Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Reactivation from the Ni–B state in [NiFe] hydrogenase of Ralstonia eutropha is controlled by reduction of the superoxidised proximal cluster†
Valentin
Radu
a,
Stefan
Frielingsdorf
b,
Oliver
Lenz
b and
Lars J. C.
Jeuken
*a
aSchool of Biomedical Sciences, The Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, UK. E-mail: L.J.C.Jeuken@leeds.ac.uk
bInstitut für Chemie, Sekretariat PC14, Technische Universität Berlin, Straße des 17. Juni 135, 10623 Berlin, Germany
First published on 4th January 2016
Abstract
The tolerance towards oxic conditions of O2-tolerant [NiFe] hydrogenases has been attributed to an unusual [4Fe–3S] cluster that lies proximal to the [NiFe] active site. Upon exposure to oxygen, this cluster converts to a superoxidised (5+) state, which is believed to secure the formation of the so-called Ni–B state that is rapidly reactivated under reducing conditions. Here, the reductive reactivation of the membrane-bound [NiFe]-hydrogenase (MBH) from Ralstonia eutropha in a native-like lipid membrane was characterised and compared to a variant that instead carries a typical [4Fe–4S] proximal cluster. Reactivation from the Ni–B state was faster in the [4Fe–4S] variant, suggesting that the reactivation rate in MBH is limited by the reduction of the superoxidised [4Fe–3S] cluster. We propose that the [4Fe–3S] cluster plays a major role in protecting MBH by blocking the reversal of electron transfer to the [NiFe] active site, which would produce damaging radical oxygen species.
Hydrogenases are metalloenzymes that catalyze the interconversion between H2 and H+s. Their high activities coupled with high affinities for H2 have attracted interest in their reaction mechanism.1–3 Based on the metal content of the active site, hydrogenases are grouped into two main classes: [NiFe] and [FeFe] hydrogenases.4 Both classes serve as models for designing sustainable synthetic catalysts for fuel cells and hydrogen production.5–9 Maintaining activity in the presence of oxygen is key for these functions10 and a sub-class of “O2-tolerant” [NiFe] hydrogenases have therefore been of particular interest.11,12 “O2-tolerant” [NiFe] hydrogenases can be clearly differentiated from the so-called “standard” [NiFe] hydrogenases which, like [FeFe] hydrogenases, are instantly and almost completely inactivated by O2. Among the most studied O2-tolerant [NiFe] hydrogenases are the membrane-bound hydrogenases (MBHs) from Ralstonia eutropha, Ralstonia metallidurans, Aquifex aeolicus, Hydrogenovibrio marinus, and Escherichia coli.
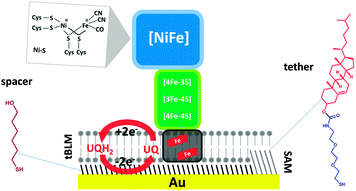
[NiFe] hydrogenases are composed of at least one large subunit (α) and a small subunit (β).1,4 The active site has the same configuration in all [NiFe] hydrogenases and it is deeply buried into the large subunit (Scheme 1).
When exposed to O2, the active sites of standard [NiFe] hydrogenases are converted to a mixture of inactive states called “Ni–A” and “Ni–B”.13–18 These inactive states are reconverted to the active Ni–S state in reducing conditions, but the reactivation of the enzyme from the Ni–A state, also called the “unready” state, can take hours. In contrast, the Ni–B or “ready” state is reduced to the active state in a matter of seconds. While the ligand bridging the Fe and the Ni atom in the Ni–B state has been assigned to an OH−, the structure of Ni–A remains unclear.
