Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Collagen labelling with an azide-proline chemical reporter in live cells†
Beatrice
Amgarten
a,
Rakesh
Rajan
b,
Nuria
Martínez-Sáez
a,
Bruno L.
Oliveira
c,
Inês S.
Albuquerque
d,
Roger A.
Brooks
b,
David G.
Reid
a,
Melinda J.
Duer
a and
Gonçalo J. L.
Bernardes
*ad
aDepartment of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK. E-mail: gb453@cam.ac.uk
bDivision of Trauma & Orthopaedic Surgery, University of Cambridge, Addenbrooke's Hospital, Cambridge CB2 0QQ, UK
cAthinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, USA
dInstituto de Medicina Molecular, Faculdade de Medicina da Universidade de Lisboa, Av. Prof. Egas Moniz, 1649-028 Lisboa, Portugal. E-mail: gbernardes@medicina.ulisboa.pt; Web: http://gbernardes-lab.com
First published on 18th November 2014
Abstract
We have developed a strategy for selective imaging of collagen in live foetal ovine osteoblasts. Our approach involves the incorporation of an azide-tagged proline in the biosynthesis of collagen followed by labelling using a strain-promoted [3+2] azide–alkyne cycloaddition reaction.
Collagen is the most abundant mammalian protein, accounting for one third of all proteins and representing the major component of the extracellular matrix (ECM).1 The basic structure of collagen consists of three single strands containing the repeating units Gly–Xaa–Yaa which form a triple helix, where X and Y are predominantly proline and hydroxyproline, respectively (both constitute approximately 23% of the total of amino acids in collagen).2,3 Due to its high abundance in collagen, proline has been widely exploited as a marker of collagen biosynthesis by measuring incorporation of either radioactive or non-radioactive isotopically-labelled proline (3H-Pro, 14C-Pro, 13C-Pro, 15N-Pro, cis-18F-Pro) as well as incorporation of heavy atoms (13C and 15N) directly in tissues.3–5 Collagen plays a key role in many pathogenic processes, as for example fibrosis6 and tumour growth and metastasis,7 to mention the most prominent. Thus, we undertook to exploit bioorthogonal strategies that would allow non-invasive imaging of collagen structures and provide a powerful imaging tool to monitor and study diseases where abnormal collagen production is a main feature, including cirrhosis, diabetic nephropathy or idiopathic pulmonary fibrosis.
Recent advances in bioorthogonal chemical reactions enables the design of probes tailored for imaging tagged macromolecules.8,9 Using this approach, tagging and imaging of temporal and spatial variations of the glycome,10 lipidome,11 as well as subsets of the proteome12 have been successfully achieved. Proteome tagging can be facilitated through the incorporation of non-canonical amino acids bearing reactive handles using the host cell's machinery. Subsequent labelling with a fluorescent dye or radionuclide by means of bioorthogonal reactions allows the visualisation of newly synthesised proteins. Reactive metabolic precursors applied previously for this purpose include the methionine analogues azidohomoalanine13 and homopropargylglycine14 as well as the phenylalanine analogue ethynylphenylalanine.15
Azido-proline (N3-Pro) has been used for residue-specific labelling of collagen model peptides by functionalization with triazolyl moieties.16–18 Moreover, the incorporation fluoro-proline was shown to enhance thermal stability of synthetic collagen structures.19 Importantly, the introduction of modified proline analogues still allows a collagen triple helix to form. Together these examples highlight the potential of introducing proline-based chemical reporters for the bioorthogonal imaging of collagen. Here, we report the metabolic incorporation of N3-Pro as a chemical reporter for the imaging of collagen using dibenzooctyne (DIBO)20 as a fluorescent probe and exploiting a strain promoted [3+2] azide–alkyne cycloaddition (SPAAC) bioorthogonal reaction (Fig. 1). The metabolic incorporation of a reporter group by foetal ovine osteoblasts was achieved by supplementation of the growth medium with cis-4-azido-L-proline (ESI† for synthesis details).
