Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In vitro apoptosis-inducing effect and gene expression profiles of mixed ligand Cu(II) complexes derived from 4-acyl pyrazolones on human lung cancer cells†
R. N. Jadeja *a,
K. M. Vyasb,
K. K. Upadhyayc and
R. V. Devkarc
*a,
K. M. Vyasb,
K. K. Upadhyayc and
R. V. Devkarc
aDepartment of Chemistry, Faculty of Science, The M. S. University of Baroda, Vadodara 390 002, India. E-mail: rajendra_jadeja@yahoo.com; rjadeja-chem@msubaroda.ac.in; Tel: +91-265-2795552 ext 30
bDiscipline of Chemistry, School of Basic Sciences, Indian Institute of Technology (IIT) Indore, Indore 452 017, Madhya Pradesh, India
cDivision of Phytotherapeutics and Metabolic Endocrinology, Faculty of Science, The M.S. University of Baroda, Vadodara 390 002, Gujarat, India
First published on 17th March 2017
Abstract
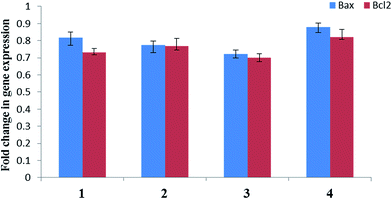
In order to study the less understood anti-tumor mechanism of pyrazolone based mixed ligand complexes, four Cu(II) mixed ligand complexes [Cu(L1)(Phen)NO3] (1), [Cu(L2)(Bipy)NO3] (2), [Cu(L2)(Phen)NO3] (3) and [Cu(L2)(Bipy)NO3] (4) [L1 = 3-methyl-5-oxo-1-phenyl-4,5-dihydro-1H-pyrazole-4-carbaldehyde; L2 = 4-(1-naphthoyl)-3-methyl-1-(p-tolyl)-1H-pyrazol-5(4H)-one] have been synthesized and fully characterized. Single crystal X-ray diffraction analysis of three complexes indicated mononuclear structures exhibiting square pyramidal geometries. The inhibitory effects of the complexes on the cell population growth of the human lung carcinoma (A549) and human lung epithelial cell lines (L132) were determined by MTT assay, DAPI staining and DCFDA staining, which assessed mitochondrial dysfunction, nuclear condensation and intracellular oxidative stress, respectively. The expression levels of Bax (pro-apoptotic) and BCl2 (anti-apoptotic) genes were also studied in A549 cells wherein, both genes showed moderate downregulation after treatment with all the test complexes.
Introduction
Cancer is a life threatening group of diseases, in which deregulated proliferation of cells attacks and disrupts the surrounding tissues, leading to dysfunctional metabolic, angiogenetic and immunologic functions.1 According to World Health Organization (WHO), lung cancer, the most common cause of cancer-related mortality in humans, was responsible for 1.56 million deaths in 2012.2 Surgery, radiotherapy and chemotherapy have been widely used to treat lung cancer. However, decreasing the mortality and improving the life quality of lung cancer patients remain a huge therapeutic challenge.Although a large number of anticancer drugs were launched, their limited success, the drug resistance problems and severe side effects indicate that there is an imperative need of novel strategies for cancer management.3 Thus, increasing efforts are being made towards developing novel such agents, with higher safety margins, possibly targeting novel pathways which exploit the particular biology/biochemistry of tumour cells.4
Metal complexes have been considered as potential source of anticancer drugs for years to overcome toxicity and drug-resistance and to achieve higher selectivity and better activity in comparison with platinum-based therapeutics.5 Platinum-based complexes such as cisplatin, carboplatin and oxaliplatin have been widely used as chemotherapeutic drugs but these drugs always accompany with serious side effects and drug resistance. Thus, it is very important to develop non-platinum based anticancer drugs with high efficacy and low toxicity.1,6,7 Up to now, many transition-metal based complexes have been investigated in the development of non-platinum-based drugs. Amongst all, copper complexes are the best alternative to a group of platinum anticancer drugs with potential application for alternative treatments in tumours e.g., (1) copper complexes with homoscorpionate tridentate tris(pyrazolyl)borate and auxiliary monodentate phosphine ligands have been reported with similar cytotoxicity profile both in cisplatin-sensitive and resistant cell lines.8a (2) Water-soluble bis(1,10-phenanthroline) octanedioate Cu(II) complexes have shown greater in vivo drug tolerance compared to cisplatin,8b etc.
A large number of heterocyclic compounds with five or six member rings have been fruitfully incorporated into new drugs and therapeutic agents.9–11 Pyrazolones, especially, 4-acyl pyrazolones and its derivatives has been widely studied that forms a key pharmacophore in numerous compounds of biological importance.12–22 Attention has been paid to pyrazole-based metal complexes due to the advantages including stability, easy synthesis, easy alteration, and abundant bio-activity.23 These compounds have antibacterial,24,25 antiviral26 and anticancer activity.27,28 Consequently, the design and preparation of novel 4-acyl pyrazolone based metal complexes is still a notable scientific challenge.22 Pt(II) complexes with the pyrazole based ligands have shown relatively high cytotoxic activity against different cell lines, which is comparable with the activity of cisplatin and much higher than the activity of carboplatin.29 Recently, water soluble Pt(II) complexes bearing acyl pyrazolones have been reported with better cytotoxicity compared to cisplatin.30 Acylpyrazolone-Cu(II) had been shown to be able to induce the apoptosis in KB cells and KBv200 cells via reactive oxygen species (ROS)-independent mitochondrial pathway.31
Following our interest in the design and development of new mixed-ligand transition metal complexes with good anti-cancer activity,32–34 four mixed-ligand Cu(II) complexes were synthesized by the reactions of copper nitrate, different 4-acylpyrazolones and polypyridyls as secondary ligands. The complexes were characterized using different analytical and spectroscopic techniques including single crystal studies. Antitumor activity of these complexes were investigated on A549 lung carcinoma cell lines by MTT assay, DAPI staining and DCFDA staining, which suggested that the complexes possess good anticancer property. The expression levels of Bax (pro-apoptotic) and BCl2 (anti-apoptotic) genes were also studied in A549 cells using all the synthesized complexes.
