. Kuchenreuther, C. S. Grady-Smith, A. S. Bingham, S. J. George, S. P. Cramer and J. R. Swartz, PLoS One, 2010, e15491 Search PubMed.
- M. K. Akhtar and P. R. Jones, Appl. Microbiol. Biotechnol., 2008, 78, 853–862 CrossRef CAS PubMed.
- G. von Abendroth, S. Stripp, A. Silakov, C. Croux, P. Soucaille, L. Girbal and T. Happe, Int. J. Hydrogen Energy, 2008, 33, 6076–6081 CrossRef CAS.
- J. Noth, D. Krawietz, A. Hemschemeier and T. Happe, J. Biol. Chem., 2013, 288, 4368–4377 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- W. W. Fish, Methods Enzymol., 1988, 158, 357–364 CAS.
- E. Reijerse, F. Lendzian, R. Isaacson and W. Lubitz, J. Magn. Reson., 2012, 214, 237–243 CrossRef CAS PubMed.
- A. Hemschemeier, A. Melis and T. Happe, Photosynth. Res., 2009, 102, 523–540 CrossRef CAS PubMed.
- A. Adamska, A. Silakov, C. Lambertz, O. Rüdiger, T. Happe, E. Reijerse and W. Lubitz, Angew. Chem., Int. Ed., 2012, 51, 11458–11462 CrossRef CAS PubMed.
- P. Rodriguez-Maciá, A. Dutta, W. Lubitz, W. J. Shaw and O. Rüdiger, Angew. Chem., Int. Ed., 2015, 54, 12303–12307 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tables showing the mutagenic primers used in this study, the iron content of apoHydA1 and holoHydA1 variants after 1 h and 30 h of in vitro maturation, and a summary of the relevant redox states. Figures showing spectroscopic characterization of apoHydA1 “A” and “D” variants, in vivo maturation and oxidation screening of holoHydA1 variants, annotated FTIR spectra of holoHydA1 variants, comparison of normalized FTIR spectra of holoHydA1 variants after 1 h and 30 h of in vitro maturation, in vitro maturation followed by FTIR over 30 h, and a comparison of cyclic voltammograms of holoHydA1 variants from pH 5 to pH 10. See DOI: 10.1039/c7sc03444j |
| This journal is © The Royal Society of Chemistry 2017 |