A survey of indicator parameters to monitor regrowth in unchlorinated drinking water†
Paul W. J. J.
van der Wielen
*a,
Geo
Bakker
b,
Adrie
Atsma
b,
Maarten
Lut
c,
Guus
Roeselers
d and
Bendert
de Graaf
b
aKWR Watercycle Research Institute, P.O. Box 1072, 3430 BB, Nieuwegein, The Netherlands. E-mail: paul.van.der.wielen@kwrwater.nl
bVitens, Zwolle, The Netherlands
cOasen, Gouda, The Netherlands
dTNO, Zeist, The Netherlands
First published on 8th March 2016
Abstract
The objective of our study was to explore microbiological parameters that are suitable as indicators for regrowth in distribution systems that receive unchlorinated drinking water in the Netherlands. Treated water and distributed water at two locations in the distribution system of 28 treatment plants were analyzed for a range of biological parameters. The results demonstrated that Aeromonas, HPCs, and Mycobacterium were significantly (more often) higher and ATP was significantly (more often) lower in the distributed water than in the treated water, whereas cell numbers remained stable during distribution. Average fungal 18S rRNA gene copies did not differ significantly between the treated and the distributed water, but fungal numbers were significantly more often higher in the distributed water than in the corresponding treated water. These results demonstrate that the distribution system has a clear effect on HPCs, Aeromonas, Mycobacterium, ATP and, to a lesser extent, on fungi in drinking water, but not on cell numbers. Furthermore, we noticed that ATP in the treated water is a better predictor for the regrowth level in the distribution system than total, membrane-intact or HNA cell numbers. In contrast to previous studies, we conclude that ATP and cell numbers seem to be poor indicators for regrowth in the drinking water environment, whereas HPCs, Aeromonas, Mycobacterium and, to a lesser extent, fungi appear to be more reliable indicators to monitor regrowth.
Water impactDrinking water companies assess regrowth in drinking water distribution systems by monitoring microbial parameters. This study demonstrates that general microbiological parameters (ATP and cell numbers) are poor indicators for regrowth in distribution systems, whereas parameters targeting more specific microbial groups (heterotrophic or Aeromonas plate counts, Mycobacterium spp. and fungi) are reliable indicators to monitor regrowth in distribution systems. |
Introduction
The drinking water environment provides an ecosystem for (re)growth of microorganisms. Growth of these microorganisms can occur in drinking water during the relatively short transportation time to consumer (days) or on surfaces that are in contact with drinking water (e.g. pipes, sediment). Moreover, microbial growth in drinking water can be problematic, because it may result in multiplication of opportunistic pathogenic bacteria like Legionella pneumophila,1 nontuberculous mycobacteria,2,3 or Pseudomonas aeruginosa.3,4 In addition, microbial growth can also result in esthetic problems, e.g. deteriorated taste and odor,5 growth of invertebrates,6,7 and/or technical problems, e.g. corrosion of pipe materials.8 Consequently, growth of microorganisms in drinking water (distribution system) is normally controlled by maintaining a disinfectant residual in drinking water and/or by limiting the concentration of growth-promoting compounds, such as easily assimilable organic carbon (AOC), in water that enters the distribution system. The latter is the case in the Netherlands.Drinking water companies in many countries monitor microbiological drinking water quality according to their national legislation. In most countries, heterotrophic plate counts (HPCs) in treated and distributed drinking water are determined as an indicator for microbial water quality and regrowth. The Drinking Water Decree in the Netherlands also states that HPCs have to be determined in treated and distributed drinking water, but an additional parameter for regrowth, Aeromonas plate counts (Aeromonas), is also included in the Drinking Water Decree.9
Although monitoring of HPCs and Aeromonas has proven their value as indicators for the microbiological quality of drinking water in the Netherlands, there are some limitations related to the use of these parameters. HPCs and Aeromonas constitute only a small fraction (<1%) of the total microbial community in drinking water,10–12 and these methods are based on cultivation on selective agar media, which is time-consuming. Therefore, alternative parameters that can be determined relatively fast and easily, like adenosine triphosphate concentration (ATP) or cell counts, have been proposed as indicators for regrowth or as monitoring parameters for drinking water quality.12,13 Despite the attractive characteristics of ATP and cell count assays in drinking water, it remains uncertain how well these parameters relate to regrowth in unchlorinated drinking water in the Netherlands, since biofilm growth on pipe walls and sediments contribute to regrowth in drinking water distribution systems as well.14–17
The objective of our study is to investigate whether traditional parameters (HPCs and Aeromonas plate counts) and/or alternative parameters (ATP concentration, cell counts, 16S rRNA gene copy numbers of Mycobacterium spp., 18S rRNA gene copy numbers of fungi and community composition) in drinking water can be used as indicators for microbial regrowth in distribution systems that are fed with drinking water without a disinfectant residual.
Materials and methods
Sample campaign
The sample campaign took place from May till September 2012. The treated water and distributed drinking water at two locations (selected randomly) in a distribution system, taken from a kitchen cold tap, were sampled from 28 different treatment plants in the Netherlands, which use groundwater as a source for drinking water production. Treatment differed between plants, but in general consisted of marble filtration for oxic groundwater treatment or aeration followed by one- or two-step rapid sand filtration with or without softening for anoxic groundwater treatment.Drinking water samples (1500 ml) were taken from the kitchen cold water tap of two houses connected to the distribution system of each treatment plant. Taps were flushed until temperature was constant for 30 seconds, followed by an additional 15 min of flushing before samples were taken to ensure that the drinking water sample came from the distribution system. In addition, a sample was also taken from the treated water at each treatment plant.
