Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Copper-mediated arylsulfanylations and arylselanylations of pyrimidine or 7-deazapurine nucleosides and nucleotides†
Filip
Botha‡
a,
Michaela
Slavíčková
a,
Radek
Pohl
a and
Michal
Hocek
 *ab
*ab
aInstitute of Organic Chemistry and Biochemistry, Academy of Science Czech Republic, Gilead Sciences and IOCB Research Center, Flemingovo nám. 2, 16610 Prague 6, Czech Republic. E-mail: hocek@uochb.cas.cz
bDepartment of Organic Chemistry, Faculty of Science, Charles University in Prague, Hlavova 8, 12843 Prague 2, Czech Republic
First published on 29th September 2016
Abstract
The syntheses of 5-arylsulfanyl- or 5-arylselanylpyrimidine and 7-arylsulfanyl- or 7-arylselanyl-7-deazapurine nucleosides and nucleotides were developed by the Cu-mediated sulfanylations or selanylations of the corresponding 5-iodopyrimidine or 7-iodo-7-deazapurine nucleosides or nucleotides with diaryldisulfides or -diselenides. The reactions were also applicable for direct modifications of 2′-deoxycytidine triphosphate and the resulting 5-arylsulfanyl or 5-arylselanyl-dCTP served as substrates for the polymerase synthesis of modified DNA bearing arylsulfanyl or arylselanyl groups in the major groove.
Introduction
DNA molecules bearing modifications in the major groove have found diverse applications mainly in bioanalysis and chemical biology.1 5-Substituted pyrimidine and 7-substituted 7-deazapurine 2′-deoxyribonucleoside 5′-O-triphosphates (dNTPs) are good substrates for DNA polymerases in the enzymatic synthesis of base-modified DNA.2,3 Modified dNTPs are mostly synthesized by the triphosphorylation of the corresponding modified nucleosides4 but this approach can fail in the case of some reactive modifications not compatible with the triphosphorylation methodology. Therefore, direct methods of functionalization of dNTPs are desirable but are inherently difficult due to the lability of dNTPs which are prone to hydrolysis. So far, the only reported reactions suitable for modification of dNTPs have been the aqueous cross-coupling reactions of halogenated dNTPs,3,5 thiol–maleimide addition,6 some amide-forming reactions of 5-aminoalkylethynyl-dUTP,7 hydrazone-formation,8 Diels–Alder9 and the CuAAC click reaction.10 The Suzuki–Miyaura cross-coupling reaction with arylboronic acids11 and the Sonogashira reactions with terminal acetylenes12 are the most general and useful reactions used in the synthesis of base-modified dNTPs. In addition, several examples of the Heck coupling13 with acrylates, as well as the Stille reaction14 with aryl- or alkenylstannanes were recently reported. To the best of our knowledge, no method for direct attachment of a heteroatom to dNTPs has been published.5-Alkylsulfanyl- or arylsulfanyl-pyrimidine nucleosides were reported to inhibit thymidylate kinase15 and slightly destabilized DNA duplexes,16 whereas saturated 5-phenylsulfanyl-thymidine analogues were used17 as radical precursors for photochemical generation of thymine in DNA. Some 7-arylsulfanyl-7-deazaadenosine analogues displayed18 weak cytostatic effects, while the corresponding 7-S-substituted 7-deazaguanine derivatives have never been reported. 5-Selenylated pyrimidine nucleotides inhibit thymidylate synthase19 and have been utilized20 for modification of DNA or RNA for X-ray crystallography and 5-(phenylselenylmethyl)uracil was used21 as a T radical precursor for DNA crosslinking. Also the related 5-(phenyltelluranyl)uracil nucleoside has been prepared22 and, after incorporation into DNA, it was used for X-ray and STM imaging. However, no selenylated 7-deazapurines have been known so far. Therefore, we report here the synthesis of the arylsulfanyl and arylselanyl derivatives of pyrimidine and 7-deazapurine nucleosides and nucleotides and their potential for polymerase incorporation into DNA.