We and others have proposed that an unusual proximal [4Fe–3S] cluster that is only found in O2-tolerant [NiFe] hydrogenases provides a reductive environment that ensures the formation of the fast-reactivating Ni–B state.17,19–22 The tolerance towards O2 has also been ascribed to their high affinity for H2, outcompeting and thus excluding O2 from the active site. The KM for H2 of O2-tolerant [NiFe] hydrogenases can be three orders of magnitude smaller than the O2 inhibition constant 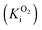 .18,20,21
.18,20,21
MBHs contain a membrane-integral subunit, cytochrome b, which couples H2 oxidation activity to the reduction of the quinone pool.12,23–28 The cytochrome b subunit also anchors the protein complex to the cytoplasmic membrane, in which MBH is known to form oligomers (for simplicity, the higher oligomeric state is not shown in Scheme 1). A distal [4Fe–4S] cluster of MBH is located close to the surface of the small subunit and it has been proposed that this enables intermolecular electron transfer within the dipartite or tripartite supercomplex in the membrane.23,26
Electrochemical techniques have been very useful for testing the catalytic properties of hydrogenases via mediated or direct electron transfer. Protein film electrochemistry (PFE) has been especially helpful in elucidating inhibition mechanisms. PFE monitors the catalytic turnover of an enzyme directly adsorbed on the electrode while the redox state of the enzyme is controlled via the applied potential. PFE studies have shown that under H2, O2-tolerant [NiFe] hydrogenases recover activity quickly after being exposed to oxidizing potentials (anaerobic inactivation) or O2.18–21,29–32 So far, most PFE studies have employed the heterodimeric hydrogenase module only, which is a water soluble heterodimeric (αβ) sub-complex of the MBHs. We have recently shown that the full heterotrimeric membrane-bound [NiFe] hydrogenase (MBH) from R. eutropha in equilibrium with the quinone pool displays enhanced tolerance to oxidizing conditions compared to the heterodimeric sub-complex.33 For instance, no inactivation was observed in the presence of H2 under (anaerobic) oxidative redox conditions where PFE of the hydrogenase module showed Ni–B formation.
In our system, the MBH from R. eutropha, along with its native lipid environment in the cytoplasmic membrane, is tethered to the electrode via a mixed self-assembled monolayer (SAM) (Scheme 1) containing a cholesterol-based tether (EO3-cholesteryl). The tethers are mixed with 6-mercaptohexanol spacer molecules, which phase-separate on the surface, leaving space for transmembrane proteins such as MBH. A tethered bilayer lipid membrane (tBLM), containing E. coli polar lipids, quinones, and cytoplasmic membrane extracts of R. eutropha, is then formed on top of the SAM by self assembly. In this system, the redox-active MBH is in equilibrium with quinones added in the tBLM, either ubiquinone-10 or menaquinone-7. The redox state of the quinone pool is controlled via the electrode potential.
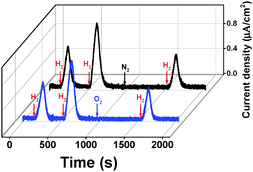
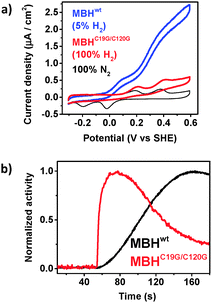
In the absence of H2, MBH is known to reside in the Ni–B state, which is thought to protect the active site from irreversible damage.19 To confirm that the Ni–B state is formed in MBH in our setup, multiple H2-saturated aliquots of buffer were injected under anaerobic conditions while the electrode potential was poised at 0.5 V vs. SHE (Fig. 1, black trace). The transient exposure leads to the peak-shaped signals shown in the chronoamperograms in Fig. 1. Upon injection of hydrogen, MBH reactivates, leading to a rise in oxidative current. As the concentration of the injected gas decays exponentially with time (while the electrochemical cell is stirred and flushed with another gas, like N2), the oxidative current returns to the baseline as H2 is flushed out.34 The current increase and decay reveal information about the (re)activation and inactivation kinetics, respectively. We note that the enhanced oxygen tolerance of the full heterotrimeric MBH in our system enables, for the first time, to detect the reactivation under oxidative conditions, whereas in previous PFE experiments this was not possible as reactivation required reductive (or mild oxidative) conditions.
