Dendrimer conjugates for light-activated delivery of antisense oligonucleotides†
Abstract
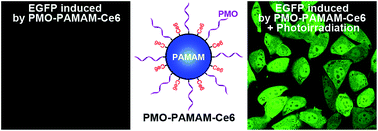
Therapeutic oligonucleotides (ONs), such as splice switching ONs (SSOs), provide opportunities for treating serious, life-threatening diseases. However, the development of ONs as therapeutic agents has progressed slowly, because cytosolic delivery of ONs into the cytosol and nucleus remains a major hurdle. Photochemical internalization (PCI), a promising strategy for endosomal escape, was introduced to disrupt the endosomal membrane using light and a photosensitizer. Here we constructed poly(amido amine) (PAMAM) dendrimer conjugates to simultaneously deliver SSOs and photosensitizers into endo/lysosomal compartments of cancer cells. After photo-irradiation, considerable ONs were observed to diffuse into the cytosol and accumulate in the nucleus. Furthermore, the PCI mediated cytosolic delivery of SSOs effectively enhanced their nuclear splice switching activity.

 Please wait while we load your content...
Please wait while we load your content...