Results and discussion
Synthesis
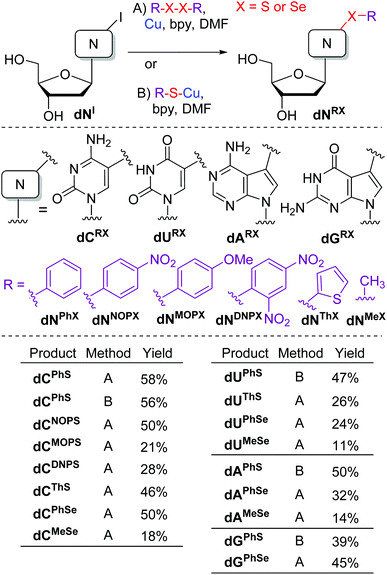
Previously, 5-(alkylsulfanyl)pyrimidine bases or nucleosides were prepared by alkylation of 5-mercaptouracil,23 reactions of toxic 5-(chloromercuri)pyrimidines with disulfides,24 or more recently by Pd-catalyzed coupling of 5-bromopyrimidine derivatives with thiols,25 whereas 7-arylsulfanyl-7-deazapurines were prepared by Cu-mediated S–H sulfenylations.18 The 5-selenylated pyrimidines were prepared by Mn-mediated C–H selenylations20 or electrophilic aromatic selenylation.26Inspired by the work of Taniguchi27 on the copper-catalyzed reactions of diaryldisulfides or diaryldiselenides with iodoarenes, we started our study by testing the reactions of unprotected halogenated nucleosides.5 The reaction conditions were first tested on 5-iodo-2′-deoxycytidine (dCI) in reaction with diphenyldisulfide. The Cu-catalyzed (10 mol% of CuI) reactions in the presence or absence of Mg27 gave complex mixtures of products. Therefore, we used stoichiometric amounts of copper powder in the presence of 2,2′-bipyridine (bpy). The reactions were performed at 80–110 °C in DMF (Scheme 1). Under these conditions (Method A), the desired 5-phenylsulfanyl-2′-deoxycytidine was formed as the major product (in addition to small amounts of dehalogenated 2′-deoxycytidine) and isolated in a good yield of 58%. A similar conversion and yield were achieved when using pre-generated phenylsulfanylcuprate (Method B).
The same reaction of nucleoside dCI (Method A) was then performed with a small series of diaryldisulfides to obtain the corresponding 5-(4-nitrophenyl)sulfanyl (dCNOPS), 5-(4-methoxyphenyl)sulfanyl (dCMOPS), 5-(2,4-dinitrophenyl)sulfanyl (dCDNPS) and 5-(2-thienylsulfanyl) (dCThS) 2′-deoxycytidines in moderate yields (21–50%). The reaction (Method A) with diphenyldiselenide gave the 5-(phenylselanyl)cytosine nucleoside dCPhSe in good 50% yield, whereas the corresponding 5-(methylselanyl)C nucleoside dCMeSe was only obtained in low 18% yield.
Then we tested other iodinated nucleosides (Scheme 1). The reaction of 5-iodo-2′-deoxyuridine (dUI) with PhSCu (Method B) provided the 5-substituted dUPhS nucleoside in 47% yield, whereas the reactions with dithienyldisulfide or diselenides (Method A) gave the other corresponding 5-arylsulfanyl- or phenyl- or methylselanyl uracil nucleosides (dUThS, dUPhSe and dUMeSe) in low yields. The reactions of 7-iodo-7-deazaadenine dAI and -7-deazaguanine dGI nucleosides with PhSCu (Method B) gave the 7-(phenylsulfanyl)deazapurine nucleosides dAPhS and dGPhS in acceptable 50 or 39% yields, whereas the reactions with diphenyldiselenide furnished the corresponding phenylselanyl nucleosides dAPhSe and dGPhSe in moderate yields. Again, the reaction with dimethyldiselenide gave very low conversion and dAMeSe was isolated only in 14% yield. Apparently the reactivity of dimethyldiselenide is very low and the methylsulfanylation is of very limited synthetic applicability.
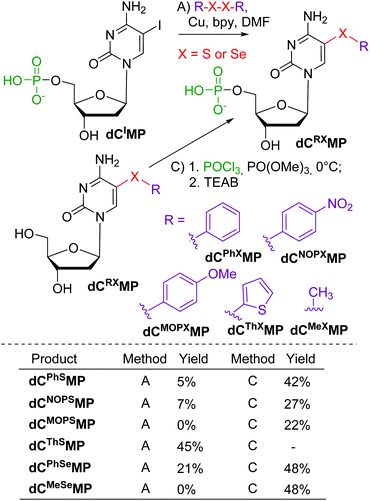
Next, we tested the reactions of nucleotides and started with stable nucleoside 5′-O-monophosphates (dNMPs). The model iodinated dCIMP was tested in reactions with diaryldisulfides or diselenides (Scheme 2, Method A). Most of these reactions gave very low conversions and only two products, dCThSMP and dCPhSeMP, were isolated in acceptable yields. On the other hand, the phosphorylation of the 5-arylsulfanyl- or arylselanyl-cytosine nucleosides gave the desired modified nucleotides in better yields (22–48%).
