Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enzymatic synthesis and reverse transcription of RNAs incorporating 2′-O-carbamoyl uridine triphosphate†
Yoshiaki
Masaki
,
Hyugo
Ito
,
Yuki
Oda
,
Kazufumi
Yamazaki
,
Nobuhiro
Tago
,
Kentaro
Ohno
,
Nozomi
Ishii
,
Hirosuke
Tsunoda
,
Takashi
Kanamori
,
Akihiro
Ohkubo
,
Mitsuo
Sekine
and
Kohji
Seio
*
Department of Life Science and Technology, Tokyo Institute of Technology, J2-16, 4259 Nagatsuta-cho, Midori-ku, Yokohama, Japan. E-mail: kseio@bio.titech.ac.jp
First published on 29th September 2016
Abstract
Enzymatic synthesis and the reverse transcription of RNAs containing 2′-O-carbamoyl uridine were evaluated. A mild acidic deprotection procedure allowed the synthesis of 2′-O-carbamoyl uridine triphosphate (UcmTP). UcmTP was incorporated correctly into long RNAs, and its fidelity during reverse transcription using SuperScript III was sufficient for RNA aptamer selection.
Chemical modifications on RNA and DNA aptamers improve their biostability and affinity with targets of interest.1–11 To introduce modifications, chemical or enzymatic synthesis of RNA aptamers is utilized. Chemical synthesis can introduce a wide variety of chemical modifications into RNA aptamers that are selected using canonical nucleoside triphosphates (NTPs). However, because the chemical modifications sometimes diminish the activity of the initial unmodified RNA aptamer, optimal chemical modifications should be identified after structure–activity analyses for many synthetic RNA aptamer derivatives. On the other hand, because enzymatic synthesis is compatible with RNA aptamer selection, the selected aptamers are expected to have an optimized structure without additional optimization. To expand the usefulness of this enzymatic synthesis strategy, it is important to develop modified nucleosides that can be substrates of the polymerases used in the aptamer selection procedure.
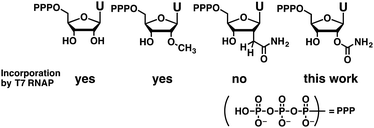
Among various chemical modifications,2,10,12 those at the 2′-position are promising because they can improve the resistance of oligonucleotides toward both enzymatic and chemical degradation.13–16 In addition, 2′-modifications can change RNA structure flexibility,17 hydration,18–20 and base pairing selectivity.21 Despite their importance, only a limited number of 2′-modified NTPs can be applied for enzymatic syntheses.10 For example, natural T7 RNA polymerase or the Y639F mutant22 can incorporate modified nucleosides having single heavy atom substituents at the 2′-position such as 2′-fluoro,23 2′-amino,23 and 2′-thiol.24 The Y639F mutant allows incorporation of larger 2′-O-methyl25 or 2′-hydroxymethyl groups26 that have two heavy atoms at the 2′-position. In the case of substituents having three heavy atoms, incorporation of 2′-azido nucleosides by Y639F/H784A T7 RNA polymerase is reported.27 However, incorporation of a 2′-C-carbamoylmethyl group, which is a four heavy-atom substituent, was unsuccessful (Fig. 1).26
Here, we report that a new NTP having a 2′-O-carbamoyl group, which is a four heavy-atom substituent, can be incorporated into RNA by a natural T7 RNA polymerase. Previously, we reported the chemical synthesis of RNA containing 2′-O-carbamoyl uridine (Ucm). The introduction of Ucm improved nuclease resistance and unexpectedly enhanced G–U base pair formation.14,21 Molecular dynamics simulations suggested the additional hydrogen bonding of a 2′-O-carbamoyl moiety with the 2-amino group on guanosine (Fig. S1–S3, ESI†).17,21,28–33 This unique effect of Ucm might facilitate the formation of various RNA secondary structures including G–U mismatches. Here, we report the synthesis of 2′-O-carbamoyl uridine triphosphate (UcmTP) and its applicability toward aptamer selection.