In the case of MBHwt donating electrons to ubiquinone, reactivation after the second injection was 1.5 times faster compared to the first reactivation based on the slope of the linear domain of the current increase at a potential of 0.5 V vs. SHE (normalised rate at 1st reactivation: 92.2 (±13.9) × 10−4 s−1; 2nd reactivation: 136.6 (±11.5) × 10−4 s−1; n = 9; the time between injections was approximately 400 s). Because the H2 depletion rate is identical after both injections, the peak height after the second injection is thus higher (Fig. S1, ESI†).
To further confirm that the inactive state in our setup represents the Ni–B state, O2 was injected into the cell solution (via an air-saturated aliquot of buffer) after two injections of substrate (Fig. 1, blue trace). The O2 injection ensures the total conversion of the enzyme to the Ni–B state.17,21 A third aliquot of H2-saturated buffer was injected into the cell solution after the complete depletion of O2. A control experiment was recorded replacing O2 with N2 (Fig. 1, black trace). The reactivation kinetics after O2 treatment was identical to that after N2 injection, confirming that the Ni–B state is indeed responsible for the observed kinetics.
The reactivation rate decreases as the time of the oxidative poise between the 2nd and 3rd injection increases (Fig. S2, ESI†). The reactivation rate after the 1st H2 injection is only marginally faster than that of the 3rd injection after prolonged oxidative poise, in line with the expectation that the MBH mainly resides in the Ni–B state in anaerobic electron-deficient conditions. The inactivation kinetics indicates that the accumulation of Ni–B is complete approximately 400 s after all H2 is removed from solution.
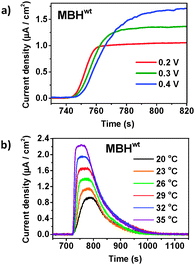
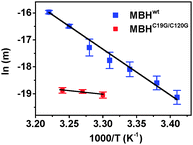
Having confirmed that inactivation is due to Ni–B formation, we studied the reactivation kinetics by analysing the slope of current increase after injection of H2. First, the potential dependence of the reactivation kinetics was investigated using menaquinone-containing tBLMs (Fig. 2a). Menaquinol has a lower oxidation potential, which enables studies over a wider potential window (0.2–0.6 V). Reactivation rates were only marginally slower at higher potential (a 17% decrease of the slope of the reactivation trace was observed when increasing the potential from 0.2 V to 0.4 V). A similar potential dependence of Ni–B reactivation was observed with the hydrophilic heterodimeric forms of the MBHs from A. aeolicus and E. coli.31,35 Next, we studied the temperature dependence and higher temperatures significantly increased the reactivation rate (Fig. 2b). The temperature profiles allowed the determination of the activation energy (Ea) from Arrhenius plots using the slope of the reactivation trace as the temperature dependent variable (Fig. 3). Ea was determined to be 141.5 (±5.0) kJ mol−1 (n = 3) at 0.3 V, in line with values obtained with Hyd-1 for which Ea varied from a value of 56.8 kJ mol−1 at 0.035 V to 96.3 kJ mol−1 at 0.235 V.35
To determine how the reactivation kinetics is dependent on the unusual [4Fe–3S] promixal cluster, these experiments were repeated with an MBH variant (MBHC19G/C120G). The variant carries a [4Fe–4S] cluster proximal to the [NiFe] site, thus resembling the [FeS] cluster configuration of standard O2-sensitive [NiFe] hydrogenases.32 MBHC19G/C120G is known to share similar catalytic properties with MBHwt and the same active site structure as any [NiFe] hydrogenase.32 As previously observed for this variant (Supplementary Table 1 in ref. 32), the activity of MBHC19G/C120G was 4 to 5 times lower compared to MBHwt in our setup (Fig. 4a), which is attributed to the lower expression level of the variant, an unfavourable potential of the engineered [4Fe–4S] cluster and/or a higher sensitivity towards oxygen, irreversibly deactivating more of the MBH during extraction from R. eutropha. Importantly and unexpectedly, it was found that MBHC19G/C120G reactivates 7 times faster than the MBHwt (Fig. 