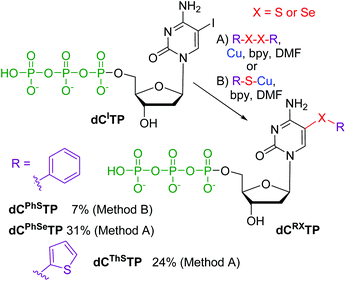
Finally, we tested the reactions for direct modification of hydrolytically labile dNTPs (Scheme 3). Thus the iodinated triphosphate dCITP was reacted with PhSCu (Method B) to give the desired 5-(phenylsulfanyl)-dCTP (dCPhSTP) in low 7% yield. Better conversions were achieved when using reactions with diaryldisulfides or diselenides (Method A). The desired dCThSTP and dCPhSeTP were obtained in good yields of 24 and 31% (which are fully comparable to the typical yields of the cross-coupling reactions of dNTPs11–14).
Polymerase incorporation of modified nucleotides
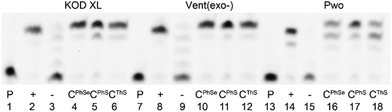
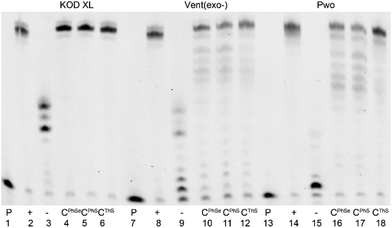
The three new 5-S- or Se-linked dNTPs (dCPhSTP, dCThSTP and dCPhSeTP) were then tested as substrates for DNA polymerases. At first we tested them in a primer extension (PEX) reaction with KOD XL, Vent(exo-) or Pwo polymerases, 15-mer primer248-sh and a 19-mer template tempoligo1C (for sequences, see Table 1). Fig. 1 shows the PAGE analysis of the PEX reactions. While KOD XL and Vent(exo-) polymerases gave quite clean bands of the 19-mer oligonucleotide (ON) products bearing one modified dCRX nucleotide, Pwo gave a mixture of the full-lengths and a truncated product.| Oligo | Sequence |
|---|---|
| Primer248-sh | 5′-CATGGGCGGCATGGG-3′ |
| Tempoligo1C | 5′-CCCGCCCATGCCGCCCATG-3′ |
| TempPrb4baseII | 5′-CTAGCATGAGCTCAGTCCCATGCCGCCCATG-3′ |
| PrimerLT25TH | 5′-CAAGGACAAAATACCTGTATTCCTT-3′ |
| PrimerL20 | 5′-GACATCATGAGAGACATCGC-3′ |
| TempFVL-A | 5′-GACATCATGAGAGACATCGCCTCTGGGCTAATAGGACTACTTCTAATCTGTAAGAGCAGATCCCTGGACAGGCAAGGAATACAGGTATTTTGTCCTTG-3′ |
Then we tested the same nucleotides (dCPhSTP, dCThSTP and dCPhSeTP) in a more challenging PEX reaction using a 31-mer template tempPrb4baseII (Fig. 2). This PEX reaction leads to a 31-mer DNA containing four modified dCRX nucleotides. KOD XL was found to be the best polymerase which gave clean full-length products in all three cases, whereas the other two enzymes gave less clean products containing minor amounts of truncated products. The PEX products were characterized by MALDI-TOF analysis (Table 2).
| ssDNA | M (calc.) (Da) | M (found) [M or M + H]+ (Da) |
|---|---|---|
| ON4ThS | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 073.2 073.2 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 075.1 075.1 |
| ON4PhS | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 049.3 049.3 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050.8 050.8 |
| ON4PhSe | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 241.1 241.1 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240.2 240.2 |
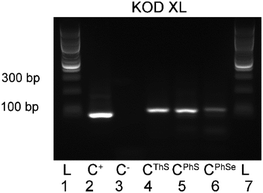
Finally, we tested the nucleotides (dCPhSTP, dCThSTP and dCPhSeTP) in PCR amplification using a 98-mer template (tempFLV-A). Fig. 3 shows that all three dNTPs were good substrates of KOD XL polymerase in PCR reaction and gave the corresponding full-length amplified products (double-stranded DNA with modification in both strands). The yield of PCR with dCPhSeTP was further improved by the addition of Mg2+ or a higher concentration of the modified nucleotide (see Fig. S1–S3 in the ESI†).