Microbiological analyses
The total ATP concentration was determined in duplicate in all the drinking water samples by measuring the amount of light produced in a luciferin–luciferase assay as previously described by van der Wielen & van der Kooij.12 In a previous study, it was shown and explained that free ATP is negligible in drinking water samples from the Netherlands,12 meaning that total ATP concentrations determined in this study are a reliable measure for active biomass. The total and membrane-intact cell counts were determined using a flow cytometer, as described previously.3 The number of high nucleic acid (HNA) cells, which has been suggested to be related to more active bacteria that respond to dynamic changes,18 and the number of low nucleic acid (LNA) cells, which has been suggested to be related to less active bacteria that remain more constant during changes,18 were enumerated by applying gates to two cluster patterns obtained after flow cytometer analysis which are indicative of HNA and LNA cells.18 HPCs and Aeromonas were determined according to the Dutch Drinking Water Decree.9 This means that the samples were analyzed according to ISO 6222:1999 for HPCs and Aeromonas plate counts were determined as previously described.12DNA isolation
A volume of 1000 ml was filtered through a 25-mm polycarbonate filter (0.22 μm pore size, type GTTP; Millipore, The Netherlands). The filter and a DNA fragment of an internal control were added to phosphate/MT buffer in a Lysing Matrix E tube of the FastDNA® SPIN Kit for Soil (Qbiogene Inc, Carlsbad, CA) and stored at −20 °C. The internal control was used to determine the recovery efficiency of DNA isolation and PCR analysis19 in the quantitative PCR (qPCR) analyses. DNA was isolated according to the supplier's protocol and the eluted DNA was subsequently kept at −80 °C.t-RFLP analysis
DNA was amplified with bacterial 16S rRNA specific primer 8F,20 containing the modification described by Heuer et al.,21 in combination with 1392R.22 The 8F primer was fluorescently labelled with FAM. PCR reactions were performed using 10 μl of DNA template in a reaction volume of 50 μl using amplitaq Gold (Invitrogen, The Netherlands) with the following PCR program: 5 min at 94 °C; 40 cycles of 45 s at 94 °C, 1 min at 57 °C and 2 min at 72 °C; 7 min at 72 °C. Subsequently, PCR products were purified and concentrated using DNA clean and concentrator columns (Zymo Research, The Netherlands), digested with the HhaI restriction enzyme for 6 hours at 36 °C and purified and concentrated again using the same clean and concentrator columns. Purified digestion products (5 μL) were mixed with 15 μL of loading buffer Hi-di formamide and 1 μl of internal standard GeneScan-1000 ROX (Applied Biosystems, the Netherlands). After heating for 3 min at 95 °C and subsequent cooling on ice, fluorescently labelled terminal restriction fragments (T-RFs) were size separated on an ABI Prism 310 genetic analyzer (Applied Biosystems, the Netherlands) in GeneScan analysis mode (Applied Biosystems, the Netherlands). T-RFLP electropherograms were analyzed with BioNumerics 6.0 software (Applied Maths, Belgium). Unweighted pair group method with arithmetic mean (UPGMA) cluster analysis was based on the densitometric curve-based Pearson correlation coefficient between the T-RF band patterns of the drinking water samples.Quantitative PCR analyses
The gene copy number of nontuberculous mycobacteria and fungi were determined in duplicate with previously developed qPCR analyses for drinking water samples.23 These qPCR methods target the 16S rRNA gene of Mycobacterium spp. and the 18S rRNA gene of fungi. Reaction mixtures for PCR analyses contained 25 μl of 2× IQTM SYBR® Green Supermix (Bio-Rad Laboratories BV, the Netherlands), 0.2 μmol l−1 each primer (and if applicable, probe), 0.4 mg ml−1 bovine serum albumin and 10 μl DNA template in a total volume of 50 μl. Amplification, detection and data analysis were performed in an iCycler CFX Real-Time detection system (Bio-Rad laboratories BV, the Netherlands). Primer/probe sequences and amplification programs were according to van der Wielen et al.23 The PCR cycle after which the fluorescence signal of the amplified DNA is detected (threshold cycle Ct), was used to quantify the gene copy concentration. Quantification was based on comparison of the sample Ct value with the Ct values of a calibration curve based on known copy numbers of the respective gene from the different microorganisms.Statistical analysis
Two statistical methods were done to test the hypothesis whether each parameter was significantly lower or higher in distributed drinking water than in treated water at the plant. In the first method, the values of each microbiological parameter from all the treated water samples were compared with the values from all the distributed water samples using the Mann–Whitney test. In the second method, a Wilcoxon signed-rank test was performed to determine whether microbiological parameters increased or decreased during transport in the distribution system of each of the 28 treatment plants (two locations for each plant). The microbiological parameters were determined at 56 locations in the distribution system of 28 plants. For each microbiological parameter, the difference between a distributed drinking water sample and the corresponding treated water sample (total of 56 cases) were ranked and statistically evaluated using the Wilcoxon signed-rank test. Differences for both methods were considered statistically significant at p < 0.05.Results
Traditional parameters
The geometric mean ×/ geometric standard deviation of HPCs obtained from unchlorinated treated and distributed drinking water of 28 different treatment plants are shown in Table 1, whereas the HPC values for the individual samples are shown in Fig. S1.† HPCs varied between <1 and 17 cfu ml−1 in the treated water with a geometric mean of 1.8 ×/ 2.4 cfu ml−1 and between <1 and 85 cfu ml−1 with a geometric mean of 3.9 ×/ 3.8 cfu ml−1 in the water samples taken from the distribution system. Statistical analysis demonstrated that the HPC values in the distributed drinking water were significantly higher than those in the treated water (p < 0.05). However, this statistical result comes from all the pooled samples from 28 different treatment plants and it is possible that this statistical difference is caused by (large) differences between treated and distributed water in only a few treatment plants. Additional analysis was, therefore, done in which each distributed sample was compared with its corresponding treated water sample. In 37 cases (66.1%), HPCs were higher and in 11 cases (19.6%), HPCs were lower in the distributed drinking water sample than in the corresponding treated water sample (Table 2). The remaining eight cases had equal numbers in treated and distributed water (n = 3; 5.4%) or HPC numbers were below the detection limit of 1 cfu ml−1 in both samples (n = 5; 8.9%). The Wilcoxon-rank test revealed that there were significantly more cases with higher HPCs in distributed water than in treated water than cases with lower or equal numbers in distributed water (p < 0.05).| Treated water | Distributed water | ||||