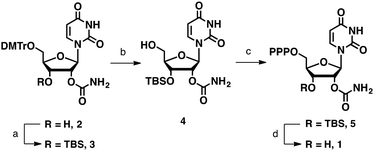
Although the 2′-O-carbamoyl group remains intact under neutral or weakly acidic conditions, it migrates to the adjacent 3′-hydroxyl group under basic conditions such as 28% ammonium hydroxide treatment (Fig. S4, ESI†). Thus, for the synthesis of UcmTP (Scheme 1), we protected the 3′-hydroxyl group with tert-butyldimethylsilyl ether (TBS) and removed this group under non-basic conditions. 5′-O-Dimethoxytrityl-2′-O-carbamoyl uridine (2) was synthesized by following a synthetic scheme reported previously.21 After TBS-protection to give 3 and dimethoxytrityl-deprotection from 3, the 5′-free derivative 4 was synthesized. Subsequently, the 5′-triphosphate moiety of 5 was introduced by Eckstein's method.34 Then, removal of the TBS group of 5 was tested using fluoride sources such as tetrabutylammonium fluoride and triethylammonium trifluoride. However, they gave only complexed mixtures (see Table S1 for detailed conditions, ESI†).
Previously, Kawahara et al. found that the 2′-O-TBS group on oligoribonucleotides could be removed by an 8% acetic acid aqueous solution because of the neighboring effect of the internucleotidic phosphate.35 Thus, we speculated that the 3′-O-TBS group on NTPs could also be deprotected under weakly acidic conditions because of the presence of the 5′-triphosphate. After screening various weakly acidic conditions, we found that pH 3.0 citrate buffer was suitable for deprotection of the TBS group (Fig. S5, ESI†). Under this mild condition, we could synthesize UcmTP (1) in 78% yield without any detectable migration product.
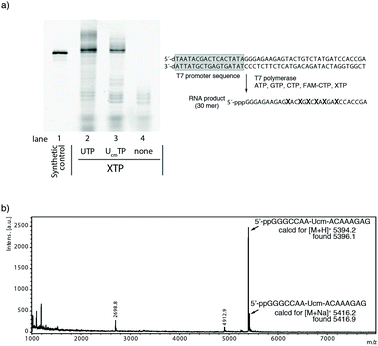
Initial transcription experiments were performed with T7 RNA polymerase. We used a synthetic RNA labeled at the 5′-terminus with fluorescein as a reference product (Fig. 2a, lane 1). RNA transcription using ATP, GTP, CTP (20% FAM-CTP), and UTP was performed as a positive control reaction (Fig. 2a, lane 2). For a negative control, the transcription reaction was performed in the absence of UTP. This reaction showed possible mis-incorporated products when 1 was not incorporated by T7 RNA polymerase (Fig. 2a lane 4). As expected, lane 4 did not show any detectable full-length product.
Lane 3 in Fig. 2a shows the product of the RNA transcription reaction using ATP, GTP, CTP (20% FAM-CTP), and 1. Surprisingly, 1 was incorporated smoothly even by wild-type T7 RNA polymerase. The ratio of the full-length products between lanes 2 and 3, estimated by the gel band intensities, was 0.76. This result indicated that 1 could be used as a substrate for T7 RNA polymerase. The 24% decrease of the full-length product might be explained by steric repulsion with the T7 polymerase active site because the X-ray structure indicated the presence of only a small space around the 2′-oxygen atom.36,37
To confirm the incorporation of 1, we synthesized a shorter 15mer RNA (5′-pppGGGCCAA-Ucm-ACAAAGA-3′), purified it by gel electrophoresis, and determined the molecular weight of the main product by MALDI-TOF mass analysis. As shown in Fig. 2b, the mass data suggested that the molecular weight of the main product was 5396.1. This corresponded to ppGGGCCAA-Ucm-ACAAAGAG which contains additional guanosine residues at the 3′-terminus38 and lacks the gamma phosphate of the 5′-guanosine triphosphate.
To confirm the sequences of RNAs synthesized by enzymatic incorporation of 1, we attempted to sequence each RNA after its conversion into the corresponding cDNA. For this purpose, the fidelity of reverse transcription of RNA containing Ucm was evaluated. The RNA template containing Ucm was chemically synthesized according to a method reported previously (Fig. 3a).21 Single nucleotide extension experiments were performed with SuperScript III under standard conditions with one of the four natural deoxy (d)NTPs (Fig. 3a). After the enzymatic reaction, the products were analyzed by polyacrylamide gel electrophoresis. Lane 1 is the FAM-labeled cDNA primer. Lanes 2 to 5 are the positive controls reverse transcribed with the natural RNA template using one of the four natural dNTPs. Lanes 6 to 9 are experiments with the RNA template containing a Ucm residue. Interestingly, dATP was selectively incorporated opposite the template Ucm residue (lane 6 versus lanes 7, 8, and 9).