4b). The normalised rate of current increase for MBHC19G/C120G, after H2 injection, was 1027.7 (±41.7) × 10−4 s−1 (n = 11). To determine if the faster reactivation kinetics is due to a lower activation energy, the temperature dependence of the kinetics was determined. As the variant is less active, it was not possible to accurately probe the reactivation at low potentials (the lower currents led to unacceptable signal-to-noise ratios). It was observed that the variant was also less thermostable. Nonetheless, in spite of the smaller temperature window, it is clear that the reactivation energy is significantly lower (Fig. 3). At 0.5 V, Ea was 22 ± 8 kJ mol−1 (n = 4), which is remarkably similar to that determined for H2 oxidation.36,37
These results suggest that the reactivation kinetics of MBHwt is rate limited by the reduction of the superoxidized [4Fe–3S]5+ cluster. The relatively slow reduction of the [4Fe–3S]5+ cluster compared to that of other [FeS] clusters can be explained by the chemical reorganisation that is coupled to this step. In contrast to MBHwt, the reactivation kinetics of MBHC19G/C120G was independent of the time of exposure to oxidizing electrode potentials (i.e., the inactivation time, Fig. S3, ESI†). We propose that the inactivation kinetics of MBHwt (Fig. S2, ESI†) are determined by the superoxidation of the [4Fe–3S] cluster, which is absent in MBHC19G/C120G as the standard [4Fe–4S] cluster cannot be superoxidised.
It has been previously observed that standard [NiFe] hydrogenases like the ones from Allochromatium vinosum and Desulfovibrio gigas display a much slower anaerobic inactivation compared to O2-tolerant [NiFe] hydrogenases like the ones from R. eutropha and A. aeolicus.19,21 In the case of O2-tolerant hydrogenases, protection of the active site, achieved by Ni–B formation, is related to the formation of the superoxidised cluster.24,35,38 When the reduction of the superoxidised proximal cluster is slow, as observed here, it follows that the [FeS] relay cannot function as an efficient electron hub for the active site. We therefore hypothesize that the “locking” of the proximal cluster in the superoxidized state prevents the irreversible damage to the active site under aerobic conditions by prohibiting electron transfer to the [NiFe] active site forming radical oxygen species. The temporary shutdown of the electron relay responsible for disabling the active site reactivity would impede the irreversible deterioration of the protein and implicitly the complete loss of enzymatic activity. In contrast, standard [NiFe] hydrogenases do not shut down the electron relay and continuous electron transfer to the active site could form damaging radicals under aerobic conditions, possibly resulting in the formation of the Ni–A state and other irreversible inactive states.39
In conclusion, we have confirmed that in the absence of H2 and under electron-deficient conditions, the full heterotrimeric MBH in our experimental system forms the Ni–B state, as previously extensively shown for the hydrogenase module (a water soluble heterodimeric (αβ) sub-complex of the membrane-bound hydrogenases). The MBH variant carrying a standard [4Fe–4S] proximal cluster reactivated many times faster than wild-type MBH, suggesting that the reduction of the superoxidised proximal cluster determines the kinetics of Ni–B reactivation. Active site protection in oxidative conditions is proposed to be achieved by the formation of the superoxidized state of the proximal cluster, which interrupts the functioning of the [FeS] electron relay and prevents the formation of radical oxygen species at the [NiFe] site.
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013)/ERC Grant no. 280518 (V. R. and L. J. C. J.) and from the DFG cluster of Excellence “Unifying Concepts in Catalysis” (S. F. and O. L.).
Notes and references
- W. Lubitz, H. Ogata, O. Rüdiger and E. Reijerse, Chem. Rev., 2014, 114, 4081 CrossRef CAS PubMed.
- A. K. Jones, E. Sillery, S. P. J. Albracht and F. A. Armstrong, Chem. Commun., 2002, 866 RSC.