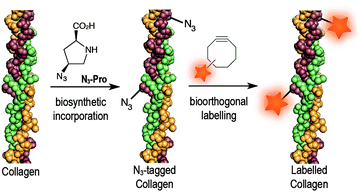 | ||
| Fig. 1 Incorporation of N3-Pro chemical reporter into collagen and labelling using a strain-promoted azide–alkyne cycloaddition reaction with the fluorescent probe DIBO. | ||
We chose ovine osteoblasts as our model system to exploit the bioorthogonal imaging of collagen in live cells because they have been shown to behave similarly to human osteoblasts in terms of viability, and osteocalcin and interleukin-6 production, compared to other animal derived cells, making them a suitable model system for our proposed investigations.21 Furthermore, since foetal ovine osteoblasts are not auxotrophic for proline – meaning that they are able to synthesise proline themselves – the levels of incorporation in the presence of the chemical reporter N3-Pro is expected to be lower than in pulse-labelling. However, here we make use of a low, physiological concentration for osteoblast culturing which makes it relevant for applications in in vivo models.
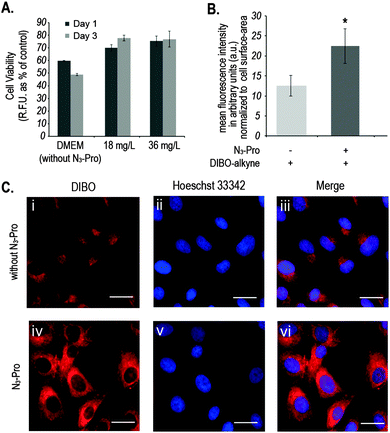
We started by analysing the effect on cell viability of culturing foetal ovine osteoblasts in the presence of 18 mg L−1 or 36 mg L−1 of N3-Pro. The presence of N3-Pro had no effect on cell viability at either concentration (Fig. 2A and Fig. S1, ESI†). To assess incorporation of N3-Pro into collagen, amino acid analysis by HPLC and infrared spectroscopy on harvested ECM grown in the absence or presence of N3-Pro was performed. Amino acid analysis provided evidence of N3-Pro incorporation in the ECM (Fig. S2, ESI†). Infrared spectral analysis further supports incorporation of N3-Pro into ECM as a band attributed to the stretching of the N3 group could be detected in the labelled ECM samples as opposed to the unlabelled (Fig. S3, ESI†).
Next, we used fluorescence microscopy to image N3-Pro tagged collagen in foetal ovine osteoblasts after SPAAC reaction with the fluorescent probe DIBO (Fig. 2B and C). Cells were grown in the presence or absence of 36 mg L−1 of N3-Pro, fixed and stained with 5 μM DIBO Alexa Fluor 555 for 60 min at room temperature and counterstained with the nuclear dye Hoechst 33342 (Fig. 2C). We found that foetal ovine osteoblasts cultured with N3-Pro and bioorthogonally labelled with DIBO showed a nearly 2-fold increase in fluorescence (Fig. 2B and C). Confocal microscopy on stained foetal ovine osteoblast showed a dotted structure that likely corresponds to non-secreted pro-collagen (Fig. S4, ESI†) showing that the cells take up the azide-modified proline.
Fluorescent images showed, however, a relatively high background staining in the samples cultured in the absence of N3-Pro (Fig. 2C, i–iii), indicating unspecific DIBO labelling. The DIBO cyclooctyne fluorescent probe has been successfully applied in bioorthogonal approaches based on SPAAC, although only in imaging molecules densely presented on the cell surface.22 Given the observed unspecific staining, we decided to analyse whether DIBO was reacting with intracellular proteins, by in-gel fluorescence (Fig. S5, ESI†) and mass spectrometry analysis (Fig. S6, ESI†) of the stained whole cell lysate. Despite previously reported selectivity of cyclooctynes including DIBO for azides over thiols,20 mass spectrometry analysis of some of the most prominent bands in the gel revealed unspecific labelling of intracellular structures such as actin (Fig. S6, ESI†), suggesting unspecific reaction of the strained alkyne probe with sulfhydryl-containing biomolecules.