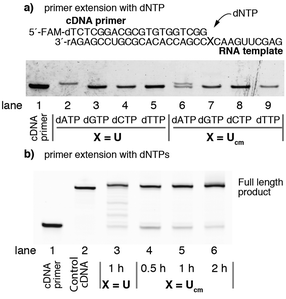 | ||
| Fig. 3 The fidelity of reverse transcription by SuperScript III. (a) Single nucleotide insertion of dNTP. (b) Primer extension with dNTPs. | ||
The reverse transcription experiments using the full RNA sequence with the four natural dNTPs are shown in Fig. 3b. Although the efficiency of reverse transcription on the Ucm containing template was lower than the natural RNA template (lane 3 versus lane 5), the prolonged reaction time reduced the residual unreacted cDNA primer (lane 6).
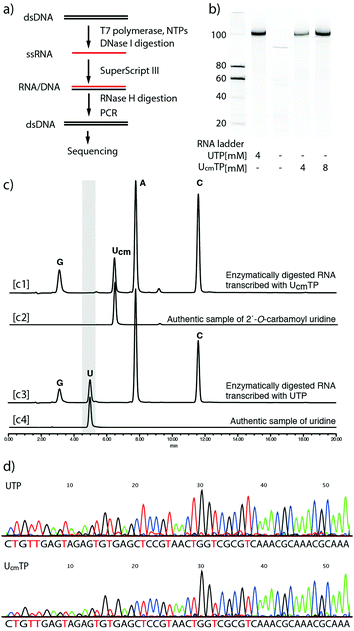
To evaluate the applicability of this method to the synthesis of longer RNA aptamers, and the fidelity of reverse transcription, we used T7 RNA polymerase to enzymatically synthesize a 99mer of RNA containing Ucm residues. The sequence was a known RNA aptamer, Spinach.39 Spinach RNA induces fluorescence upon binding of (Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-1,2-dimethyl-1H-imidazol-5(4H)-one. First, the T7 RNA polymerase-mediated transcription reaction was performed with 1 and the chemically synthesized DNA duplex coding the Spinach sequence (the entire sequence is shown in the ESI†). The reaction mixtures were treated with DNase I to remove the DNA duplex, and then analyzed by gel electrophoresis (Fig. 4b). The first and second right lanes of Fig. 4b clearly showed that the 99mer RNA was obtained. The RNA product transcribed with UcmTP showed comparable binding affinities to DFHBI with a canonical Spinach aptamer (Fig. S6, ESI†). To confirm the incorporation of 1, a portion of the RNA transcript was hydrolyzed enzymatically by snake venom phosphodiesterase and alkaline phosphatase. The completely digested mixtures were applied to reversed phase-HPLC. The result showed a pattern of A, G, C, and Ucm nucleosides without any detectable U (Fig. 4c). This clearly indicated that the RNA transcript was not generated by undetectable amounts of UTP contamination. Next, the remaining RNA transcripts were reverse transcribed by SuperScript III, and the polymerase chain reaction was performed to amplify the reverse transcribed cDNA, which was then sequenced (Fig. 4d). The sequence data of the transcript obtained by using 1 (lower panel) clearly shows a pattern similar to that obtained from the transcript using UTP (upper panel). Thus, the transcript sequence was confirmed. We also performed a transcription reaction with the DNA template containing randomized 20 nucleotides (N20) and confirmed the products by gel electrophoresis (Fig. S7, ESI†). From these results, we concluded that UcmTP could be applicable for RNA aptamer selection.
In summary, we have developed a new synthetic method for NTP 1 having a base-labile moiety. This method could be useful for the synthesis of more complex base-labile NTPs. We demonstrated the applicability of using 1 for transcription by T7 RNA polymerase and reverse transcription. Compound 1 was incorporated specifically opposite to the A base and was sufficient to transcribe the 99mer of structured RNA. It should be noted that the carbamoyl modification of 1 is the sterically largest substituent on the 2′-hydroxyl group that is thus far allowed by T7 polymerase. Moreover, dATP was incorporated correctly against the Ucm residue during reverse transcription. This precise incorporation of 1 by T7 RNA polymerase and high fidelity in reverse transcription make 1 an excellent candidate for the modified NTP, and useful for the enzymatic selection of modified RNA aptamers.