|---|---|---|---|---|---|
| Geometric mean | Geometric stdev | Geometric mean | Geometric stdev | p-value | |
| HPCs (cfu ml−1) | 1.8 | 2.4 | 3.9 | 3.8 | <0.05 |
| Aeromonas (cfu 100 ml−1) | 1.3 | 2.2 | 6.6 | 11.0 | <0.05 |
| ATP (ng L−1) | 2.3 | 2.0 | 1.5 | 1.5 | <0.05 |
| Total cells (N ml−1) | 1.2 × 105 | 2.5 | 1.1 × 105 | 2.7 | >0.05 |
| Membrane-intact cells (N ml−1) | 1.1 × 105 | 2.5 | 1.0 × 105 | 2.7 | >0.05 |
| LNA cells (N ml−1) | 8.2 × 104 | 2.4 | 8.0 × 104 | 2.8 | >0.05 |
| HNA cells (N ml−1) | 2.9 × 104 | 2.7 | 2.6 × 104 | 2.8 | >0.05 |
| Mycobacterium spp. (gc l−1) | 1.7 × 105 | 2.4 | 4.4 × 105 | 2.8 | <0.05 |
| Fungi (gc l−1) | 1.1 × 105 | 2.8 | 1.5 × 105 | 3.1 | >0.05 |
| Higher | Lower | Equal | <DLa | |
|---|---|---|---|---|
| a DL: both samples (distributed and the corresponding treated water) below the detection limit. | ||||
| HPCs (cfu ml−1) | 66.1% | 19.6% | 5.4% | 8.9% |
| Aeromonas (cfu 100 ml−1) | 41.1% | 7.1% | 1.8% | 50% |
| ATP (ng l−1) | 14.2% | 71.4% | 3.6% | 10.8% |
| Total cells (N l−1) | 46.4% | 46.4% | 7.2% | 0.0% |
| Membrane-intact cells (N l−1) | 42.9% | 48.2% | 8.9% | 0.0% |
| LNA cells (N l−1) | 46.4% | 44.6% | 8.9% | 0.0% |
| HNA cells (N l−1) | 42.9% | 50.0% | 7.1% | 0.0% |
| Mycobacterium spp. (gc l−1) | 84.0% | 14.0% | 2.0% | 0.0% |
| Fungi (gc l−1) | 70.8% | 29.2% | 0.0% | 0.0% |
The Aeromonas plate counts varied in general between <10 and 30 cfu 100 ml−1 in the treated water with a geometric mean of 1.3 ×/ 2.2 cfu 100 ml−1 and between <10 and 1300 cfu 100 ml−1 with a geometric mean of 6.6 ×/ 11.0 in the distributed drinking water (Table 1; Fig. S2†). Aeromonas numbers were significantly higher in the distributed water samples than in the treated water samples (p < 0.05). A comparison between each distributed drinking water sample and its corresponding treated water sample showed that in 28 cases (50%), Aeromonas plate counts in the distributed water sample and the corresponding treated water sample were below the detection limit, and for these cases it cannot be determined whether Aeromonas increased or decreased during distribution. In 23 cases (41.1%), higher Aeromonas counts in the distributed drinking water were observed than in the corresponding treated water (Table 2). In four cases (7.1%), Aeromonas counts in distributed water were lower than in treated water and in one case (1.8%), Aeromonas counts were equal. There were significantly more cases with higher Aeromonas counts in distributed drinking water compared to treated water than cases with equal or lower Aeromonas counts in distributed water compared to treated drinking water (p < 0.05). We conclude from these HPC and Aeromonas results that, in general, regrowth of these bacterial species occurs in drinking water distribution systems that are fed with unchlorinated drinking water produced from groundwater.
ATP and cell counts
The ATP concentrations in the treated water varied between <1 to 7.6 ± 0.1 ng ATP l−1, whereas in the distributed water, concentrations ranged from <1 to 5.0 ± 0.2 ng ATP l−1 (Fig. S3†). The geometric means were 2.3 ×/ 2.0 and 1.5 ×/ 1.5 ng l−1 for treated and distributed water, respectively, which differed significantly (p < 0.05)(Table 1). The ATP concentration in the distributed drinking water sample was lower than in the corresponding treated water sample in 40 cases (71.4%), whereas the ATP concentration in the distributed drinking water was higher than in the treated water in eight cases (14.2%) (Table 2). The ATP concentration in the distributed drinking water and in the treated water was equal (n = 2; 3.6%) or below the detection limit of 1 ng ATP l−1 (n = 6; 10.7%) in the remaining eight cases. The Wilcoxon-rank test demonstrated that there were significantly more cases where the ATP concentration in the distributed water was lower compared to the corresponding treated water than cases with higher or equal ATP concentrations in the distributed water (p < 0.05). We conclude from these results that the ATP concentrations decreased during transport in the distribution systems studied. The decrease in ATP concentration during water distribution was observed irrespective of the distance of the location from the treatment plant (data not shown).Total cell counts varied between 1.2 × 104 to 5.4 × 105 cells ml−1 (geometric mean: 1.2 × 105 ×/ 2.5 cells ml−1) in the treated water and did not differ significantly (p < 0.05) from the total cell counts in the distributed water, which varied in general between 1.3 × 104 to 3.9 × 105 cells ml−1 (geometric mean: 1.1 × 105 ×/ 2.7 cells ml−1) (Table 1; Fig. S4†). In 26 cases (46.4%), total cell counts were higher and in 26 other cases (46.4%), total cell counts were lower in the sample from the distribution system than in the corresponding treated water sample. The remaining four cases (7.1%) had equal cell counts in the distributed and treated water. These differences were not statistically significant (p > 0.05). Both statistical analyses, thus, indicated that total cell counts did not show a consistent change during distribution of drinking water from these 28 different plants. Moreover, total cell counts did only increase or decrease slightly (less than two times higher or lower) during distribution at most locations. Only at three locations (plants 10, 12 and 24) were cell counts more than two times higher or lower in the distributed drinking water sample than in the treated water sample (plants 10, 12 and 24). Besides total cells, membrane-intact, HNA and LNA cells were also enumerated with flow cytometry (Tables 1 and 2, Fig. S5–S7†). In general, 83 to 97% of the cells had an intact membrane and 54 to 95% were LNA cells. Strong significant correlations (p < 0.05) were observed between total cells counts and membrane-intact cell counts (R2 = 0.99), LNA cell counts (R2 = 0.97) and HNA cell counts (R2 = 0.75), demonstrating that membrane-intact, LNA and HNA cell counts followed the same trend as total cell counts. Moreover, the statistical results of the two statistical analyses demonstrated that membrane-intact, LNA and HNA cells counts did not differ as well between the treated and distributed drinking water samples (p < 0.05). We conclude from these results that total, membrane-intact and HNA cell counts remain fairly stable during drinking water distribution at these 28 plants.