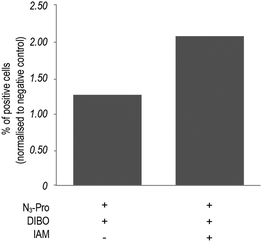
In addition, bovine serum albumin (BSA), which is used as a media supplement and blocking agent for the staining protocol (Fig. S6, ESI†) was also detected. Bertozzi and co-workers also reported the same issue in the in vivo imaging of glycoconjugates using DIFO.23 In an attempt to reduce unspecific labelling, we pre-incubated fixed samples with the thiol alkylating agent iodoacetamide (IAM) prior to staining with DIBO.24 Indeed, in-gel fluorescence showed a reduction of unspecific staining and an increase of background to signal ratio (Fig. S5, ESI†), which was further confirmed by flow cytometry analysis (Fig. 3 and Fig. S7, ESI†). Our observations indicate that the use of the fluorescent probe DIBO in SPAAC bioorthogonal approaches for imaging of molecules that are not densely presented on the cell surface meets with some limitations, namely, its high reactivity towards cellular and plasma sulfhydryl containing biomolecules limiting its application to challenging structures namely intracellular targets.
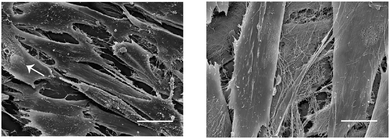
Finally, we wished to demonstrate that our approach did not interfere with the normal growth of ECM in foetal ovine osteoblasts. Scanning electron microscopy images (SEM) of cells cultured in the presence of N3-Pro confirmed adequate integrity (Fig. 4).
In summary, we report the metabolic incorporation of N3-Pro into collagen and its use as a chemical reporter for bioorthogonal labelling using a SPAAC strategy in live foetal ovine osteoblasts. Our data provides proof of principle for the use of proline derivatives equipped with orthogonal handles, such as the azide here reported, to tag and label collagen structures. The azide derivative was successfully incorporated and labelled with the fluorescent probe DIBO. Although our approach suffered from unspecific reaction of the strained alkyne DIBO with sulfhydryl rich proteins such as actin or albumin, this was partially prevented by pre-incubation with IAM. We are now exploring the incorporation of prolines equipped with different handles that would allow a more efficient bioorthogonal labelling of collagen.
Our strategy could be particularly useful for the diagnosis and monitoring of pathogenic accumulations of ECM. The development of new therapies for fibrotic diseases (e.g. cirrhosis or idiopathic pulmonary fibrosis characterized by excessive deposition of collagen) has been impeded by difficulties in assessing efficacy, since sensitive biomarkers of fibrogenesis/fibrolysis, either in cells or animal models, are not available.6 The cellular model described here could be used for example for high-throughput screening of therapeutic agents by fluorescent microplate-based analysis and help in the identification of lead anti-fibrotic drugs from a series of candidates.
We thank the EU (Erasmus studentship to B.A.; Marie Curie CIG to G.J.L.B.), FCT Portugal (FCT Investigator to G.J.L.B.), the EPSRC and the National Institute of Health Research for funding. We also thank Karin Müller for help with confocal microscopy and SEM and Dr Filipa Cruz for critical reading of the manuscript. G.J.L.B. is a Royal Society University Research Fellow.
Notes and references
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929–958 CrossRef CAS PubMed.
- K. Gelse, E. Pöschl and T. Aigner, Adv. Drug Delivery Rev., 2003, 55, 1531–1546 CrossRef CAS PubMed.
- K. Smith and M. J. Rennie, Curr. Opin. Clin. Nutr. Metab. Care, 2007, 10, 582–590 CrossRef CAS PubMed.