- F. A. Armstrong, N. A. Belsey, J. A. Cracknell, G. Goldet, A. Parkin, E. Reisner, K. A. Vincent and A. F. Wait, Chem. Soc. Rev., 2009, 38, 36 RSC.
- P. M. Vignais and B. Billoud, Chem. Rev., 2007, 107, 4206 CrossRef CAS PubMed.
- M. Hambourger, M. Gervaldo, D. Svedruzic, P. W. King, D. Gust, M. Ghirardi, A. L. Moore and T. M. Moore, J. Am. Chem. Soc., 2008, 130, 2015 CrossRef CAS PubMed.
- A. Le Goff, V. Artero, B. Jousselme, P. D. Tran, N. Guillet, R. Métayé, A. Fihri, S. Palacin and M. Fontecave, Science, 2009, 326, 1384 CrossRef CAS PubMed.
- L.-C. Song, J.-P. Li, Z.-J. Xie and H.-B. Song, Inorg. Chem., 2013, 52, 11618 CrossRef CAS PubMed.
- G. M. Chambers, R. Angamuthu, D. L. Gray and T. B. Rauchfuss, Organometallics, 2013, 32, 6324 CrossRef CAS.
- S. Roy, T. L. Groy and A. K. Jones, Dalton Trans., 2013, 42, 3843 RSC.
- D. W. Wakerley and E. Reisner, Energy Environ. Sci., 2015, 8, 2283 CAS.
- K. A. Vincent, A. Parkin and F. A. Armstrong, Chem. Rev., 2007, 107, 4366 CrossRef CAS PubMed.
- J. Fritsch, O. Lenz and B. Friedrich, Nat. Rev. Microbiol., 2013, 11, 106 CrossRef CAS PubMed.
- A. K. Jones, S. E. Lamle, H. R. Pershad, K. A. Vincent, S. P. J. Albracht and F. A. Armstrong, J. Am. Chem. Soc., 2003, 125, 8505 CrossRef CAS PubMed.
- S. E. Lamle, S. P. J. Albracht and F. A. Armstrong, J. Am. Chem. Soc., 2004, 126, 14899 CrossRef CAS PubMed.
- S. E. Lamle, S. P. J. Albracht and F. A. Armstrong, J. Am. Chem. Soc., 2005, 127, 6595 CrossRef CAS PubMed.
- H. Ogata, S. Hirota, A. Nakahara, H. Komori, N. Shibata, T. Kato, K. Kano and Y. Higuchi, Structure, 2005, 13, 1635 CrossRef CAS PubMed.
- M. Saggu, I. Zebger, M. Ludwig, O. Lenz, B. Friedrich, P. Hildebrandt and F. Lendzian, J. Biol. Chem., 2009, 284, 16264 CrossRef CAS PubMed.
- J. A. Cracknell, A. F. Wait, O. Lenz, B. Friedrich and F. A. Armstrong, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 20681 CrossRef PubMed.
- K. A. Vincent, A. Parkin, O. Lenz, S. P. J. Albracht, J. C. Fontecilla-Camps, R. Cammack, B. Friedrich and F. A. Armstrong, J. Am. Chem. Soc., 2005, 127, 18179 CrossRef CAS PubMed.
- M. Ludwig, J. A. Cracknell, K. A. Vincent, F. A. Armstrong and O. Lenz, J. Biol. Chem., 2009, 284, 465 CrossRef CAS PubMed.
- M.-E. Pandelia, V. Fourmond, P. Tron-Infossi, E. Lojou, P. Bertrand, C. Leger, M.-T. Giudici-Orticoni and W. Lubitz, J. Am. Chem. Soc., 2010, 132, 6991 CrossRef CAS PubMed.
- M. J. Lukey, M. M. Roessler, A. Parkin, R. M. Evans, R. A. Davies, O. Lenz, B. Friedrich, F. Sargent and F. A. Armstrong, J. Am. Chem. Soc., 2011, 133, 16881 CrossRef CAS PubMed.