t-RFLP
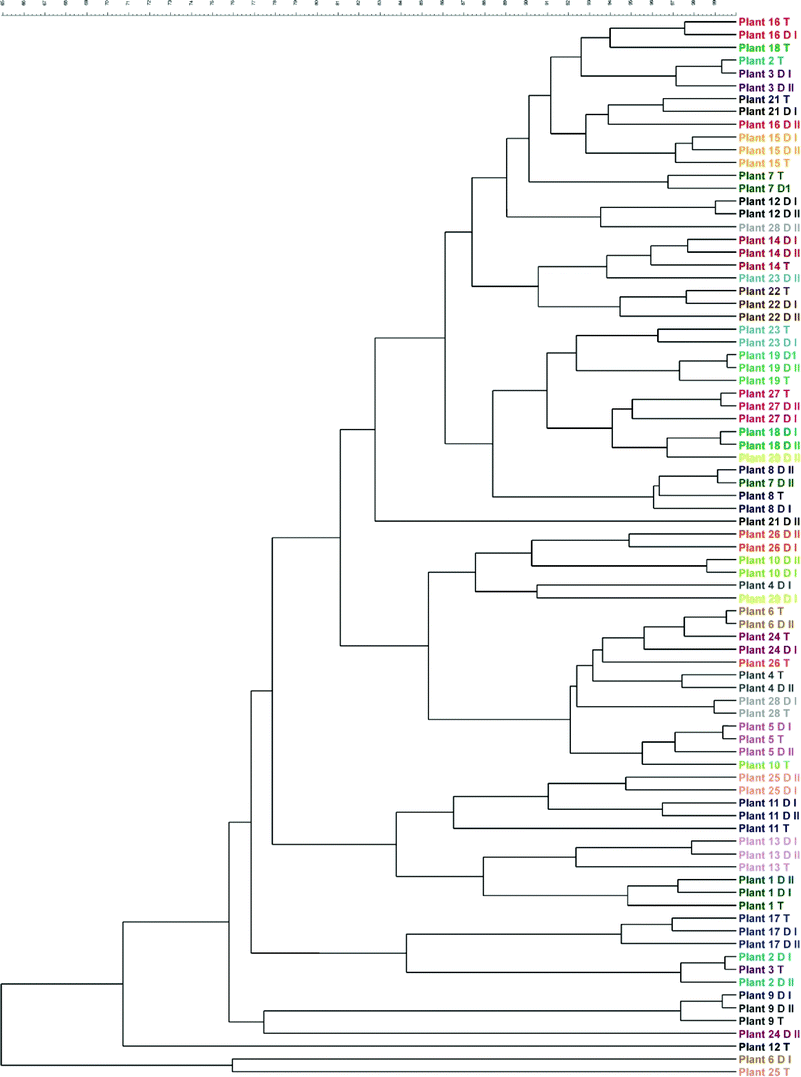
The dominant bacterial communities in the treated water and distributed water of each treatment plant were also compared using abundance analysis (densitometric curve-based Pearson correlation coefficient) of the t-RFLP banding patterns (Fig. 1). The dominant bacterial composition in the treated water and in the water at the two locations in the distribution system were similar at ten treatment plants (35.7%). At nine treatment plants (32.1%), the bacterial communities in the treated water were similar to the bacterial community in one of the two corresponding distributed drinking water samples. Finally, the bacterial communities in the two distributed drinking water samples were similar, but differed from the bacterial community in the corresponding treated water at nine plants (32.1%). Remarkably, none of the treatment plants showed different bacterial communities among all three samples analysed (i.e. treated water and the two distributed drinking water samples). Thus, in general, the dominant bacterial community composition does not seem to change considerably during unchlorinated drinking water distribution. Furthermore, the dominant bacterial communities in the water samples differed considerably between plants, indicating that the drinking water of each treatment plant harbors a unique bacterial community.Nontuberculous mycobacteria and fungi
The treated water sampled from plants 7 and 10 and one of the two distributed drinking water samples from plants 6, 9, 19 and 24 did not result in reliable gene copy numbers. Therefore, the treated and/or distributed drinking water samples of these plants were not included in the analysis. The 16S rRNA gene of Mycobacterium spp. was detected in all the drinking water samples and gene copy (gc) numbers varied in general between 4.7 × 104 and 1.4 × 106 gc l−1 with a geometric mean of 1.7×105 ×/ 2.4 gc l−1 in the treated water and between 7.6 × 104 and 3.1 × 106 gc l−1 with a geometric mean of 4.4 × 105 ×/ 2.8 gc l−1 in the distributed water (Table 1; Fig. S8†). Statistical analysis demonstrated that the Mycobacterium numbers in the distributed drinking water were significantly higher than in treated water (p < 0.05). In 42 cases (84.0%), Mycobacterium numbers were higher in the distributed drinking water than in the corresponding treated water (Table 2). The other eight cases had lower (n = 7; 14.0%) or equal (n = 1; 2%) Mycobacterium numbers in the distributed water compared to the corresponding treated water. Significantly, more cases had higher Mycobacterium numbers in the distributed drinking water compared to the corresponding treated water than cases with lower or equal Mycobacterium numbers in the distributed water compared to the corresponding treated water (p < 0.05).Fungi were detected as well in all the drinking water samples using qPCR and the 18S rRNA gene copy numbers between the treated and distributed water did not differ significantly (p < 0.05) (Table 1). The numbers varied between 3.0 × 104 and 1.5 × 106 gc l−1 (geometric mean: 1.1 × 105 ×/ 2.8 gc l−1) in the treated water and between 1.3 × 104 and 4.5 × 106 gc l−1 (geometric mean: 1.5 × 105 ×/ 3.1 gc l−1) in the distributed water (Table 1; Fig. S9†). Fungal numbers were higher in the distributed drinking water than in the corresponding treated water in 34 cases (70.8%) and lower than the corresponding treated water in the remaining 14 cases (29.2%) (Table 2). The Wilcoxon rank test showed significantly more cases with higher fungal numbers in the distributed water compared to the corresponding treated water than cases with lower fungal numbers in the distributed compared to the corresponding treated water (p < 0.05). Thus, in general, regrowth of nontuberculous mycobacteria and fungi occurs in the drinking water distribution systems that were studied.