Results and discussion
Synthesis and characterization
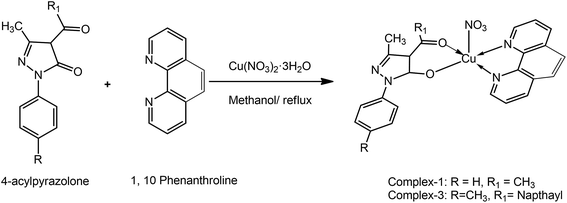
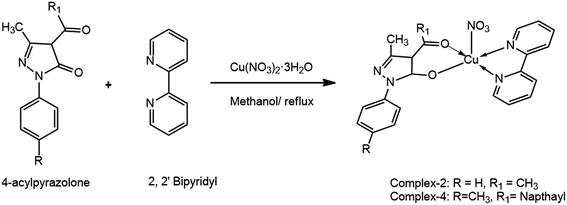
The acyl pyrazolone ligands have been synthesized according to our previously reported procedure.32–34 The NMR and mass spectra of ligands are given in ESI (Fig. S1 and S2†). The complexes have been synthesized using methanolic solution of 4-acyl pyrazolone, 2,2′-bipyridyl/1,10-phenanthrolin and Cu(NO3)2 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio (Schemes 1 and 2). Our primary aim was to get the ternary mixed ligand Cu(II) complexes with pyrazolone as primary ligand and 2,2′-bipyridyl/1,10-phenanthrolin as a secondary ligand. The stoichiometry of the reaction was controlled in such a way that no ML2 complex formation takes place with the either of the ligands. All the complexes are coloured and moisture free crystalline solids. The different spectra of the complexes are given in ESI (Fig. S3–S5†). The complexes are insoluble in water and non-polar solvents, but soluble in MeOH, EtOH, DMF, DMSO, acetonitrile and chlorinated solvents. The 4-acyl pyrazolone ligands act as a bi-dentate O, O donor towards the Cu(II) core. Molar conductance values of the complexes in DMF (10−3 M solution at 25 °C) indicate that the complexes are electrically non-conducting in nature.35 Results of chemical analyses are in good agreement with those of the calculated ones (see Experimental section).
1 molar ratio (Schemes 1 and 2). Our primary aim was to get the ternary mixed ligand Cu(II) complexes with pyrazolone as primary ligand and 2,2′-bipyridyl/1,10-phenanthrolin as a secondary ligand. The stoichiometry of the reaction was controlled in such a way that no ML2 complex formation takes place with the either of the ligands. All the complexes are coloured and moisture free crystalline solids. The different spectra of the complexes are given in ESI (Fig. S3–S5†). The complexes are insoluble in water and non-polar solvents, but soluble in MeOH, EtOH, DMF, DMSO, acetonitrile and chlorinated solvents. The 4-acyl pyrazolone ligands act as a bi-dentate O, O donor towards the Cu(II) core. Molar conductance values of the complexes in DMF (10−3 M solution at 25 °C) indicate that the complexes are electrically non-conducting in nature.35 Results of chemical analyses are in good agreement with those of the calculated ones (see Experimental section).
The FT-IR spectra of complexes exhibit the band in the range 1560–1600 cm−1, which can be assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N (cyclic). All the complexes show absorption in the region 3000–2900 cm−1, which may be due to vC–H. Furthermore, comparison of IR spectra of the ligands with those of their Cu(II) complexes, indicates that the ligand is coordinated to Cu(II) by two sites, that is the ligand is bi-dentate. The band due to vC
N (cyclic). All the complexes show absorption in the region 3000–2900 cm−1, which may be due to vC–H. Furthermore, comparison of IR spectra of the ligands with those of their Cu(II) complexes, indicates that the ligand is coordinated to Cu(II) by two sites, that is the ligand is bi-dentate. The band due to vC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O is completely missing in the spectra of the complexes, suggesting enolization of the ligand on complexation. This is supported by the fact that no band for vOH is observed in the infrared spectra of the ligand and its Cu(II) complex. Instead, a band due to vC–O was observed for the complex, which supports the observation of its enolization during coordination. This fact suggests that the ligand remains in the keto form in the solid state, but in solution both the keto and enol forms remain in equilibrium. Deprotonation occurs from the enol form on complexation. In addition, the new band near 580 cm−1 is assigned to Cu–N bond. The new band in the range 410–440 cm−1 is assigned to Cu–O bond. The wave no. of cyclic C
O is completely missing in the spectra of the complexes, suggesting enolization of the ligand on complexation. This is supported by the fact that no band for vOH is observed in the infrared spectra of the ligand and its Cu(II) complex. Instead, a band due to vC–O was observed for the complex, which supports the observation of its enolization during coordination. This fact suggests that the ligand remains in the keto form in the solid state, but in solution both the keto and enol forms remain in equilibrium. Deprotonation occurs from the enol form on complexation. In addition, the new band near 580 cm−1 is assigned to Cu–N bond. The new band in the range 410–440 cm−1 is assigned to Cu–O bond. The wave no. of cyclic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N does not change in all the complexes, indicates no coordination via C
N does not change in all the complexes, indicates no coordination via C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N cyclic. By comparing the IR spectra of ligands and metal complexes, it is easy to predict that the ligand binds to the metal ion via O and O i.e. bi-dentate manner. The IR spectral data of the ligands and complexes are given in Experimental section.
N cyclic. By comparing the IR spectra of ligands and metal complexes, it is easy to predict that the ligand binds to the metal ion via O and O i.e. bi-dentate manner. The IR spectral data of the ligands and complexes are given in Experimental section.
The visible spectra for the complexes under investigation were measured in DMF. In DMF, the complexes displayed absorbance maxima in the 670–720 nm range. For five-coordinate Cu(II) complexes, this spectral feature is typical for Square Planar (SP) or distorted SP geometries, which generally exhibit a band in the 550–660 nm range (dxz, dyz → dx−y); the geometry around each Cu is SP or distorted SP. In contrast, Trigonal Bi-pyramidal (TBP) Cu(II) complexes usually show a maximum at λ > 800 nm (dxz, dx−y → dz) with a higher energy shoulder. Thus, in DMF, the geometry about the Cu(II) centre in the complexes is closer to SP. These data are consistent with the reported literature for the square pyramidal Cu(II) complexes.36
The synthesized complexes are air stable and have higher thermal stability, which was confirmed by thermo-gravimetric analysis. Up to 200 °C mass loss is not observed, indicating the absence of lattice water molecule in the complexes. The complexes slowly started to decompose within the range of 200–350 °C, corresponds to the loss of nitrate molecules. At temp 340 °C there is a loss of one ligand molecule and then at temp 450 °C the second ligand molecule is decomposed. The final product of the thermal decomposition CuO was determined by elemental analysis.