Conclusions
In conclusion, we developed a new method for the direct functionalization of 5-iodopyrimidine and 7-iodo-7-deazapurine nucleosides and nucleotides based on Cu-mediated arylsulfanylation or aryl/alkylselenylation. The reactions are even applicable for modification of fragile halogenated dNTPs. The S- or Se-modified dNTPs are good substrates for DNA polymerases and can be used as building blocks for the enzymatic synthesis of modified ONs or DNA.In this way, an aryl substituent can be attached to the nucleobase (in nucleoside, nucleotides or DNA) through a flexible sp3-hybridized sulfide or selenide linkage, which in principle offers a possibility for further transformations (e.g. oxidations). The arylsulfanyl or arylselenyl group can also serve as a radical precursor and the selenyl substituents can be used for X-ray crystallography of nucleic acids. The aryl group can be functionalized (e.g. NO2 or MeO groups) so the approach can be potentially used for redox28 or fluorescent29 labelling of nucleic acids. Research along these lines is ongoing.
Experimental
For the full Experimental part, procedures and characterization of all compounds, see the ESI.†Acknowledgements
This work was supported by the Academy of Sciences of the Czech Republic (RVO: 61388963 and Praemium Academiae to M. H.), the Czech Science Foundation (14-04289S to F. B., M. S. and M. H.), and Gilead Sciences, Inc. (Foster City, CA, U. S. A.).Notes and references
- Modified Nucleic Acids, Nucleic Acids and Molecular Biology Series, ed. K. Nakatani and Y. Tor, Springer, 2016, vol. 31, pp. 1–276 Search PubMed.
- Reviews: (a) A. Hottin and A. Marx, Acc. Chem. Res., 2016, 49, 418–427 CrossRef CAS PubMed; (b) M. Hollenstein, Molecules, 2012, 17, 13569–13591 CrossRef CAS PubMed.
- Reviews: (a) M. Hocek, J. Org. Chem., 2014, 79, 9914–9921 CrossRef CAS PubMed; (b) J. Dadová, H. Cahová and M. Hocek, Polymerase Synthesis of Base-Modified DNA, in Modified Nucleic Acids, ed. K. Nakatani and Y. Tor, Springer International Publishing, Cham, 2016, pp. 123–144 Search PubMed.
- (a) J. Ludwig, Acta Biochim. Biophys. Acad. Sci. Hung., 1981, 16, 131–133 CAS; (b) T. Kovacs and L. Otvos, Tetrahedron Lett., 1988, 29, 4525–4528 CrossRef CAS.
- Reviews on the cross-coupling reactions of unprotected nucleosides and nucleotides: (a) K. H. Shaughnessy, Molecules, 2015, 20, 9419–9454 CrossRef CAS PubMed; (b) G. Hervé, G. Sartori, G. Enderlin, G. Mackenzie and C. Len, RSC Adv., 2014, 4, 18558–18594 RSC.
- M. Welter, D. Verga and A. Marx, Angew. Chem., Int. Ed., 2016, 55, 10131–10135 CrossRef CAS PubMed.
- (a) A. Baccaro, A. L. Steck and A. Marx, Angew. Chem., Int. Ed., 2012, 51, 254–257 CrossRef CAS PubMed; (b) X. Ren, M. Gerowska, A. H. El-Sagheer and T. Brown, Bioorg. Med. Chem., 2014, 22, 4384–4390 CrossRef CAS PubMed.
- V. Raindlová, R. Pohl, B. Klepetářová, L. Havran, E. Šimková, P. Horáková, H. Pivoňková, M. Fojta and M. Hocek, ChemPlusChem, 2012, 77, 652–662 CrossRef.
- (a) V. Borsenberger, M. Kukwikila and S. Howorka, Org. Biomol. Chem., 2009, 7, 3826–3835 RSC; (b) V. Borsenberger and S. Howorka, Nucleic Acids Res., 2009, 37, 1477–1485 CrossRef CAS PubMed.
- (a) X. Yang, C. Dai, A. D. Molina and B. Wang, Chem. Commun., 2010, 46, 1073–1075 RSC; (b) Y. Cheng, C. Dai, H. Peng, S. Zheng, S. Jin and B. Wang, Chem. – Asian J., 2011, 6, 2747–2752 CrossRef CAS PubMed.
- P. Capek, H. Cahová, R. Pohl, M. Hocek, C. Gloeckner and A. Marx, Chem. – Eur. J., 2007, 13, 6196–6203 CrossRef CAS PubMed.
- L. H. Thoresen, G. S. Jiao, W. C. Haaland, M. L. Metzker and K. Burgess, Chem. – Eur. J., 2003, 9, 4603–4610 CrossRef CAS PubMed.