Discussion
Regrowth in a drinking water distribution system occurs on pipe walls (biofilm formation), sediments and/or drinking water,14–17,24 and growth in each of these three different niches can induce water quality problems. The parameters that have been investigated in our study have been identified in all these three niches, but seem to differ in the preferred niche they occupy, with fungi being dominantly present in pipe wall biofilms, Aeromonas and HPCs in sediments and Mycobacterium spp., ATP and cell numbers in pipe wall biofilms and sediments.15,24 Microbial growth in pipe wall biofilms, sediments and water will result in increased microbial numbers in drinking water and, therefore, regrowth indicators measured in drinking water should reflect growth in each of these three niches. In the past, ATP and cell counts have been proposed as alternative parameters for HPCs to determine microbial regrowth in drinking water distribution system.11–13,18,25,26 The advantages of using ATP and cell counts over HPCs are that ATP and cell counts can be determined relatively fast (seconds to minutes) and are better indicators for active biomass (ATP) or total cell counts than HPCs, since only a small percentage of bacterial cells in drinking water can be cultivated by HPCs.10–12 In this study, we have demonstrated that ATP concentrations and cell counts in drinking water can differ between 28 treatment plants, but that these parameters decrease or remain stable during transport in the distribution system of most treatment plants. Hence, these results dispute whether monitoring these parameters in the distribution system can be used as reliable indicators for regrowth in the drinking water distribution system. The results from the t-RLFP analysis further support this disputation, since t-RFLP profiles demonstrated for most cases that the dominant bacterial community composition remained stable during transport of drinking water from the treatment plant to locations in the distribution system. This observation was also confirmed by our accompanying study where bacterial communities from the same samples were analyzed by next generation sequencing of the 16S rRNA gene.27The observation that ATP concentrations decreased, but total cell counts and membrane-intact cells remained relatively stable during drinking water distribution, demonstrates that the ATP per membrane-intact cell ratio decreases in the distributed drinking water. This decrease in ATP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cell ratio indicates that the dominant membrane-intact microbial cells lose activity when transported in the distribution system. It has been reported that microbial cells in the treated water originate from process(es) in the treatment train,27,28 which is mostly marble or rapid sand filtration at the treatment plants studied. The loss of cell activity in microorganisms during transport from treatment plants to consumers' tap is likely due to the different environmental conditions in treatment filters compared to the drinking water distribution system.
cell ratio indicates that the dominant membrane-intact microbial cells lose activity when transported in the distribution system. It has been reported that microbial cells in the treated water originate from process(es) in the treatment train,27,28 which is mostly marble or rapid sand filtration at the treatment plants studied. The loss of cell activity in microorganisms during transport from treatment plants to consumers' tap is likely due to the different environmental conditions in treatment filters compared to the drinking water distribution system.
We observed for most treatment plants that, in contrast to ATP and cell counts, additional microbiological parameters (HPCs, Aeromonas, Mycobacterium spp. and, to a lesser extent, fungi) were higher in the distributed drinking water than in the treated water, suggesting that these organisms were able to multiply in the drinking water distribution system. Others have studied bacterial community compositions in biofilms and bulk water from distribution systems in Denmark and Germany.29,30 The results from those studies showed that the bacterial community in biofilms is different from the bacterial community in drinking water. Also, in our accompanying study, we observed that the bacterial community in drinking water was different from that in biofilms using next generation sequencing of the 16S rRNA gene.27 These results indicate that regrowth in drinking water distribution systems mainly takes place in biofilms on pipe walls and sediments, as concluded before.14–17 Previous studies have shown that Aeromonas, nontuberculous Mycobacterium and fungi are often detected in biofilms.31–36 It is, therefore, likely that Aeromonas, mycobacteria and fungi in drinking water have their niche in biofilms on pipe walls and/or sediments. However, exchange of bacteria from the biofilm to the water phase is apparently so low that it does not alter the dominant bacterial community during transport of water from the treatment plant to locations in the distribution system.
Regrowth monitoring in the distribution system is not a goal in itself, but it is done and/or required by legislation to indicate regrowth problems, so that causative measures can be taken. Most of the regrowth problems seem to relate to biofilms, since opportunistic pathogens like L. pneumophila are known to multiply in biofilms,37 animal vertebrates graze (in)directly on biofilms,6,7 and microbial induced corrosion is caused by growth of certain bacteria (e.g. sulphate reducing bacteria) in biofilms.8,38 The results from our study showed that monitoring the dominant bacterial community in unchlorinated distributed drinking water produced from groundwater using ATP measurement, cell counting or bacterial community analysis will mainly give information about the bacterial water quality at the treatment plant. Measuring these parameters in the distribution system does not seem to give information regarding regrowth in the drinking water distribution system. Consequently, it is not recommended to perform ATP or cell count (total cells, membrane-intact cells, HNA cells) measurements in the distribution system as an indication for regrowth in distribution systems in the Netherlands. HPCs and Aeromonas plate counts of distributed drinking water, which are included in the legislation in the Netherlands, seem to be better parameters to monitor regrowth, although levels can be below the detection limit and the methods are relatively slow and time consuming. In general, gene copy numbers of Mycobacterium and fungi also increased during distribution of water. These parameters were present in all the drinking water samples and can be determined relatively fast and easily and might, therefore, be alternative indicators for regrowth. Still, other parameters (e.g. gene copy numbers of selective genes from dominant biofilm members) might even be better indicators for regrowth problems in the distribution system; however, additional research is required to identify and test such selective gene markers.