Crystal structure description of the complexes
The green block shaped single crystals of three of the synthesized complexes (1, 3 and 4) suitable for X-ray diffraction studies, were obtained by slow evaporation in methanol. A suitable crystal was mounted on a glass fibre for data collection at 298(2) K. The crystal structures of the complexes were solved by single-crystal XRD in the space group![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) 1 of the monoclinic system. The crystallographic data of these complexes are listed in Table 1. The important bond lengths and bond angles of the complexes are listed in Table S1 (ESI†). The molecular structures of the synthesized complexes together with the atom-numbering scheme are illustrated in Fig. 1.
1 of the monoclinic system. The crystallographic data of these complexes are listed in Table 1. The important bond lengths and bond angles of the complexes are listed in Table S1 (ESI†). The molecular structures of the synthesized complexes together with the atom-numbering scheme are illustrated in Fig. 1.
| Complex | 1 | 3 | 4 |
|---|---|---|---|
| Chemical formula | C23H17CuN5O5 | C34H26CuN5O5 | C32H25CuN5O5 |
| Form. weight | 506.97 | 648.15 | 623.12 |
| a (Å) | 8.8175(3) | 11.9167(2) | 11.31171(18) |
| b (Å) | 12.8309(4) | 13.7410(2) | 13.8659(3) |
| c (Å) | 19.0229(8) | 18.0275(4) | 18.2058(3) |
| α (°) | 90.00 | 90.00 | 90.00 |
| β (°) | 102.082(4) | 100.6935(18) | 99.5069(17) |
| γ (°) | 90.00 | 90.00 | 90.00 |
| Z | 4 | 4 | 4 |
| V (Å3) | 2104.50(13) | 2900.69(9) | 2816.31(9) |
| Refl. Collected | 8325 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 617 617 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 488 |
| Ind. refl. | 3238 | 4818 | 4690 |
| R(int) | 0.0244 | 0.0235 | 0.0209 |
| No. of param. | 307 | 408 | 390 |
| Crystal system | Monoclinic | Monoclinic | Monoclinic |
| Space group | ![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) 1 1 |
![[P with combining macron]](https://www.rsc.org/images/entities/char_0050_0304.gif) 1 1 |
![[P with combining macron]](https://www.rsc.org/images/entities/char_0050_0304.gif) 1 1 |
| ρcalcd (g cm−3) | 1.6000 | 1.484 | 1.470 |
| Abs coeff, μ (cm−1) | 1.888 | 1.507 | 1.527 |
| F(000) | 1031.3468 | 1137 | 1284 |
| Temp (°C) | 23 | 23 | 23 |
| GOF on F2 | 1.0428 | 1.041 | 0.832 |
| R1/wR2 ([I > 2σ(I)] | 0.0546/0.1643 | 0.0488/0.1368 | 0.0443/0.1740 |
| R1/wR2 (all data) | 0.0656/0.1826 | 0.0553/0.1427 | 0.0504/0.1918 |
| CCDC | 1488810 | 1488811 | 1488812 |
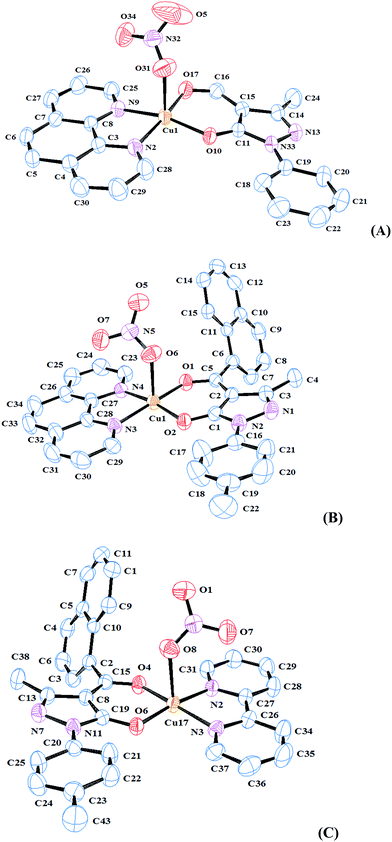 | ||
| Fig. 1 ORTEP views of the (A) complex 1, (B) complex 3 and (C) complex 4 with displacement ellipsoids drawn at 50%. | ||
As theses complexes are pentacoordinated, τ values have been calculated for all complexes. Therefore, we can find out the degree of distortion of the polyhedron from square pyramidal to trigonal bi-pyramidal according to our previous reports.31–33 The τ values are near 0 for all complexes [0.06 for complex 1, 0.001 for complex 3 and 0.07 for complex 4] revealing square pyramidal geometry.
The deviation from a perfect TBP as well as SP structure may be due to the difference in the electro-negativities of all the different coordinating atoms in the equatorial as well as axial sites, resulting in different bond lengths and bond angles. A comparison of the bond lengths of the central metal-donor atoms shows that the two oxygen donors and two nitrogen donors are almost similar in the nature of coordination. This has a net effect that the five bond lengths are almost similar, resulting in SP structure. Similarly, the adjacent five and six membered chelate rings might also be responsible for distortion from either of the ideal geometries.
As shown in Fig. 1, the crystallographically independent Cu(II) ion in all three complexes is pentacoordinated by three nitrogen atoms (two bipy-N/phen-N) and three oxygen donors (two pyrazolone-O and one nitrate-O) in all the three complexes. The coordination sphere is of the type Cu–N2–N9–O10–O17–O31 (complex 1)/Cu–N3–N4–O1–O2–O6 (complex 3)/Cu–N2–N3–O4–O6–O8 (complex 4), exhibiting a geometry close to square pyramidal (SP). The coordination geometry of the complexes can best be described as square pyramidal (SP) with O10, O17, N2 and N9 (complex 1)/O1, O2, N3 and N4 (complex 3)/O4, O6, N2 and N3 (complex 4) forming the plane, and O31 (complex 1)/O6 (complex 3)/O8 (complex 4) occupying axial position, with a slight deviation at the axial site. The crystal packing of complexes 1 and 4 involves weak π–π interactions, which play a decisive role in cohesion of molecules (Table S2†). C–H–π interactions are also present in the crystal structures (Table S3†).