- S. Frielingsdorf, T. Schubert, A. Pohlmann, O. Lenz and B. Friedrich, Biochemistry, 2011, 50, 10836 CrossRef CAS PubMed.
- J. Fritsch, P. Scheerer, S. Frielingsdorf, S. Kroschinsky, B. Friedrich, O. Lenz and C. M. T. Spahn, Nature, 2011, 479, 249 CrossRef CAS PubMed.
- Y. Shomura, K.-S. Yoon, H. Nishihara and Y. Higuchi, Nature, 2011, 479, 253 CrossRef CAS PubMed.
- A. Volbeda, C. Darnault, A. Parkin, F. Sargent, F. A. Armstrong and J. C. Fontecilla-Camps, Structure, 2013, 21, 184 CrossRef CAS PubMed.
- A. Volbeda, P. Amara, C. Darnault, J.-M. Mouesca, A. Parkin, M. M. Roessler, F. A. Armstrong and J. C. Fontecilla-Camps, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 5305 CrossRef CAS PubMed.
- H. Ogata, P. Kellers and W. Lubitz, J. Mol. Biol., 2010, 402, 428 CrossRef CAS PubMed.
- G. Goldet, A. F. Wait, J. A. Cracknell, K. A. Vincent, M. Ludwig, O. Lenz, B. Friedrich and F. A. Armstrong, J. Am. Chem. Soc., 2008, 130, 11106 CrossRef CAS PubMed.
- M. J. Lukey, A. Parkin, M. M. Roessler, B. Murphy, J. Harmer, T. Palmer, F. Sargent and F. A. Armstrong, J. Biol. Chem., 2010, 285, 3928 CrossRef CAS PubMed.
- V. Fourmond, P. Infossi, M.-T. Giudici-Orticoni, P. Bertrand and C. Leger, J. Am. Chem. Soc., 2010, 132, 4848 CrossRef CAS PubMed.
- T. Goris, A. F. Wait, M. Saggu, J. Fritsch, N. Heidary, M. Stein, I. Zebger, F. Lendzian, F. A. Armstrong, B. Friedrich and O. Lenz, Nat. Chem. Biol., 2011, 7, 310 CrossRef CAS PubMed . Correction published: Nat. Chem. Biol., 2011, 7, 648.
- V. Radu, S. Frielingsdorf, S. D. Evans, O. Lenz and L. J. C. Jeuken, J. Am. Chem. Soc., 2014, 136, 8512 CrossRef CAS PubMed.
- C. Leger, S. Dementin, P. Bertrand, M. Rousset and B. Guigliarelli, J. Am. Chem. Soc., 2004, 126, 12162 CrossRef CAS PubMed.
- R. M. Evans, A. Parkin, M. M. Roessler, B. J. Murphy, H. Adamson, M. J. Lukey, F. Sargent, A. Volbeda, J. C. Fontecilla-Camps and F. A. Armstrong, J. Am. Chem. Soc., 2013, 135, 2694 CrossRef CAS PubMed.
- B. Schink and H. G. Schlegel, Biochim. Biophys. Acta, 1979, 567, 315 CrossRef CAS.
- D. L. Hallahan, V. M. Fernandez and D. O. Hall, Eur. J. Biochem., 1987, 165, 621 CrossRef CAS PubMed.
- J. Fritsch, S. Loscher, O. Sanganas, E. Siebert, I. Zebger, M. Stein, M. Ludwig, A. L. De Lacey, H. Dau, B. Friedrich, O. Lenz and M. Haumann, Biochemistry, 2011, 50, 5858 CrossRef CAS PubMed.
- A. Volbeda, L. Martin, C. Cavazza, M. Matho, B. W. Faber, W. Roseboom, S. P. Albracht, E. Garcin, M. Rousset and J. C. Fontecilla-Camps, JBIC, J. Biol. Inorg. Chem., 2005, 10, 239 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting figures and experimental details. See DOI: 10.1039/c5cc10382g |
| This journal is © The Royal Society of Chemistry 2016 |