- D. Skovgaard, A. Kjaer, K. M. Heinemeier, M. Brandt-Larsen, J. Madsen and M. Kjaer, PLoS One, 2011, 6, e16678 CAS.
- W. Y. Chow, R. Rajan, K. H. Muller, D. G. Reid, J. N. Skepper, W. C. Wong, R. A. Brooks, M. Green, D. Bihan, R. W. Farndale, D. A. Slatter, C. M. Shanahan and M. J. Duer, Science, 2014, 344, 742–746 CrossRef CAS PubMed.
- S. L. Friedman, D. Sheppard, J. S. Duffield and S. Violette, Sci. Transl. Med., 2013, 5, 167sr161 Search PubMed.
- D. M. Gilkes, G. L. Semenza and D. Wirtz, Nat. Rev. Cancer, 2014, 14, 430–439 CrossRef CAS PubMed.
- M. Grammel and H. C. Hang, Nat. Chem. Biol., 2013, 9, 475–484 CrossRef CAS PubMed.
- K. Lang and J. W. Chin, Chem. Rev., 2014, 114, 4764–4806 CrossRef CAS PubMed.
- S. T. Laughlin and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 12–17 CrossRef CAS PubMed.
- R. N. Hannoush and N. Arenas-Ramirez, ACS Chem. Biol., 2009, 4, 581–587 CrossRef CAS PubMed.
- K. E. Beatty, Mol. BioSyst., 2011, 7, 2360–2367 RSC.
- D. C. Dieterich, A. J. Link, J. Graumann, D. A. Tirrell and E. M. Schuman, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9482–9487 CrossRef CAS PubMed.
- K. E. Beatty, J. C. Liu, F. Xie, D. C. Dieterich, E. M. Schuman, Q. Wang and D. A. Tirrell, Angew. Chem., Int. Ed., 2006, 45, 7364–7367 CrossRef CAS PubMed.
- M. Grammel, P. D. Dossa, E. Taylor-Salmon and H. C. Hang, Chem. Commun., 2012, 48, 1473–1474 RSC.
- R. S. Erdmann and H. Wennemers, J. Am. Chem. Soc., 2010, 132, 13957–13959 CrossRef CAS PubMed.
- R. S. Erdmann and H. Wennemers, Org. Biomol. Chem., 2012, 10, 1982–1986 CAS.
- R. S. Erdmann and H. Wennemers, Bioorg. Med. Chem., 2013, 21, 3565–3568 CrossRef CAS PubMed.
- M. D. Shoulders, K. A. Satyshur, K. T. Forest and R. T. Raines, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 559–564 CrossRef CAS PubMed.
- X. Ning, J. Guo, M. A. Wolfert and G.-J. Boons, Angew. Chem., Int. Ed., 2008, 47, 2253–2255 CrossRef CAS PubMed.
- P. Torricelli, M. Fini, G. Giavaresi and R. Giardino, Artif. Cells, Blood Substitutes, Immobilization Biotechnol., 2003, 31, 263–277 CrossRef CAS PubMed.
- J. Dommerholt, S. Schmidt, R. Temming, L. J. A. Hendriks, F. P. J. T. Rutjes, J. C. M. van Hest, D. J. Lefeber, P. Friedl and F. L. van Delft, Angew. Chem., Int. Ed., 2010, 49, 9422–9425 CrossRef CAS PubMed.
- P. V. Chang, J. A. Prescher, E. M. Sletten, J. M. Baskin, I. A. Miller, N. J. Agard, A. Lo and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1821–1826 CrossRef CAS PubMed.
- R. van Geel, G. J. M. Pruijn, F. L. van Delft and W. C. Boelens, Bioconjugate Chem., 2012, 23, 392–398 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S7, detailed experimental procedures, IR and mass spectrometry analysis, NMR spectra. See DOI: 10.1039/c4cc07974d |
| This journal is © The Royal Society of Chemistry 2015 |