Evaluation of in vitro cytotoxicity-MTT assay
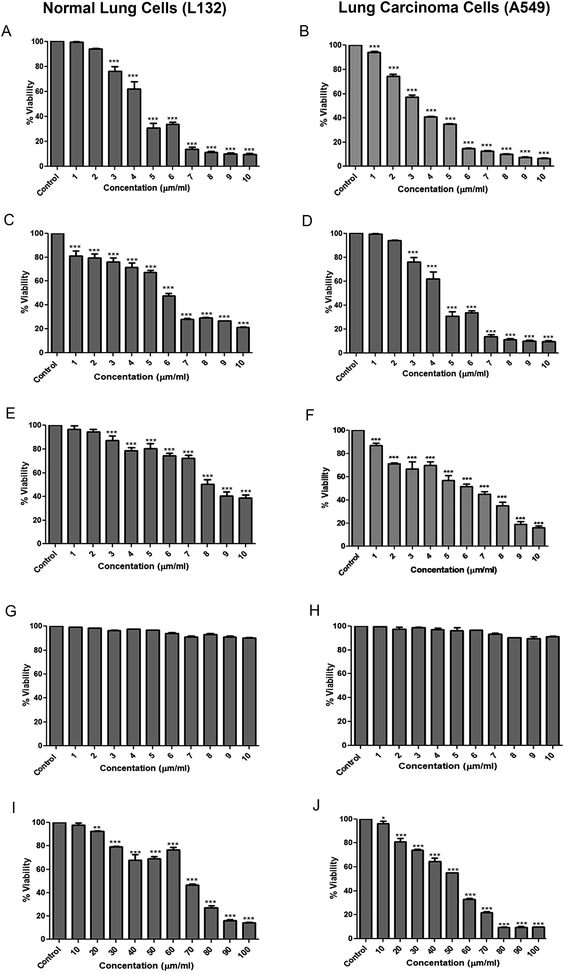
Mitochondrial dysfunctions are a preferred trigger to induce death in cancer cells by test therapeutants and hence, MTT assay is popular and widely used technique for screening of anti-cancer potential of test compounds. Complexes 1–4 were accessed for their possible cytotoxicity against A549 lung carcinoma cells and L132 human lung epithelial cells. MTT produces purple colour formazan in metabolically active cells wherein, the intensity of colour reflects the functional status of mitochondria.37,38 In this study, the rationale behind taking healthy (L132) and cancerous (A549) cell lines was to confirm specificity of the synthesized complexes against cancer cells. Results revealed that complexes 1–3 inhibited growth of A549 cells in dose dependent manner (Fig. 2; IC50 4.67, 6 and 8.2 μm, respectively). Though L132 cells had identical IC50 values, higher doses did not account for further loss of cell viability (Fig. 2). Complex 4 accounted for near normal percentage of viable cells (1–10 μm mL−1) that prompted us for further investigation using a higher dose range (10–100 μm mL−1) that recorded a more pronounced dose dependent cytotoxicity of A549 cells as compared to L132 cells (IC50 50 vs. 70 μm, respectively). Hence, it can be tentatively surmised that complexes 1–4 showed preferential cytotoxicity against A549 cells. These results are prima-facie evidence on selective cytotoxicity potential of metal complexes against cancer cells and therefore qualify them as possible anti-cancer agents.Mechanism of action
Conclusions
To confirm the formation of a new series of pyrazolone based mixed-ligand Cu(II) complexes 1–4, analytical, spectral and structural analyses were carried out. The crystal structure of three of the synthesized complexes showed that the ligands are coordinated to Cu(II) in a bi-dentate manner. The crystallographically independent Cu(II) ion is pentacoordinated by two nitrogen atoms (two bipy-N/phen-N) and three oxygen donors (two pyrazolone-O and one nitrate-O) in all the three complexes exhibiting square pyramidal geometry.The synthesized metal complexes were screened for anti-cancer activities towards human lung carcinoma (A549) and human lung epithelial cells (L132). Anti-cancer property of complexes 1–4 was established through a series of fluorescent staining and gene expression studies. Complexes 1–4 could elevate intracellular oxidative stress, alter nuclear morphology and induce apoptosis possibly via BCl2 independent pathway. These results are encouraging and warrant a detailed in vivo study to establish anti-cancer potential of these complexes. All the results of this study provide guidance for designing structurally modified pyrazolones with specific cytotoxicity towards human lung cancer cells.
Experimental
Materials
All reagents and solvents were purchased from commercial sources and were further purified by the standard methods, wherever necessary. Pyrazolones were obtained from Nutan Dye Chem. Sachin, Surat. Calcium hydroxide and Cu(NO3)2·3H2O were purchased from Sigma-Aldrich. 1,10-Phenanthroline was purchased from LobaChem, Mumbai. 2,2′-Bipyridyl was purchased from SRL Chem. Formyl chloride and 1-naphthoyl chloride were obtained from Shiva Pharmachem. Ltd., Baroda as free gift samples.Human lung carcinoma (A549) and human lung epithelial cell line (L132) were obtained from National Centre for Cell Science, Pune, India. Ethidium bromide (EB), Dulbecco's Modified Eagle Medium (DMEM), Trypsin Phosphate Versene Glucose (TPVG) solution trypsin and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were purchased from HiMedia Laboratories Pvt. Ltd. (Bombay, India). Fetal bovine serum (FBS) was purchased from Biosera (Ringmer, East Sussex UK) and dimethyl sulfoxide (DMSO) was purchased from the Sisco Research Laboratories Pvt. Ltd. (Mumbai, India). Rhodamine 123, 4′,6-diamidino-2-phenylindole (DAPI) and 2′,7′-dichlorofluoresceindiacetate (DCFDA) were purchased from Sigma (Delhi, India). Dulbecco's Modified Eagle Medium (DMEM), Trypsin Phosphate Versene Glucose (TPVG) solution trypsin and methylthiazolyldiphenyl-tetrazolium bromide (MTT) were purchased from HiMedia Laboratories Pvt. Ltd. (Bombay, India). Fetal bovine serum (FBS) was purchased from Biosera (Ringmer, East Sussex UK) and dimethyl sulfoxide (DMSO) was purchased from the Sisco Research Laboratories Pvt. Ltd. (Mumbai, India). Rhodamine 123, 4′,6-diamidino-2-phenylindole (DAPI) and 2′,7′-dichlorofluoresceindiacetate (DCFDA) were purchased from Sigma (Delhi, India).