A previous study on ATP concentrations in unchlorinated drinking water from the distribution system of six different treatment plants in the Netherlands also demonstrated that ATP did not significantly change or decrease during distribution of drinking water.12 Cell counts have also been monitored in unchlorinated drinking water of distribution systems in Switzerland and the Netherlands,11,13,26,39 but in most of these studies, samples were taken directly from the tap, public fountains or information on how samples were taken from taps is missing. Because the premise plumbing system can alter the microbiological quality in drinking water,18,40,41 such samples are probably not indicative of water quality in the distribution system. Consequently, the results from these studies cannot easily be compared to the samples analyzed in our study. Still, some of those studies observed that cell counts did not change during transport in the distribution and premises plumbing system,13,26,39 which is in agreement with the observations made in our study. Besides measuring total cell counts, flow cytometry can also determine subgroups of cells (e.g. membrane-intact, HNA and LNA cells),18,42 and it has been suggested that monitoring for these subgroups might give additional information on microbiological water quality. In our study, however, there was a strong correlation between the total cells and membrane-intact cells (R2 = 0.99), LNA cells (R2 = 0.94) or HNA cells (R2 = 0.75) and statistical analyses demonstrated that these parameters were not significantly higher in distributed water compared to treated water at the plant. The bacterial community composition in drinking water has been studied by others as well.28,29,43 The results from those studies demonstrated in general that bacterial communities in drinking water from different locations in the distribution system remained relatively constant, which is in agreement with the results from the t-RFLP analysis presented here.
Although the results from our study demonstrated that the dominant bacterial flora entering the distribution system lose cell activity during distribution, they can still serve as a substrate or food for microorganisms in biofilms or for animal invertebrates that graze on these biofilms in the distribution system,44 which will increase regrowth in distribution system. Consequently, monitoring of the treated water for (active) biomass is desirable. In that case, parameters are not used to directly demonstrate actual regrowth in the distribution system, but as indicators for the amount of (active) biomass that enters the distribution system. The concentration of (active) biomass in the treated water at the plant might then predict the amount of regrowth in the distribution system. We observed in our study that ATP and cell counts in the treated water are correlated poorly (R2 = 0.23), which poses the question whether ATP or cell counts can best be monitored in the treated water for this purpose. In a previous study, the treatment plants analyzed in our study were ranked based on routinely analyzed HPC and Aeromonas numbers in the distribution system,45 which have to be monitored as regrowth indicators according to the Drinking Water Decree in the Netherlands. This ranking (low, medium and high regrowth in the distribution system) was compared to the ATP concentration and total cell counts in the treated water that were measured in the current study (Table 3). Seven of the ten treatment plants with the highest ATP concentrations in the treated water also had the highest rank for regrowth in the distribution system (medium to high). In contrast, seven of the ten treatment plants with the highest total cell counts in the treated water had a low regrowth rank and the other three plants had a medium regrowth rank. The same observations were made with membrane-intact or HNA cells (data not shown). Mycobacterium spp. and fungi have also been determined in the treated water and those numbers were also compared with the regrowth rank (Table S1†). It was observed that seven of the ten treatment plants with the highest mycobacterial numbers in treated water also had the highest rank of regrowth. In contrast, only four of the ten plants with the highest fungal numbers in treated water had the highest regrowth rank, the other six had low regrowth ranks. Aeromonas and HPC numbers in the treated water could not reliably be compared with the regrowth rank, because the numbers were below the detection limit for several plants. These results suggest that ATP or Mycobacterium spp. in the treated water might be a better predictor than cell counts or fungi for regrowth of Aeromonas and HPCs in the distribution system. The apparent difference between ATP and cell counts can be explained by the level at which these two parameters are an indication of biomass. Several studies have demonstrated that microbial cell counts do not or poorly correlate to biomass,46–49 whereas ATP is an indication of active biomass, which correlates reasonably well with other biomass parameters under non-phosphate limiting conditions.45,46,48 Still, relatively high ATP concentrations were observed in the treated water of plants 7, 11 and 13, but the regrowth level in the distribution system of these plants were low. In addition, the relationship between ATP, cell counts, Mycobacterium spp., fungi and the regrowth rank was based on a single sample for ATP and cell counts from 28 different treatments plants. The results, therefore, can only be seen as a first indication that ATP or Mycobacterium spp. seem to be better monitoring parameters for water quality in treated water than cell counts or fungi. Additional experiments are required to confirm these possible relationships before a definite conclusion can be drawn.