We reported previously that the Ucm residue enhanced G–U base pair formation in RNA duplexes.21 The G–U base pair plays a crucial role in structure formation and the activity of various functional RNAs such as tRNA,40,41 rRNA,42–44 and group I self-splicing introns.45 In addition, most RNA secondary structure prediction software treats the G–U base pair as a canonical base pair.46,47 Because Ucm facilitates G–U base pair formation, we believe UcmTP will be useful to explore a wider range of RNA secondary structures in aptamer selection.
This work was supported by the Japan Society for the Promotion of Science (grant numbers 26810086, 15H01062, 15K13738, and 26288075). DNA sequencing was supported by the Biomaterials Analysis Division, Technical Department, Tokyo Institute of Technology.
Notes and references
- T. Chen, N. Hongdilokkul, Z. Liu, R. Adhikary, S. S. Tsuen and F. E. Romesberg, Nat. Chem., 2016, 8, 556–562 CrossRef CAS PubMed
.
- S. A. Lapa, A. V. Chudinov and E. N. Timofeev, Mol. Biotechnol., 2016, 58, 79–92 CrossRef CAS PubMed
.
- N. Tarashima, T. Sumitomo, H. Ando, K. Furukawa, T. Ishida and N. Minakawa, Org. Biomol. Chem., 2015, 13, 6949–6952 CAS
.
- Y. Imaizumi, Y. Kasahara, H. Fujita, S. Kitadume, H. Ozaki, T. Endoh, M. Kuwahara and N. Sugimoto, J. Am. Chem. Soc., 2013, 135, 9412–9419 CrossRef CAS PubMed
.
- M. Kimoto, R. Yamashige, K. Matsunaga, S. Yokoyama and I. Hirao, Nat. Biotechnol., 2013, 31, 453–457 CrossRef CAS PubMed
.
- F. Tolle and G. Mayer, Chem. Sci., 2013, 4, 60 RSC
.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537–550 CrossRef CAS PubMed
.
- E. W. Ng, D. T. Shima, P. Calias, E. T. Cunningham, Jr., D. R. Guyer and A. P. Adamis, Nat. Rev. Drug Discovery, 2006, 5, 123–132 CrossRef CAS PubMed
.
- G. Kolb, S. Reigadas, C. Boiziau, A. van Aerschot, A. Arzumanov, M. J. Gait, P. Herdewijn and J. J. Toulme, Biochemistry, 2005, 44, 2926–2933 CrossRef CAS PubMed
.
- L. H. Lauridsen, J. A. Rothnagel and R. N. Veedu, ChemBioChem, 2012, 13, 19–25 CrossRef PubMed
.
- X. M. Ren, A. H. El-Sagheer and T. Brown, Nucleic Acids Res., 2016, 44, e79 CrossRef PubMed
.
- T. Carell, C. Brandmayr, A. Hienzsch, M. Muller, D. Pearson, V. Reiter, I. Thoma, P. Thumbs and M. Wagner, Angew. Chem., Int. Ed., 2012, 51, 7110–7131 CrossRef CAS PubMed
.
- A. Demesmaeker, R. Haner, P. Martin and H. E. Moser, Acc. Chem. Res., 1995, 28, 366–374 CrossRef CAS
.
- K. Seio, M. Tokugawa, T. Kanamori, H. Tsunoda, A. Ohkubo and M. Sekine, Bioorg. Med. Chem. Lett., 2012, 22, 2470–2473 CrossRef CAS PubMed
.
- T. Yamada, Y. Masaki, N. Okaniwa, T. Kanamori, A. Ohkubo, H. Tsunoda, K. Seio and M. Sekine, Org. Biomol. Chem., 2014, 12, 6457–6464 CAS
.
- T. P. Prakash, Chem. Biodiversity, 2011, 8, 1616–1641 CAS
.
- Y. Masaki, R. Miyasaka, A. Ohkubo, K. Seio and M. Sekine, J. Phys. Chem. B, 2010, 114, 2517–2524 CrossRef CAS PubMed
.
- R. Pattanayek, L. Sethaphong, C. L. Pan, M. Prhavc, T. P. Prakash, M. Manoharan and M. Egli, J. Am. Chem. Soc., 2004, 126, 15006–15007 CrossRef CAS PubMed
.