Characterization techniques
The synthesized compounds were characterized using different techniques. Elemental analyses (C, H, N) of the synthesized compounds were performed on a model 2400 Perkin-Elmer elemental analyzer. Infrared spectra (4000–400 cm−1, KBr discs) of the samples were recorded on a model RX 1 FT-IR Perkin-Elmer spectrophotometer. The electronic spectra (in DMF at room temperature) in the range of 400–800 nm were recorded on a model Perkin Elmer Lambda 35UV-VIS spectrophotometer. LC-MS of the compounds were recorded at SAIF, Panjab University, Chandigarh. A simultaneous TG/DTA was carried out on a SII EXSTAR6000 TG/DTA 6300. The experiments were performed in N2 at a heating rate of 10 °C min−1 in the temperature range 25–750 °C using a platinum pan. Molar conductivity of 10−3 M solutions of the complexes in DMF was measured at room temperature with a model Elico CM 180 digital direct reading deluxe digital conductivity meter. Gravimetric and volumetric analysis were performed for the determination of copper after decomposition of complexes in nitric acid.Preparation of 4-acyl pyrazolones
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH
MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 1
H2O = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80
80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19).
19).L1 is brown crystalline solid. Yield 82.45% mp 95 °C. Anal. calc. for C11H10N2O2 MW: 202.21, C (65.34%), H (4.98%), N (13.85%), found: C (65.76%), H (4.68%), N (13.59%); 1H NMR (CDCl3): δ ppm 2.43 (s, 3H, PZ C–CH3), 7.28–7.81 (m, 5H, Ph), 9.53 (s, 1H, formyl). IR (KBr, cm−1): 1527 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, cyclic), 1636 (m) (C
N, cyclic), 1636 (m) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, pyrazolone ring); MS: m/z = 202.14 [C11H11N2O2]+, 201.32 [M − 1]+, 185.29, [C10H7N2O2]+, 128.51 [C5H6N2O2]+.
O, pyrazolone ring); MS: m/z = 202.14 [C11H11N2O2]+, 201.32 [M − 1]+, 185.29, [C10H7N2O2]+, 128.51 [C5H6N2O2]+.
L2 is brown crystalline solid. Yield 71.20% mp 110 °C. Anal. calc. for C22H18N2O2 MW: 342.39, C (77.17%), H (5.30%), N (8.18), found: C (77.02%), H (5.45%), N (8.41%); 1H NMR (CDCl3): δ ppm 1.66 (s, 3H, PZ C–CH3), 2.42 (s, 3H, N-TL C–CH3), 7.28–8.03 (m, 11H, Ph & naphthyl). IR (KBr, cm−1): 1554 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, cyclic), 1607 (m) (C
N, cyclic), 1607 (m) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, pyrazolone ring); MS: m/z = 342.54 [C22H18N2O2]+, 341.22 [M − 1]+, 213.32 [C12H10N2O2]+, 342.54 [C22H18N2O2]+, 127.27 [C5H6N2O2]+.
O, pyrazolone ring); MS: m/z = 342.54 [C22H18N2O2]+, 341.22 [M − 1]+, 213.32 [C12H10N2O2]+, 342.54 [C22H18N2O2]+, 127.27 [C5H6N2O2]+.
[Cu(L1)(Phen)]NO3 (1). To the solution of Cu(NO3)2·3H2O (1 mmol, 0.241 g) in methanol (5 mL), a solution of L1 (1 mmol, 0.202 g) in methanol (10 mL) was added slowly while stirring. To this, a solution of 1,10-phenanthroline (1 mmol, 0.198 g) in methanol (5 mL) was added, followed by small amount of Na-acetate. The resultant mixture was refluxed for 4 h. The solid green crystalline product obtained was filtered off, washed with hot water followed by methanol and dried. Synthesis of complex 1 is summarized in Scheme 1.
Green; yield 76.89%, mp > 250 °C. Anal. calc. for C23H19CuN5O5: MW: 508.97, C (58.28%), H (3.76%), N (13.76%), Cu (12.49%), found: C (58.35%), H (3.56%), N (13.89%), Cu (12.67%). IR (KBr, cm−1): 1566 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, cyclic), 1610 (m) (C
N, cyclic), 1610 (m) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 585 (s) (Cu–N), 422 (s) (Cu–O); λmax/nm 670 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 11.00.
O), 585 (s) (Cu–N), 422 (s) (Cu–O); λmax/nm 670 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 11.00.
[Cu(L1)(Bipy)]NO3 (2). It was prepared analogously from L1, Cu(NO3)2·3H2O and 2,2′-bipyridyl. The solid green crystalline product obtained was filtered off, washed with methanol and dried. Synthesis of complex 2 is summarized in Scheme 2.
Green; yield 68.99%, mp > 250 °C. Anal. calc. for C22H21CuN5O5: MW: 498.98, C (52.96%), H (4.24%), N (14.04%), Cu (12.74%), found: C (52.76%), H (4.58%), N (13.98%), Cu (12.83%). IR (KBr, cm−1): 1584 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, cyclic), 1519 (m) (C–O), 592 (s) (Cu–N), 432 (s) (Cu–O); λmax/nm 673 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 11.00.
N, cyclic), 1519 (m) (C–O), 592 (s) (Cu–N), 432 (s) (Cu–O); λmax/nm 673 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 11.00.
[Cu(L2)(Phen)]NO3 (3). It was prepared analogously from L2, Cu(NO3)2·3H2O and 1,10-phenanthroline. The solid green crystalline product obtained was filtered off, washed with methanol and dried. Synthesis of complex 3 is summarized in Scheme 1.
Green; yield 73.45%, mp > 250 °C. Anal. calc. for C34H27CuN5O5: MW: 649.15, C (62.91%), H (4.19%), N (10.79%), Cu (9.79%), found: C (62.60%), H (4.46%), N (10.50%), Cu (9.80%). IR (KBr, cm−1): 1599 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, cyclic), 1487 (m) (C–O), 583 (s) (Cu–N), 412 (s) (Cu–O); λmax/nm 672 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 10.00.
N, cyclic), 1487 (m) (C–O), 583 (s) (Cu–N), 412 (s) (Cu–O); λmax/nm 672 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 10.00.
[Cu(L2)(Bipy)]NO3 (4). It was prepared analogously from L2, Cu(NO3)2·3H2O and 2,2′-bipyridyl. The solid green crystalline product obtained was filtered off, washed with methanol and dried. Synthesis of complex 4 is summarized in Scheme 2.
