Open Access Article
Open Access ArticleCustom-shaped metal nanostructures based on DNA origami silhouettes†
Boxuan
Shen‡
a,
Veikko
Linko‡
b,
Kosti
Tapio
a,
Mauri A.
Kostiainen
*b and
J. Jussi
Toppari
*a
aUniversity of Jyvaskyla, Department of Physics, Nanoscience Center, P.O. Box 35, FI-40014. and University of Jyväskylä, Finland. E-mail: j.jussi.toppari@jyu.fi
bBiohybrid Materials, Department of Biotechnology and Chemical Technology, Aalto University, P.O. Box 16100, FI-00076 Aalto, Espoo, Finland. E-mail: mauri.kostiainen@aalto.fi
First published on 29th May 2015
Abstract
The DNA origami technique provides an intriguing possibility to develop customized nanostructures for various bionanotechnological purposes. One target is to create tailored bottom-up-based plasmonic devices and metamaterials based on DNA metallization or controlled attachment of nanoparticles to the DNA designs. In this article, we demonstrate an alternative approach: DNA origami nanoshapes can be utilized in creating accurate, uniform and entirely metallic (e.g. gold, silver and copper) nanostructures on silicon substrates. The technique is based on developing silhouettes of the origamis in the grown silicon dioxide layer, and subsequently using this layer as a mask for further patterning. The proposed method has a high spatial resolution, and the fabrication yields can approach 90%. The approach allows a cost-effective, parallel, large-scale patterning on a chip with fully tailored metallic nanostructures; the DNA origami shape and the applied metal can be specifically chosen for each conceivable implementation.
During the past three decades a great variety of different nanoscale objects have been constructed using DNA as a programmable building material.1,2 One of the most promising and robust methods for bottom-up fabrication with DNA is the so-called origami technique,3 which is based on folding a long single-stranded DNA scaffold into a desired shape with the help of a set of synthetic “staple” strands. It was originally designed for flat 2D single-layer structures, but since then the method has been generalized for partially double-layered tiles,4 hollow 3D objects,5 multilayer 3D origamis,6 and structures containing customized curvatures and twists.7,8 Lately, methods allowing 3D meshing of DNA structures9 and scaffold-free origamis10,11 have been demonstrated. The techniques include powerful software for designing12 and simulating13–15 the shapes of the user-defined structures. These methods together form a versatile tool-kit for the designers. Recent progress in the field of structural DNA nanotechnology16,17 has yielded a plethora of intriguing bionanotechnological applications, such as artificial ion channels,18 nanoreactors,19 gatekeepers for nanopores20–22 and drug delivery vehicles.23–25
In addition to the aforementioned applications, the superior spatial addressability of the self-assembled DNA structures can be utilized in nanoscale patterning. Reliable nanoparticle patterning on the DNA architectures is a key feature for miniaturizing electronics26,27 and developing photonic metamaterials,28 as well as for novel plasmonic nanostructures and devices.29–31 The reported assemblies include various types of DNA scaffolds decorated with complex or chiral nanoparticle geometries32–35 and DNA-templated growth of metallic nanoshapes.36–38 Lately, hollow origamis have been used as “molds” for casting metal nanoparticles, i.e., for guiding the growth of the encapsulated metallic “seed” particles into the desired nanoshapes.39,40 Furthermore, single DNA molecules or structures can be directed and anchored to the selected areas of lithographically fabricated substrates in order to form desired patterns,41–49 or alternatively, the DNA objects can be directly tiled together into well-ordered large-scale assemblies.4,50–52
In this communication, we expand the toolbox of bottom-up-based methods by presenting a novel technique for creating uniform custom-shaped metallic nano-objects directly on the silicon chip by exploiting the high spatial accuracy of DNA origami nanoarchitectures (see Fig. 1). We believe that the method can readily open up new opportunities in nanolithographic sample fabrication aiming towards metamaterials and nanoplasmonics. Through further optimization, the technique could be generalized for other substrates, as well as for larger and more complex DNA-based assemblies.