- M. Egli, G. Minasov, V. Tereshko, P. S. Pallan, M. Teplova, G. B. Inamati, E. A. Lesnik, S. R. Owens, B. S. Ross, T. P. Prakash and M. Manoharan, Biochemistry, 2005, 44, 9045–9057 CrossRef CAS PubMed
.
- M. Egli and P. S. Pallan, Chem. Biodiversity, 2010, 7, 60–89 CAS
.
- K. Seio, R. Tawarada, T. Sasami, M. Serizawa, M. Ise, A. Ohkubo and M. Sekine, Bioorg. Med. Chem., 2009, 17, 7275–7280 CrossRef CAS PubMed
.
- R. Sousa and R. Padilla, EMBO J., 1995, 14, 4609–4621 CAS
.
- H. Aurup, D. M. Williams and F. Eckstein, Biochemistry, 1992, 31, 9636–9641 CrossRef CAS PubMed
.
- K. Raines and P. A. Gottlieb, RNA, 1998, 4, 340–345 CAS
.
- R. Padilla and R. Sousa, Nucleic Acids Res., 1999, 27, 1561–1563 CrossRef CAS PubMed
.
- J. B. Pavey, A. J. Lawrence, I. A. O'Neil, S. Vortler and R. Cosstick, Org. Biomol. Chem., 2004, 2, 869–875 CAS
.
- R. Padilla and R. Sousa, Nucleic Acids Res., 2002, 30, e138 CrossRef PubMed
.
- W. D. Cornell, P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell and P. A. Kollman, J. Am. Chem. Soc., 1995, 117, 5179–5197 CrossRef CAS
.
- J. A. McDowell, L. Y. He, X. Y. Chen and D. H. Turner, Biochemistry, 1997, 36, 8030–8038 CrossRef CAS PubMed
.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed
.
- Y. Masaki, R. Miyasaka, K. Hirai, H. Tsunoda, A. Ohkubo, K. Seio and M. Sekine, Chem. Commun., 2012, 48, 7313–7315 RSC
.
- H. J. C. Berendsen, J. P. M. Postma, W. F. Vangunsteren, A. Dinola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS
.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089 CrossRef CAS
.
- J. Ludwig and F. Eckstein, J. Org. Chem., 1989, 54, 631–635 CrossRef CAS
.
- S.-i. Kawahara, T. Wada and M. Sekine, J. Am. Chem. Soc., 1996, 118, 9461–9468 CrossRef CAS
.
- Y. Huang, F. Eckstein, R. Padilla and R. Sousa, Biochemistry, 1997, 36, 8231–8242 CrossRef CAS PubMed
.
- T. A. Steitz, Curr. Opin. Struct. Biol., 2009, 19, 683–690 CrossRef CAS PubMed
.
- J. F. Milligan, D. R. Groebe, G. W. Witherell and O. C. Uhlenbeck, Nucleic Acids Res., 1987, 15, 8783–8798 CrossRef CAS PubMed
.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Science, 2011, 333, 642–646 CrossRef CAS PubMed
.
- Y. M. Hou and P. Schimmel, Nature, 1988, 333, 140–145 CrossRef CAS PubMed
.
- W. McClain and K. Foss, Science, 1988, 240, 793–796 CAS
.
- D. Gautheret, D. Konings and R. R. Gutell, RNA, 1995, 1, 807–814 CAS
.
- S. A. White, M. Nilges, A. Huang, A. T. Brunger and P. B. Moore, Biochemistry, 1992, 31, 1610–1621 CrossRef CAS PubMed
.
- M. Szymanski, M. Z. Barciszewska, V. A. Erdmann and J. Barciszewski, Mol. Biol. Evol., 2000, 17, 1194–1198 CrossRef CAS PubMed
.
- S. A. Strobel and T. R. Cech, Biochemistry, 1996, 35, 1201–1211 CrossRef CAS PubMed
.
- M. Andronescu, V. Bereg, H. H. Hoos and A. Condon, BMC Bioinf., 2008, 9, 340 CrossRef PubMed
.
- D. H. Mathews, J. Sabina, M. Zuker and D. H. Turner, J. Mol. Biol., 1999, 288, 911–940 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cc05796a |
| This journal is © The Royal Society of Chemistry 2016 |