Green; yield 72.38%, mp > 250 °C. Anal. calc. for C32H27CuN5O5: MW: 625.13, C (61.48%), H (4.35%), N (11.20%), Cu (10.17%), found: C (61.78%), H (4.54%), N (11.36%), Cu (10.89%). IR (KBr, cm−1): 1601 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, cyclic), 1472 (m) (C–O), 581 (s) (Cu–N), 417 (s) (Cu–O); λmax/nm 670 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 12.00; MS: m/z = 562 [C32H27N4O2]+, 561 [M − 1]+, 563 [M + 1]+.
N, cyclic), 1472 (m) (C–O), 581 (s) (Cu–N), 417 (s) (Cu–O); λmax/nm 670 (dxz, dyz → dx2−y2, square pyramidal); molar conductance (DMF, Ω−1 cm2 mol−1): 12.00; MS: m/z = 562 [C32H27N4O2]+, 561 [M − 1]+, 563 [M + 1]+.
Single crystal X-ray structure determination
Crystals having good morphology were chosen for three-dimensional intensity data collection. X-ray intensity data of some of the complexes were collected at room temperature on X'calibur CCD area-detector diffractometer equipped with graphite monochromated MoKα radiation (λ = 0.71073 Å). The crystals used for data collection was of suitable dimensions. The unit cell parameters were determined by least-square refinements of all reflections in both cases. All the structures were solved by direct method and refined by full-matrix least squares on F2. Data were corrected for Lorentz, polarization and multi-scan absorption correction.43 The structures were solved by direct methods using SHELXS97.44 All non-hydrogen atoms of the molecule were located in the best E-map. Full-matrix least-squares refinement was carried out using SHELXL97.44 Hydrogen atoms were placed at geometrically fixed positions and allowed to ride on the corresponding non-H atoms with C–H = 0.93–0.96 Å, and Uiso = 1.5Ueq of the attached C atom for methyl H atoms and 1.2Ueq for other H atoms. Atomic scattering factors were taken from International Tables for X-ray Crystallography (1992, Vol. C, Tables 4.2.6.8 and 6.1.1.4). An ORTEP45 view of three complexes with atomic atomic labelling is shown in Fig. 1. The geometry of the molecule has been calculated using the software PLATON46 and PARST.47Cell culture
Human lung carcinoma (A549) and human lung epithelial cells (L132) were seeded in T25 flasks (1 × 105 cells per flask) and maintained at 37 °C with 5% CO2 in water jacketed incubator (Thermo scientific, forma series II 3111, USA). Dulbecco's minimum eagle's medium (DMEM; with 10% FBS and 1% antibiotic-antimycotic solution) was used for cell culture. Cells were sub-cultured every third day by trypsinization with TPVG solution. All reagents were filtered through 0.22 μ filter purchased from Millipore Biomedical Aids Pvt. Ltd, Pune. Metal complexes 1–4 were solubilised in DMSO in a way that the total content of DMSO does not exceed 1%.Cytotoxicity studies
Cell viability was determined by MTT assay. A549 and L132 cells (7 × 103 cells per well) were maintained in 96-well culture plates as mentioned above for 24 h in absence or presence of metal complexes. Later, 10 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT, 5 mg mL−1) was added to each wells and incubated for 4 h at 37 °C. After that wells were washed with Phosphate Buffer Saline. Resulted formazan is dissolved in 150 μL per well with DMSO and measured at 540 nm using ELX800 Universal Microplate Reader.Determination of reactive oxygen species (DCFDA staining)
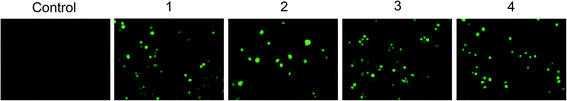
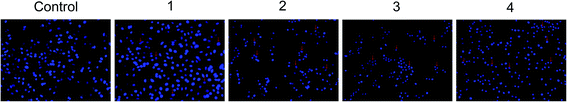
A549 cells were cultured as mentioned earlier and after 18 h of treatment with metal complexes, cells were incubated with 10 μM 2,7-dichlorodihydrofluoroscein diacetate (CM-H2-DCFDA) at 37 °C for 30 min. Control and treated cells were observed and photographed on EvosFloid cell imaging station (Life Technologies, USA).Determination of nuclear morphology (DAPI staining)
A549 cells (5 × 105 cells per well) were plated in a 6-well plate. Later, cells were treated with or without metal complexes at 37 °C for 24 h, washed with PBS and fixed with 70% ethanol, re-washed with PBS and incubated with DAPI (1 μg mL−1 in PBS) for 10 min. Cells were examined to assess changes in nuclear morphology (condensed/fragmented nuclei) and photographed on EvosFloid cell imaging station (Life Technologies, USA).Gene expression studies
Total RNA was isolated from the A549 cells using TRIzol reagent (Invitrogen). The quantity and quality of the RNA was assessed by their absorbance at 260 and 280 nm. 1 μg of total RNA was reverse-transcribed using iScriptcDNA Synthesis Kit (Bio-Rad). The primer sequences used to amplify the genes were as follows: forward primer 5′-GGCCCTTTTGCTTCAGGGTT-3′ and reverse primer 5′-GAAAAAGGCTCACCGTCGA-3′ Bax; forward primer 5′-GTCATGTGTGTGGAGAGCGT-3′ and reverse primer 5′-GGAAACACCTTGACATGCCG-3′ BCl2; forward primer 5′-GCTCTCTGCTCCTCCTGTTC-3′ and reverse primer 5′-CAGTTCCGACTCTTGCCCT-3′ GAPDH. Amplification was performed using Thermo Scientific DremTaq green PCR master mix according to the manufacturer's instructions in a thermal cycler T100 (Bio-Rad). Data were normalized to the internal control GAPDH.Statistical analysis
Data was analyzed for statistical significance using one way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison test and results were expressed as mean ± SEM using Graph Pad Prism version 3.0 for Windows, Graph Pad Software, San Diego, California, USA.Acknowledgements
Financial assistance received from the University Grants Commission in terms of major research project to RNJ, New Delhi is gratefully acknowledged. We would like to thank Head, Department of Chemistry providing necessary facilities. RVD acknowledges research funding from Gujarat Council on Science and Technology (GUJCOST/MRP/2014-15/2553) for conducting biological studies. The help from Dr. Hemant Mande & Prof. Vivek Gupta in crystallographic data are gratefully acknowledged.References
-
(a) P. C. McDonald, S. C. Chafe and S. Dedhar, Front. Cell Dev. Biol., 2016, 4, e27 CrossRef PubMed
; (b) D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed
; (c) C. T. Supuran, J. Clin. Oncol., 2012, 3, 98–103 Search PubMed
; (d) N. M. A. Gawad, N. H. Amin, M. T. Elsaadi, F. M. M. Mohamed, A. Angeli, V. De Luca, C. Capasso and C. T. Supuran, Bioorg. Med. Chem., 2016, 24, 3043–3051 CrossRef PubMed
.