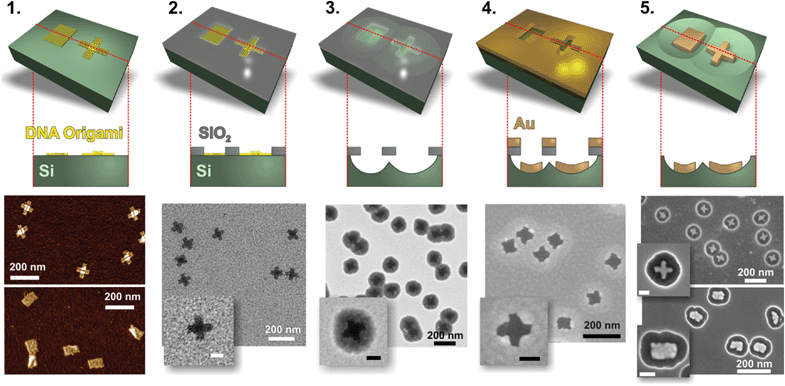 | ||
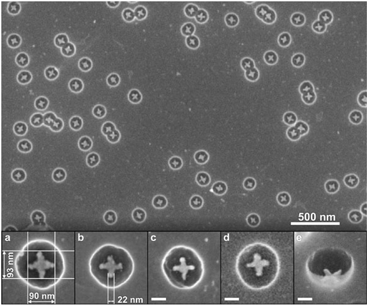
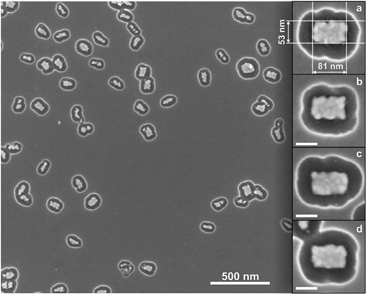
Fig. 1 Fabrication of gold nanostructures. Step 1: DNA origami structures (“Seeman tiles”4 (ST) and “Rothemund rectangles”3 (RR)) are deposited onto the silicon (Si) substrate. Note that due to the highly twisted shape of RR, some rectangles might appear slightly squeezed or rolled-up on the substrate. Step 2: Silicon dioxide (SiO2) layer is grown in a chemical vapor deposition (CVD) process on the Si chip. The oxide layer grows selectively, and thus “DNA origami silhouettes” are created. Step 3: The “silhouettes” are used as the openings in etching of the silicon underneath the SiO2 layer. The reactive ion etched (RIE) wells in the silicon are clearly visible beneath the silhouettes. Step 4: Gold is deposited onto the chip using an ultra-high vacuum (UHV) electron beam evaporator. Step 5: The SiO2 layer (with the metal on top) is removed in a HF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HCl-based lift-off procedure. This leaves the DNA origami-shaped gold nanostructures on the silicon chip. The scale bars in the insets are 50 nm. HCl-based lift-off procedure. This leaves the DNA origami-shaped gold nanostructures on the silicon chip. The scale bars in the insets are 50 nm. | ||
Briefly, our method relies on the selective growth of a thin silicon dioxide layer53 on top of the silicon substrate (with the native oxide) that supports the deposited DNA origami shapes (Fig. 1: steps 1 and 2). The SiO2 primarily grows on top of the silicon, and thus the process leaves the “DNA origami silhouettes” as openings in the formed SiO2 layer (step 2). The aforementioned layer can be used as a mask for plasma etching (RIE) the silicon beneath the opening (step 3). The procedure allows forming smooth and rounded wells in the silicon, which have the SiO2 window with the origami-shaped opening on the top (step 3). The origami silhouette can be subsequently used as a mask for depositing metal by evaporation onto the chip (step 4). Finally, the SiO2 layer can be removed by hydrofluoric acid (HF) and hydrochloride (HCl)-based wet etching, leaving just the origami-shaped metallic nanostructures on the silicon chip (step 5).