| Plant | ATP (ng L−1) | Regrowth rank* | Plant | Cell counts (N mL−1) | Regrowth ranka |
|---|---|---|---|---|---|
| a Regrowth ranks were based on the average Aeromonas plate counts and HPCs in the distribution system over a three-year period and were taken from van der Wielen & van der Kooij.44 | |||||
| 19 | 7.6 | Medium | 14 | 5.4 × 105 | Medium |
| 13 | 6.6 | Low | 22 | 4.6 × 105 | Medium |
| 7 | 6.5 | Low | 11 | 4.0 × 105 | Low |
| 16 | 5.8 | Medium | 7 | 3.5 × 105 | Low |
| 11 | 5.7 | Low | 13 | 3.0 × 105 | Low |
| 22 | 4.5 | Medium | 23 | 2.8 × 105 | Low |
| 2 | 4.2 | Medium | 27 | 2.6 × 105 | Low |
| 14 | 3.8 | Medium | 1 | 2.3 × 105 | Low |
| 25 | 3.5 | High | 16 | 1.7 × 105 | Medium |
| 21 | 3.1 | Medium | 15 | 1.7 × 105 | Low |
| 3 | 2.7 | Medium | 21 | 1.6 × 105 | Medium |
| 12 | 2.7 | Medium | 25 | 1.6 × 105 | High |
| 1 | 2.6 | Low | 12 | 1.5 × 105 | Medium |
| 27 | 2.3 | Low | 19 | 1.4 × 105 | Medium |
| 15 | 2.2 | Low | 8 | 1.1 × 105 | Low |
| 23 | 2.0 | Low | 2 | 9.1 × 104 | Medium |
| 8 | 1.8 | Low | 20 | 9.0 × 104 | Low |
| 17 | 1.6 | Medium | 17 | 8.5 × 104 | Medium |
| 18 | 1.5 | Low | 3 | 7.9 × 104 | Medium |
| 9 | 1.2 | Low | 26 | 7.8 × 104 | Low |
| 10 | 1.2 | Low | 18 | 7.5 × 104 | Low |
| 26 | 1.0 | Low | 4 | 6.4 × 104 | Low |
| 4 | <1.0 | Low | 9 | 5.6 × 104 | Low |
| 5 | <1.0 | Low | 6 | 5.0 × 104 | Low |
| 6 | <1.0 | Low | 24 | 4.2 × 104 | Low |
| 20 | <1.0 | Low | 10 | 4.0 × 104 | Low |
| 24 | <1.0 | Low | 28 | 2.6 × 104 | Low |
| 28 | <1.0 | Low | 5 | 1.2 × 104 | Low |
Conclusion
The results from our study show that ATP, bacterial population species composition and total, membrane-intact and HNA cell counts in distributed drinking water are unsuitable indicators for regrowth in the drinking water distribution system. In contrast, HPCs, Aeromonas, Mycobacterium and fungi can be used as indicators for regrowth, although it remains unclear which parameter performs best as a regrowth indicator. Although ATP concentrations in distributed drinking water is an unsuitable monitoring parameter, ATP concentrations in treated water at the plant seems to have a predictive function about the level of regrowth in the distribution system. It is therefore recommended that routine monitoring for regrowth in the drinking water distribution system should focus on (i) ATP measurement of the treated water at the plant and (ii) HPCs, Aeromonas, Mycobacterium and/or fungi in drinking water sampled from the distribution system.Acknowledgements
This research would not have been possible without the funding and support of 3M (Paul Martens/Debby Polet) and Wavin (Tinus de Lange). In addition, we would also like to thank Cees Viets (Vitens), Weren de Vet (Oasen) and Andreas Farnleiter/Georg Reischer/Thomas Lendenfeld (University of Vienna) and sample takers, laboratory technicians and process technologists from Vitens and KWR for their support.References
- C. Lawrence, M. Reyrolle, S. Dubrou, F. Forey, B. Decludt, C. Goulvestre, P. Matsiota-Bernard, J. Etienne and C. Nauciel, J. Clin. Microbiol., 1999, 37, 2652–2655 CAS.
- J. O. Falkinham, 3rd, C. D. Norton and M. W. LeChevallier, Appl. Environ. Microbiol., 2001, 67, 1225–1231 CrossRef PubMed.
- P. W. J. J. van der Wielen and D. van der Kooij, Appl. Environ. Microbiol., 2013, 79, 825–834 CrossRef CAS PubMed.
- S. C. Edberg, P. Gallo and C. Kontnick, Microb. Ecol. Health Dis., 1996, 9, 67–77 CrossRef.
- R. C. Hoehn, Water Qual. Bull., 1988, 13, 46–51 CAS.
- S. C. Christensen, E. Nissen, E. Arvin and H. J. Albrechtsen, Water Res., 2011, 45, 3215–3224 CrossRef CAS PubMed.
- J. H. van Lieverloo, W. Hoogenboezem, G. Veenendaal and D. van der Kooij, Water Res., 2012, 46, 4918–4932 CrossRef CAS PubMed.
- A. K. Camper, in Microbial Growth in Drinking-Water Supplies. Problems, Causes, Control and Research Needs, ed. D. van der Kooij and P. W. J. J. van der Wielen, IWA Publishing, London, UK, 2014, pp. 73–94 Search PubMed.
- Anonymous, Drinking Water Decree 2001 (Waterleidingbesluit 2001), Staatsblad, The Hague, The Netherlands, 2001 Search PubMed.
- J. S. Maki, S. J. LaCroix, B. S. Hopkins and J. T. Staley, Appl. Environ. Microbiol., 1986, 51, 1047–1055 CAS.
- E. Siebel, Y. Wang, T. Egli and F. Hammes, Drinking Water Eng. Sci., 2008, 1, 1–6 CrossRef.
- P. W. J. J. van der Wielen and D. van der Kooij, Water Res., 2010, 44, 4860–4867 CrossRef CAS PubMed.
- M. Berney, M. Vital, I. Hulshoff, H. U. Weilenmann, T. Egli and F. Hammes, Water Res., 2008, 42, 4010–4018 CrossRef CAS PubMed.
- D. Berry, C. Xi and L. Raskin, Curr. Opin. Biotechnol., 2006, 17, 297–302 CrossRef CAS PubMed.
- G. Liu, G. L. Bakker, S. Li, J. H. Vreeburg, J. Q. Verberk, G. J. Medema, W. T. Liu and J. C. Van Dijk, Environ. Sci. Technol., 2014, 48, 5467–5476 CrossRef CAS PubMed.
- M. N. B. Momba, R. Kfir, S. N. Venter and T. E. Cloete, Water SA, 2000, 26, 59–66 Search PubMed.
- O. M. Zacheus, M. J. Lehtola, L. K. Korhonen and P. J. Martikainen, Water Res., 2001, 35, 1757–1765 CrossRef CAS PubMed.
- E. I. Prest, F. Hammes, S. Kötzsch, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Water Res., 2013, 47, 7131–7142 CrossRef CAS PubMed.
- P. W. J. J. van der Wielen and G. Medema, Appl. Environ. Microbiol., 2010, 76, 4876–4881 CrossRef CAS PubMed.
- M. J. Ferris, G. Muyzer and D. M. Ward, Appl. Environ. Microbiol., 1996, 62, 340–346 CAS.