-
(a) B. W. Stewart and C. C. Wild, International Agency for Research on, O. World Health, World cancer report 2014 Search PubMed
; (b) G. Ma, W. Luo, J. Lu, D.-L. Ma, C.-H. Leung, Y. Wang and X. Chen, Chem.-Biol. Interact., 2016, 253, 1–9 CrossRef CAS PubMed
.
-
(a) C. T. Supuran and A. Scozzafava, Expert Opin. Ther. Pat., 2004, 14, 25 Search PubMed
; (b) J. Y. Winum, A. Maresca, F. Carta, A. Scozzafava and C. T. Supuran, Chem. Commun., 2012, 8177 RSC
; (c) A. Casini, A. Scozzafava and C. T. Supuran, Expert Opin. Ther. Pat., 2002, 12, 1307 CrossRef CAS
.
-
(a) T. Ma, D. A. Fuld, J. R. Rigas, A. E. Hagey, G. B. Gordon, E. Dmitrovsky and K. H. Dragnev, Chemotherapy, 2012, 58, 321 CrossRef CAS PubMed
; (b) N. Touisni, A. Maresca, P. C. McDonald, Y. Lou, A. Scozzafava, S. Dedhar, J. Y. Winum and C. T. Supuran, J. Med. Chem., 2011, 54, 8271 CrossRef CAS PubMed
.
-
(a) P. F. Liguori, A. Valentini, M. Palma, A. Bellusci, S. Bernardini, M. Ghedini, M. L. Panno, C. Pettinari, F. Marchetti, A. Crispini and D. Pucci, J. Chem. Soc., Dalton Trans., 2010, 39, 4205 RSC
; (b) N. S. Rukk, L. G. Kuzmina, D. V. Albov, R. S. Shamsiev, S. N. Mudretsova, G. A. Davydova, V. M. Retivov, P. A. Volkov, V. V. Kravchenko, G. N. Apryshko, A. N. Streletskii, A. Yu. Skryabina, E. A. Mironova and V. V. Zamalyutin, Polyhedron, 2015, 102, 152–162 CrossRef CAS
.
- A. M. Evangelou, Crit. Rev. Oncol. Hematol., 2002, 42, 249–265 CrossRef PubMed
.
-
(a) M. Jamshidi, R. Yousefi, S. M. Nabavizadeh, M. Rashidi, M. G. Haghighi, A. Niazi and A. A. Moosavi-Movahedi, Int. J. Biol. Macromol., 2014, 66, 86–96 CrossRef CAS PubMed
; (b) J. Zhao, L. Zhang, J. Li, T. Wua, M. Wanga, G. Xu, F. Zhang, L. Liu, J. Yang and S. Sun, Chem.-Biol. Interact., 2015, 231, 1–9 CrossRef CAS PubMed
.
-
(a) V. Gandin, F. Tisato, A. Dolmella, M. Pellei, C. Santini, M. Giorgetti, C. Marzano and M. Porchia, J. Med. Chem., 2014, 57, 4745–4760 CrossRef CAS PubMed
; (b) A. Kellett, M. O'Connor, M. McCann, O. Howe, A. Casey, P. McCarron, K. Kavanagh, M. McNamara, S. Kennedy, D. D. May, P. S. Skell, D. O'Shea and M. Devereux, MedChemComm, 2011, 2, 579–584 RSC
.
- A. Kumar and R. Kumar, Int. Res. J. Pharm., 2011, 2, 11–12 RSC
.
- N. B. Patel and F. M. Shaikh, Sci. Pharm., 2010, 78, 753–765 CrossRef CAS PubMed
.
-
(a) S. Bala, S. Kamboj and A. Kumar, J. Pharm. Res., 2010, 3, 2993–2997 CAS
; (b) A. Abbas, S. Murtaza, M. N. Tahir, S. Shamim, M. Sirajuddin, U. A. Rana, K. Naseem and H. Rafique, J. Mol. Struct., 2016, 1117, 269–275 CrossRef CAS
.
- N. Raman, A. Kulandaisamy, A. Shunmugasundaram and K. Jeyasubr-amanian, Transition Met. Chem., 2001, 26, 131–135 CrossRef CAS
.
- T. Yoshikuni, J. Mol. Catal. A: Chem., 1999, 148, 285–288 CrossRef CAS
.
- B. A. Uzoukwu, P. U. Adiukwu, S. S. Al-Juaid, P. B. Hitchcock and J. D. Smith, Inorg. Chim. Acta, 1996, 250, 173–176 CrossRef CAS
.
- Z. Y. Yang, R. D. Yang, F. S. Li and K. B. Yu, Polyhedron, 2000, 19, 2599–2604 CrossRef CAS
.
- W. F. Yang, S. G. Yuan, Y. B. Xu, Y. H. Xiao and K. M. Fang, J. Radioanal. Nucl. Chem., 2003, 256, 149–152 CrossRef CAS
.
- P. Chiba, W. Holzer, M. Landau, G. Bechmann, K. Lorenz, B. Plagens, M. Hitzler, E. Richter and G. Ecker, J. Med. Chem., 1998, 41, 4001–4011 CrossRef CAS PubMed
.
- G. C. Xu, L. Zhang, L. Liu, G. F. Liu and D. Z. Jia, Polyhedron, 2008, 27, 12–24 CrossRef CAS
.
- X. H. Wang, D. Z. Jia, Y. J. Liang, S. L. Yan, Y. Ding, L. M. Chen, Z. Shi, M. S. Zeng, G. F. Liu and L. W. Fu, Cancer Lett., 2007, 249, 256–270 CrossRef CAS PubMed
.
- R. N. Jadeja, M. Chhatrola and V. K. Gupta, Polyhedron, 2013, 63, 117–126 CrossRef CAS
.