We have demonstrated the fabrication of the metal nanoshapes using three different metals (gold, copper and silver) and two structurally different DNA objects: single-layer “Rothemund rectangle”3 (RR) (92 nm × 72 nm) and partially double-layered cross-shaped “Seeman tile”4 (ST) (two 95 nm × 30 nm layers crossing each other) (step 1, Fig. 1). The origami designs were fabricated in 1× TAE (40 mM Tris, 19 mM acetic acid, 1 mM EDTA) buffer with 12.5 mM Mg++ using 5–20 nM scaffold strand concentration and 10× excess of staple strands (IDT). The side strands for both structures were left out in order to avoid blunt-end stacking of the objects. The annealing ramps for the folding of the structures are the same as reported previously.3,4 The quality of the folding was verified with agarose gel electrophoresis and AFM imaging (tapping mode, Veeco Dimension 3100). The structures can be optionally purified (excess amount of staple strands removed/buffer exchanged) by spin-filtering (see ESI†). However, we observed that the purification step is not necessarily needed in the successful fabrication procedure.
For the substrate, we used a slightly boron-doped p-type silicon chip (6 × 6 mm), which was cleaned with hot acetone and isopropanol followed by a brief sonication (2 min) and a RIE-based (Oxford Plasmalab 80 Plus) oxygen plasma treatment (oxygen flow 50 sccm, plasma power 200 W, temperature 30 °C and time 20 min). The plasma treatment was carried out in order to induce hydroxyl (–OH) group formation on the silicon surface (negatively charged and hydrophilic substrate), and therefore to help immobilize DNA origami via Mg++ ions. 5 μl of DNA origami solution in 1× TAE buffer with 100 mM Mg++ was pipetted onto the silicon chip right after the plasma treatment. The sample was incubated in a closed chamber for 5 min, washed 3 times with 50 μl of double-distilled (dd) H2O and finally gently dried under a N2 gas flow (step 1, Fig. 1).
The Si chip with the immobilized DNA origami structures was placed in a 1.5-liter glass desiccator for 16 hours together with two small glass vials containing tetraethyl orthosilicate (TEOS, ≥99.0%, Sigma-Aldrich) and ammonium hydroxide (NH4OH, 25% NH3 in H2O, Baker Analyzed). In addition, 80 grams of silica gel, which was conditioned overnight in a humidity chamber (Weiss Climatic test chamber, 80% relative humidity, room temperature), were positioned at the bottom of the desiccator in order to improve the quality of the grown silicon dioxide layer. The aforementioned chemical vapor deposition (CVD) treatment reliably created precise origami silhouettes in the 5–10 nm thick silicon dioxide layer (step 2, Fig. 1), since the oxide predominantly grows on the bare areas of the chip.53
The sample with the formed silhouettes was etched using RIE (step 3, Fig. 1). First, in order to expose the Si surface beneath the grown silicon dioxide layer (including the formed native oxide layer), 2–4 nm of SiO2 was etched away (CHF3 flow 25 sccm, Ar flow 25 sccm, plasma power 100 W, temperature 25 °C and time 12–24 s). Subsequently, 57 nm of Si was etched (SF6 flow 100 sccm, O2 flow 8 sccm, plasma power 50 W, temperature 30 °C and time 30 s) resulting in the rounded silicon wells beneath the SiO2 windows.
The following metal deposition (gold, copper or silver) was carried out using an electron beam evaporator (step 4, Fig. 1) in an ultra-high vacuum chamber (UHV). 20 nm of metal was perpendicularly evaporated onto the sample surface at a rate of 0.04 nm s−1 (for copper) or 0.06 nm s−1 (for gold and silver). After the metal deposition, the SiO2 mask together with the metal film on top of it was removed in a lift-off procedure using HF (38%)/HCl (38%) (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution for gold or 4% HF in H2O for copper and silver. Finally, the sample was washed with ddH2O and dried under N2 flow (step 5 with gold structures is presented in Fig. 2 and 3).
1) solution for gold or 4% HF in H2O for copper and silver. Finally, the sample was washed with ddH2O and dried under N2 flow (step 5 with gold structures is presented in Fig. 2 and 3).