- J. Dadová, P. Vidláková, R. Pohl, L. Havran, M. Fojta and M. Hocek, J. Org. Chem., 2013, 78, 9627–9637 CrossRef PubMed.
- A. Krause, A. Hertl, F. Muttach and A. Jäschke, Chem. – Eur. J., 2014, 20, 16613–16619 CrossRef CAS PubMed.
- A. Hampton, R. R. Chawla and F. Kappler, J. Med. Chem., 1982, 25, 644–649 CrossRef CAS PubMed.
- M. Ahmadian, P. Zhang and D. E. Bergstrom, Nucleic Acids Res., 1998, 26, 3127–3135 CrossRef CAS PubMed.
- (a) J. M. San Pedro and M. M. Greenberg, ChemBioChem, 2013, 14, 1590–1596 CrossRef CAS PubMed; (b) M. J. Resendiz, A. Schön, E. Freire and M. M. Greenberg, J. Am. Chem. Soc., 2012, 134, 12478–12481 CrossRef CAS PubMed.
- M. Klečka, L. P. Slavětínská, E. Tloušťová, P. Džubák, M. Hajdúch and M. Hocek, MedChemComm, 2015, 6, 576–580 RSC.
- (a) R. F. Schinazi, J. Arbiser, J. J. S. Lee, T. I. Kalman and W. H. Prusoff, J. Med. Chem., 1986, 29, 1293–1295 CrossRef CAS PubMed; (b) S. Choi, T. I. Kalman and T. J. Bardos, J. Med. Chem., 1979, 22, 618–621 CrossRef CAS PubMed.
- (a) A. E. Hassan, J. Sheng, J. Jiang, W. Zhang and Z. Huang, Org. Lett., 2009, 11, 2503–2506 CrossRef CAS PubMed; (b) W. Zhang, J. Sheng, A. E. Hassan and Z. Huang, Chem. – Asian J., 2012, 7, 476–479 CrossRef CAS PubMed; (c) Z. Wen, H. E. Abdalla and H. Zhen, Sci. China: Chem., 2013, 56, 273–278 CrossRef PubMed.
- (a) I. S. Hong and M. M. Greenberg, Org. Lett., 2004, 6, 5011–5013 CrossRef CAS PubMed; (b) I. S. Hong and M. M. Greenberg, J. Am. Chem. Soc., 2005, 127, 10510–10511 CrossRef CAS PubMed; (c) I. S. Hong, H. Ding and M. M. Greenberg, J. Am. Chem. Soc., 2006, 128, 2230–2231 CrossRef CAS PubMed; (d) X. Peng, I. S. Hong, H. Li, M. M. Seidman and M. M. Greenberg, J. Am. Chem. Soc., 2008, 130, 10299–10306 CrossRef CAS PubMed.
- J. Sheng, A. E. Hassan, W. Zhang, J. Zhou, B. Xu, A. S. Soares and Z. Huang, Nucleic Acids Res., 2011, 39, 3962–3971 CrossRef CAS PubMed.
- M. P. Kotick, C. Szantay and T. J. Bardos, J. Org. Chem., 1969, 34, 3806–3813 CrossRef CAS PubMed.
- D. E. Bergstrom, P. Beal, J. Jenson and X. Lin, J. Org. Chem., 1991, 56, 5598–5602 CrossRef CAS.
- D. G. Kananovich, A. Reino, K. Ilmarinen, M. Roomuskos, M. Karelsson and M. Loop, Org. Biomol. Chem., 2014, 12, 5634–5644 CAS.
- M. Abdo, Y. Zhang, V. L. Schramm and S. Knapp, Org. Lett., 2010, 13, 2982–2985 CrossRef PubMed.
- (a) N. Taniguchi and T. Onami, Synlett, 2003, 829–832 CrossRef CAS; (b) N. Taniguchi and T. Onami, J. Org. Chem., 2004, 69, 915–920 CrossRef CAS PubMed.
- J. Balintová, M. Plucnara, P. Vidláková, R. Pohl, L. Havran, M. Fojta and M. Hocek, Chem. – Eur. J., 2013, 19, 12720–12731 CrossRef PubMed.
- D. Dziuba, P. Jurkiewicz, M. Cebecauer, M. Hof and M. Hocek, Angew. Chem., Int. Ed., 2016, 55, 174–178 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental part, and copies of spectra. See DOI: 10.1039/c6ob01917j |
| ‡ Passed away on July 11, 2016. |
| This journal is © The Royal Society of Chemistry 2016 |