- K. M. Vyas, R. N. Jadeja, J. Patel and R. V. Gupta, Polyhedron, 2013, 65, 262–274 CrossRef CAS
.
-
(a) B. Shaabani, A. A. Khandar, M. Dusek, M. Pojarova and K. Mahmoudi, Inorg. Chim. Acta, 2013, 394, 563–568 CrossRef CAS
; (b) Y. Li, J. Zhaod, C.-C. He, L. Zhanga, S.-R. Sun and G.-C. Xu, J. Inorg. Biochem., 2015, 150, 28–37 CrossRef CAS PubMed
.
- M. F. Brana, A. Gradillas, A. G. Ovalles, B. Lopez, N. Acero, F. Llinares and D. M. Mingarro, Bioorg. Med. Chem., 2006, 14, 9–16 CrossRef CAS PubMed
.
- D. Castagnolo, F. Manetti, M. Radi, B. Bechi, M. Pagano, A. De Logu, R. Meleddu, M. Saddi and M. Botta, Bioorg. Med. Chem., 2009, 17, 5716–5721 CrossRef CAS PubMed
.
- A. E. Hassan, A. H. Moustafa, M. M. Tolbah, H. F. Zohdy and A. Z. Haikal, Nucleic Acids, 2012, 31, 783–800 CrossRef CAS PubMed
.
- A. Kimata, H. Nakagawa, R. Ohyama, T. Fukuuchi, S. Ohta, T. Suzuki and N. Miyata, J. Med. Chem., 2007, 50, 5053–5056 CrossRef CAS PubMed
.
- X. H. Wang, X. K. Wang, Y. J. Liang, Z. Shi, J. Y. Zhang, L. M. Chen and L. W. Fu, Chin. J. Cancer, 2010, 29, 980–987 CrossRef CAS PubMed
.
- M. H. Norman, L. Liu, M. Lee, N. Xi, I. Fellows, N. D. D'Angelo, C. Dominguez, K. Rex, S. F. Bellon, T. S. Kim and I. Dussault, J. Med. Chem., 2012, 55, 1858–1867 CrossRef CAS PubMed
.
-
(a) E. Budzisz, I.-P. Lorenz, P. Mayer, P. Paneth, L. Szatkowski, U. Krajewska, M. Rozalskid and M. Miernicka, New J. Chem., 2008, 32, 2238–2244 RSC
; (b) J. S. Casas, E. E. Castellano, J. Ellena, M. S. Garcıa-Tasende, M. L. Pérez-Parallé, A. Sánchez, A. Sánchez-González, J. Sordo and A. Touceda, J. Inorg. Biochem., 2008, 102, 33–45 CrossRef CAS PubMed
; (c) E. Budzisz, M. Malecka, B. K. Keppler, V. B. Arion, G. Andrijewski, U. Krajewska and M. Rozalski, Eur. J. Inorg. Chem., 2007, 3728–3735 Search PubMed
.
- S. A. De Pascali, D. Migoni, M. Monari, C. Pettinari, F. Marchetti, A. Muscella and F. P. Fanizzi, Eur. J. Inorg. Chem., 2014, 1249–1259 CrossRef CAS
.
- X. H. Wang, D. Z. Jia, Y. J. Liang, S. L. Yan, Y. Ding, L. M. Chen, Z. Shi, M. S. Zeng, G. F. Liu and L. W. Fu, Cancer Lett., 2007, 249, 256–270 CrossRef CAS PubMed
.
- K. M. Vyas, R. V. Devkar, A. Prajapati and R. N. Jadeja, Chem.-Biol. Interact., 2015, 240, 250–266 CrossRef CAS PubMed
.
- K. M. Vyas, R. N. Jadeja, D. Patel, R. V. Devkar and V. K. Gupta, Polyhedron, 2014, 80, 20–33 CrossRef CAS
.
- K. M. Vyas, R. N. Jadeja, D. Patel, R. V. Devkar and V. K. Gupta, Polyhedron, 2013, 65, 262–274 CrossRef CAS
.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81–122 CrossRef CAS
.
- S. S. Massoud, F. A. Mautner, M. A. M. Abu-Youssef and N. M. Shuaib, Polyhedron, 1999, 18, 2061–2067 CrossRef CAS
.
- D. Madamwar, D. K. Patel, S. N. Desai, K. K. Upadhyay and R. V. Devkar, EXCLI J., 2015, 14, 527–539 Search PubMed
.
- M. C. Thounaojam, R. N. Jadeja, M. Valodkar, P. S. Nagar, R. V. Devkar and S. Thakore, Food Chem. Toxicol., 2011, 49(11), 2990–2996 CrossRef CAS PubMed
.
- H. Kim, G. R. Lee, J. Kim, J. Y. Baek, Y. J. Jo, S. E. Hong and H. M. Kim, Free Radical Biol. Med., 2016, 91, 264–274 CrossRef CAS PubMed
.
- T. W. Sedlak, Z. N. Oltvai, E. Yang, K. Wang, L. H. Boise, C. B. Thompson and S. J. Korsmeyer, Proc. Natl. Acad. Sci. U. S. A., 1995, 92(17), 7834–7838 CrossRef CAS
.
- J. M. Adams and S. Cory, Science, 1998, 281, 1322–1326 CrossRef CAS PubMed
.
- B. Rostkowska-Nadolska, M. Fraczek, W. Gawron and M. Latocha, Acta Biochim. Pol., 2009, 56, 235–242 CAS
.
- Bruker, SMART (Version 5.631), SAINT (Version 6.45) and SADABS (Version 2.05), Bruker AXS Inc., Madison, Wisconsin, USA, 2003 Search PubMed
.
- G. M. Sheldrick, SHELXS97 and SHELXL97, University of Gottingen, Germany, 1997 Search PubMed
.
- C. K. Johnson, ORTEP II; Report ORNL-5138, Oak Ridge National Laboratory, Oak Ridge, TN, 1976 Search PubMed
.
- A. L. Spek, PLATON for Windows. September 1999 Version, University of Utrecht, Netherlands, 1999 Search PubMed
.
- M. Nardelli, J. Appl. Crystallogr., 1995, 28, 659 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Tables S1–S3 and Fig. S1–S5. CCDC 1488810–1488812 contain the supplementary crystallographic data for complexes 1, 3 and 4, respectively. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ra01025g |
| This journal is © The Royal Society of Chemistry 2017 |