Fig. 4 shows the feasibility of the proposed fabrication method: it illustrates ST patterns made out of different metals, i.e. gold, copper and silver. It is noteworthy to mention that the shape of the metallic ST structure becomes slightly rounded in the case of silver and copper deposition due to the native oxidation of these metals. The ready samples were imaged with AFM (Veeco Dimension 3100) or SEM (Raith eLine).
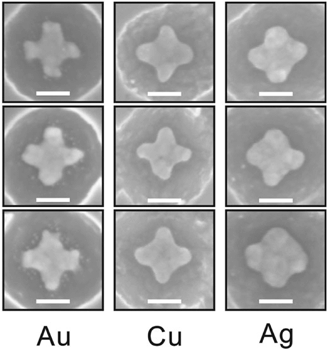 | ||
| Fig. 4 SEM images of the ST shapes made out of gold (Au), copper (Cu) and silver (Ag). The scale bars are 50 nm. | ||
For ST-patterning (Fig. 2), 86 ± 3% of all the observed particles were correctly formed metallic (gold) crosses, and the yield for the RR-based objects (Fig. 3) was 65 ± 2% (see the ESI† for the details). The adopted solution shapes for both origamis are twisted to some extent (especially RR, see the ESI†), but the twisted origamis can be straightened when they land onto the substrate. However, some structures do not adopt the desired orientation on the silicon substrate after their deposition, and thus the yield accordingly decreased. By choosing twist-corrected and more rigid origami structures for patterning one could presumably increase the success rate.
In addition, it was observed that RR-based gold nanostructures had an average width of 37 ± 10 nm and an average length of 80 ± 6 nm (see the ESI† for the statistics of the dimensions). For the gold ST-patterns the average length in both directions was 89 ± 6 nm and the average width of the arm was 23 ± 7 nm. The length of ST-patterned metallic structures is close to the length of the origami (on average only ∼7% smaller), but the width is about 25% smaller compared to the original origami design. In the case of the RR-based metallic structures, the length corresponds to that of the origami structure (∼15% smaller), but the width is on average about 48% smaller than designed, mainly because the twisted RR origami tends to roll up around its longer axis on the silicon substrate. Moreover, there are two plausible explanations for the slightly reduced dimensions of the gold nanoshapes. One is that although the silicon dioxide layer mainly grows perpendicular to the Si substrate, it can also grow on the walls of the silhouettes, thus resulting in a decreased size of the opening. The other is that the evaporated gold may adhere or cluster onto the edges of the mask, which moderately reduces the size of the silhouettes. Nevertheless, the obtained yields indicate that the proposed technique is highly reproducible (additional SEM images of the gold nanostructures in the ESI†).
Conclusions
As a conclusion, we report a novel high-throughput technique for fabricating uniform and tailored metallic nanostructures on a silicon chip. The fabrication method exploits the high spatial addressability of the tailored DNA nanostructures. We have demonstrated the feasibility and modularity of the technique by utilizing two structurally different scaffolded DNA origamis for creating the origami silhouettes, and three different metals for deposition (gold, copper and silver, Fig. 4). The advantage of the method is that, in principle, one can use any kind of origami shape (dimers, multimers and even larger arrays are equally accessible) and any metal that survives HF or HF/HCl etching. However, the fabrication of large and complex structures might require more anisotropic etch profiles of silicon, which can be achieved by utilizing advanced lithographic techniques such as a deep reactive ion etching (DRIE).54In general, one has to pay extra attention to the actual solution shape of the DNA origami in order to avoid non-uniform size distribution of the fabricated metal nanostructures. One intriguing possibility to increase the impact of the method would be to use scaffold-free origamis10,11 for fabricating any desired patterns in a cost-effective manner, i.e. by utilizing just one set of staple strands. We believe that our method could be equally extended to other substrates – such as sandwiched Si-based multilayered substrates – by completely removing the silicon layer once the metal pattern has been formed. The aforementioned approach could readily facilitate the fabrication on the transparent surfaces and thus the characterization of plasmonic properties of the created nanoshapes.