- H. Heuer, M. Krsek, P. Baker, K. Smalla and E. M. Wellington, Appl. Environ. Microbiol., 1997, 63, 3233–3241 CAS.
- R. I. Amann, W. Ludwig and K. H. Schleifer, Microbiol. Rev., 1995, 59, 143–169 CAS.
- P. W. J. J. van der Wielen, R. Italiaander, B. A. Wullings, L. Heijnen and D. van der Kooij, in Microbial Growth in Drinking-Water Supplies. Problems, Causes, Control and Research Needs, ed. D. van der Kooij and P. W. J. J. van der Wielen, IWA Publishing, London, UK, 2014, pp. 177–205 Search PubMed.
- P. W. J. J. van der Wielen and M. Lut, Water Sci. Technol.: Water Supply, 2016 DOI:10.2166/ws.2016.023.
- D. Van der Kooij and H. R. Veenendaal, in Microbial Growth in Drinking Water Supplies. Problems, Causes, Controls and Research Needs, ed. D. Van der Kooij and P. W. J. J. Van der Wielen, IWA Publishing, London, UK, 2014, pp. 291–337 Search PubMed.
- M. Vital, M. Dignum, A. Magic-Knezev, P. Ross, L. Rietveld and F. Hammes, Water Res., 2012, 46, 4665–4676 CrossRef CAS PubMed.
- G. Roeselers, J. Coolen, P. W. J. J. van der Wielen, M. C. Jaspers, A. Atsma, B. de Graaf and F. Schuren, Environ. Microbiol., 2015, 17, 2505–2514 CrossRef PubMed.
- A. J. Pinto, C. Xi and L. Raskin, Environ. Sci. Technol., 2012, 46, 8851–8859 CrossRef CAS PubMed.
- K. Henne, L. Kahlisch, I. Brettar and M. G. Höfle, Appl. Environ. Microbiol., 2012, 78, 3530–3538 CrossRef CAS PubMed.
- A. C. Martiny, H.-J. Albrechtsen, E. Arvin and S. Molin, Appl. Environ. Microbiol., 2005, 71, 8611–8617 CrossRef CAS PubMed.
- E. J. Anaissie, S. L. Stratton, M. C. Dignani, C. K. Lee, R. C. Summerbell, J. H. Rex, T. P. Monson and T. J. Walsh, Blood, 2003, 101, 2542–2546 CrossRef CAS PubMed.
- C. Chauret, C. Volk, R. Creason, J. Jarosh, J. Robinson and C. Warnes, Can. J. Microbiol., 2001, 47, 782–786 CrossRef CAS PubMed.
- J. O. Falkinham, 3rd, M. D. Iseman, P. de Haas and D. van Soolingen, J. Water Health, 2008, 6, 209–213 Search PubMed.
- L. M. Feazel, L. K. Baumgartner, K. L. Peterson, D. N. Frank, J. K. Harris and N. R. Pace, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 16393–16399 CrossRef CAS PubMed.
- G. Hageskal, N. Lima and I. Skaar, Mycol. Res., 2009, 113, 165–172 CrossRef PubMed.
- P. Holmes and L. M. Nicolls, Water Environ. J., 1995, 9, 464–469 CrossRef CAS.
- M. W. Kuiper, B. A. Wullings, A. D. L. Akkermans, R. R. Beumer and D. van der Kooij, Appl. Environ. Microbiol., 2004, 70, 6826–6833 CrossRef CAS PubMed.
- A. D. Seth and R. G. J. Edyvean, Int. Biodeterior. Biodegrad., 2006, 58, 108–111 CrossRef CAS.
- K. Lautenschlager, C. Hwang, W.-T. Liu, N. Boon, O. Köster, H. Vrouwenvelder, T. Egli and F. Hammes, Water Res., 2013, 47, 3015–3025 CrossRef CAS PubMed.
- D. J. Beale, D. R. Marlow, E. A. Palombo and P. D. Morrison, Characterisation of water network biofilms using a novel non-invasive metabolic approach, paper presented at IWA Leading Edge, Strategic Asset Management, Sydney, Australia, 2013 Search PubMed.
- H. C. Flemming, B. Bendinger, M. Exner, J. Gebel, T. Kistemann, G. Schaule, U. Szewzyk and J. Wingender, in Microbial Growth in Drinking-Water Supplies. Problems, Causes, Control and Research Needs., ed. D. van der Kooij and P. W. J. J. van der Wielen, IWA Publishing, London, UK, 2014, pp. 207–238 Search PubMed.
- F. Hammes, M. Berney and T. Egli, Adv. Biochem. Eng./Biotechnol., 2011, 124, 123–150 CrossRef PubMed.
- S. Eichler, R. Christen, C. Holtje, P. Westphal, J. Botel, I. Brettar, A. Mehling and M. G. Hofle, Appl. Environ. Microbiol., 2006, 72, 1858–1872 CrossRef CAS PubMed.
- J. H. M. van Lieverloo, W. Hoogenboezem, G. Veenendaal and D. van der Kooij, in Microbial Growth in Drinking-Water Supplies. Problems, Causes, Control and Research Needs., ed. D. van der Kooij and P. W. J. J. van der Wielen, IWA Publishing, London, UK, 2014, pp. 239–260 Search PubMed.
- P. W. J. J. van der Wielen and D. van der Kooij, H2O, 2011, vol. 2011, pp. 34–36 Search PubMed.
- J. C. Fry, in Methods in Aquatic Microbiology, ed. B. Austin, John Wiley & Sons Ltd., London, England, 1988, pp. 27–72 Search PubMed.
- D. M. Karl, Microbiol. Rev., 1980, 44, 739–796 CAS.
- M. Loferer-Krößbacher, J. Klima and R. Psenner, Appl. Environ. Microbiol., 1998, 64, 688–694 Search PubMed.
- F. B. van Es and L. A. Meyer-Reil, Adv. Microb. Ecol., 1982, 6, 111–169 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ew00007j |
| This journal is © The Royal Society of Chemistry 2016 |