In contrast to previously reported DNA-templated metallic shapes,36–38 our method is easy, cost-effective and it allows uniform, regular and accurate structures. Compared to the recently reported innovative DNA mold approach,39,40 our method allows the use of a variety of metals and still it provides a similar patterning resolution. In addition, it might allow an easier route to fabricate specific nanoshapes or even larger origami-based arrays. However, our technique is substrate-based and thus, creating nanoparticles in a solution-phase is not accessible. Despite that, well-ordered large-scale parallel patterning could be realized e.g. by exploiting electric fields for directing the origami shapes on the chip,49 and subsequently transferring the created array to the target substrate.55
To date, DNA- and substrate-based molecular lithography approaches have not fully contemplated the possibilities of fabricating metallic nanoshapes. Rather, previous studies cover either a positive- or negative-tone decoration of silicon and silicon oxide,53,56,57 or DNA-assisted graphene patterning.58 However, our straightforward method offers a novel and attractive way to combine bottom-up-based molecular self-assembly with standard top-down lithographic techniques. As a result, the proposed method facilitates the user-defined fabrication of metallic nanoshapes for a great variety of applications.
Acknowledgements
This work was supported by the Academy of Finland (grants 218182, 263526, 258309, 263504, 267497, 273645). In addition, V. L. and M. A. K. acknowledge financial support from Biocentrum Helsinki, Emil Aaltonen Foundation and EU EMRP (SIB61). This work was carried out under the Academy of Finland's Centres of Excellence Programme (2014–2019).Notes and references
- N. C. Seeman, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS
.
- N. C. Seeman, Nano Lett., 2010, 10, 1971–1978 CrossRef CAS PubMed
.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed
.
- W. Liu, H. Zhong, R. Wang and N. C. Seeman, Angew. Chem., Int. Ed., 2011, 50, 264–267 CrossRef CAS PubMed
.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. P. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73–76 CrossRef CAS PubMed
.
- S. M. Douglas, H. Dietz, T. Liedl, B. Högberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 CrossRef CAS PubMed
.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS PubMed
.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, Science, 2011, 332, 342–346 CrossRef CAS PubMed
.
- D. Han, S. Pal, Y. Yang, S. Jiang, J. Nangreave, Y. Liu and H. Yan, Science, 2013, 339, 1412–1415 CrossRef CAS PubMed
.
- B. Wei, M. Dai and P. Yin, Nature, 2012, 485, 623–626 CrossRef CAS PubMed
.
- Y. Ke, L. L. Ong, W. M. Shih and P. Yin, Science, 2012, 338, 1177–1183 CrossRef CAS PubMed
.
- S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church and W. M. Shih, Nucleic Acids Res., 2009, 37, 5001–5006 CrossRef CAS PubMed
.
- C. E. Castro, F. Kilchherr, D.-N. Kim, E. L. Shiao, T. Wauer, P. Wortmann, M. Bathe and H. Dietz, Nat. Methods, 2011, 8, 221–229 CrossRef CAS PubMed
.
- D.-N. Kim, F. Kilchherr, H. Dietz and M. Bathe, Nucleic Acids Res., 2012, 40, 2862–2868 CrossRef CAS PubMed
.
- K. Pan, D.-N. Kim, F. Zhang, M. R. Adendorff, H. Yan and M. Bathe, Nat. Commun., 2014, 5, 5578 CrossRef CAS PubMed
.
- V. Linko and H. Dietz, Curr. Opin. Biotechnol., 2013, 24, 555–561 CrossRef CAS PubMed
.
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed
.
- M. Langecker, V. Arnaut, T. G. Martin, J. List, S. Renner, M. Mayer, H. Dietz and F. C. Simmel, Science, 2012, 338, 932–936 CrossRef CAS PubMed
.
- V. Linko, M. Eerikäinen and M. A. Kostiainen, Chem. Commun., 2015, 51, 5351–5354 RSC
.
- N. A. W. Bell, C. R. Engst, M. Ablay, G. Divitini, C. Ducati, T. Liedl and U. F. Keyser, Nano Lett., 2012, 12, 512–517 CrossRef CAS PubMed
.
- R. Wei, T. G. Martin, U. Rant and H. Dietz, Angew. Chem., Int. Ed., 2012, 51, 4864–4867 CrossRef CAS PubMed
.
- C. Plesa, A. N. Ananth, V. Linko, C. Gülcher, A. J. Katan, H. Dietz and C. Dekker, ACS Nano, 2014, 8, 35–43 CrossRef CAS PubMed
.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed
.
- J. Mikkilä, A.-P. Eskelinen, E. H. Niemelä, V. Linko, M. J. Frilander, P. Törmä and M. A. Kostiainen, Nano Lett., 2014, 14, 2196–2200 CrossRef PubMed
.
- S. D. Perrault and W. M. Shih, ACS Nano, 2014, 8, 5132–5140 CrossRef CAS PubMed
.
- H. T. Maune, S.-P. Han, R. D. Barish, M. Bockrath, W. A. Goddard III, P. W. K. Rothemund and E. Winfree, Nat. Nanotechnol., 2010, 5, 61–66 CrossRef CAS PubMed
.
- V. Linko and J. J. Toppari, J. Self-Assem. Mol. Electron., 2013, 1, 101–124 CrossRef CAS
.
- C. M. Soukoulis and M. Wegener, Nat. Photonics, 2011, 5, 523–530 CAS
.
- M. R. Jones, K. D. Osberg, R. J. Macfarlane, M. R. Langille and C. A. Mirkin, Chem. Rev., 2011, 111, 3736–3827 CrossRef CAS PubMed
.
- S. J. Tan, M. J. Campolongo, D. Luo and W. Cheng, Nat. Nanotechnol., 2011, 6, 268–276 CrossRef CAS PubMed
.
- J. Chao, Y. Lin, H. Liu, L. Wang and C. Fan, Mater. Today, 2015 DOI:10.1016/j.mattod.2015.01.018
.
- J. Sharma, R. Chhabra, C. S. Andersen, K. V. Gothelf, H. Yan and Y. Liu, J. Am. Chem. Soc., 2008, 130, 7820–7821 CrossRef CAS PubMed
.
- A. J. Mastroianni, S. A. Claridge and A. P. Alivisatos, J. Am. Chem. Soc., 2009, 131, 8455–8459 CrossRef CAS PubMed
.
- A. Kuzyk, R. Schreiber, Z. Fan, G. Pardatscher, E.-M. Roller, A. Högele, F. C. Simmel, A. O. Govorov and T. Liedl, Nature, 2012, 483, 311–314 CrossRef CAS PubMed
.
- E.-M. Roller, L. K. Khorashad, M. Fedoruk, R. Schreiber, A. O. Govorov and T. Liedl, Nano Lett., 2015, 15, 1368–1373 CrossRef CAS PubMed
.
- M. Pilo-Pais, S. Goldberg, E. Samano, T. H. LaBean and G. Finkelstein, Nano Lett., 2011, 11, 3489–3492 CrossRef CAS PubMed
.
- R. Schreiber, S. Kempter, S. Höller, V. Schüller, D. Schiffels, S. S. Simmel, P. C. Nickels and T. Liedl, Small, 2011, 7, 1795–1799 CrossRef CAS PubMed
.
- J. Liu, Y. Geng, E. Pound, S. Gyawali, J. R. Ashton, J. Hickey, A. T. Woolley and J. N. Harb, ACS Nano, 2011, 5, 2240–2247 CrossRef CAS PubMed
.
- S. Helmi, C. Ziegler, D. J. Kauert and R. Seidel, Nano Lett., 2014, 14, 6693–6698 CrossRef CAS PubMed
.
- W. Sun, E. Boulais, Y. Hakobyan, W. L. Wang, A. Guan, M. Bathe and P. Yin, Science, 2014, 346, 1258361 CrossRef PubMed
.
- A. Kuzyk, B. Yurke, J. J. Toppari, V. Linko and P. Törmä, Small, 2008, 4, 447–450 CrossRef CAS PubMed
.
- A. E. Gerdon, S. S. Oh, K. Hsieh, Y. Ke, H. Yan and H. T. Soh, Small, 2009, 5, 1942–1946 CrossRef CAS PubMed
.
- R. J. Kershner, L. D. Bozano, C. M. Micheel, A. M. Hung, A. R. Fornof, J. N. Cha, C. T. Rettner, M. Bersani, J. Frommer, P. W. K. Rothemund and G. M. Wallraff, Nat. Nanotechnol., 2009, 4, 557–561 CrossRef CAS PubMed
.
- V. Linko, S.-T. Paasonen, A. Kuzyk, P. Törmä and J. J. Toppari, Small, 2009, 5, 2382–2386 CrossRef CAS PubMed
.
- A. M. Hung, C. M. Micheel, L. D. Bozano, L. W. Osterbur, G. M. Wallraff and J. N. Cha, Nat. Nanotechnol., 2010, 5, 121–126 CrossRef CAS PubMed
.
- V. Linko, J. Leppiniemi, B. Shen, E. Niskanen, V. P. Hytönen and J. J. Toppari, Nanoscale, 2011, 3, 3788–3792 RSC
.
- V. Linko, J. Leppiniemi, S.-T. Paasonen, V. P. Hytönen and J. J. Toppari, Nanotechnology, 2011, 22, 275610 CrossRef PubMed
.
- A. Gopinath and P. W. K. Rothemund, ACS Nano, 2014, 8, 12030–12040 CrossRef CAS PubMed
.
- B. Shen, V. Linko, H. Dietz and J. J. Toppari, Electrophoresis, 2015, 36, 255–262 CrossRef CAS PubMed
.
- A. A. Rafat, T. Pirzer, M. B. Scheible, A. Kostina and F. C. Simmel, Angew. Chem., Int. Ed., 2014, 53, 7665–7668 CrossRef PubMed
.
- S. Woo and P. W. K. Rothemund, Nat. Commun., 2014, 5, 4889 CrossRef CAS PubMed
.
- Y. Ke, L. L. Ong, W. Sun, J. Song, M. Dong, W. M. Shih and P. Yin, Nat. Chem., 2014, 6, 994–1002 CrossRef CAS PubMed
.
- S. P. Surwade, F. Zhou, B. Wei, W. Sun, A. Powell, C. O'Donnell, P. Yin and H. Liu, J. Am. Chem. Soc., 2013, 135, 6778–6781 CrossRef CAS PubMed
.
- F. Marty, L. Rousseau, B. Saadany, B. Mercier, O. Français, Y. Mita and T. Bourouina, Microelectron. J., 2005, 36, 673–677 CrossRef CAS PubMed
.
- T. K. Hakala, V. Linko, A.-P. Eskelinen, J. J. Toppari, A. Kuzyk and P. Törmä, Small, 2009, 5, 2683–2686 CrossRef CAS PubMed
.
- S. P. Surwade, S. Zhao and H. Liu, J. Am. Chem. Soc., 2011, 133, 11868–11871 CrossRef CAS PubMed
.
- F. Zhou, B. Michael, S. P. Surwade, K. B. Ricardo, S. Zhao and H. Liu, Chem. Mater., 2015, 27, 1692–1698 CrossRef CAS
.
- Z. Jin, W. Sun, Y. Ke, C. J. Shih, G. L. C. Paulus, Q. H. Wang, B. Mu, P. Yin and M. S. Strano, Nat. Commun., 2013, 4, 1663 CrossRef PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available: CanDo-simulated solution shape of a rectangular origami. Fabrication and purification of DNA origamis. Experimental details, instrumentation and fabrication procedures for metallic nanostructures. Statistics of the dimensions of the gold nanostructures. Additional SEM images of the gold nanostructures. See DOI: 10.1039/c5nr02300a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2015 |