Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Crystal structure, stability and Ago2 affinity of phosphorodithioate-modified RNAs†
Pradeep S.
Pallan
a,
Xianbin
Yang
*b,
Malgorzata
Sierant
c,
N. Dinuka
Abeydeera
b,
Tom
Hassell
d,
Carlos
Martinez
d,
Magdalena
Janicka
c,
Barbara
Nawrot
c and
Martin
Egli
*a
aDepartment of Biochemistry, Vanderbilt University, School of Medicine, Nashville, TN 37232, USA. E-mail: martin.egli@vanderbilt.edu; Fax: +1-615-322-7122; Tel: +1-615-343-8070
bAM Biotechnologies LLC, 12521 Gulf Freeway, Houston, TX 77034, USA. E-mail: xianbin.yang@thioaptamer.com; Fax: +1-832-476-0294; Tel: +1-832-379-2175
cDepartment of Bioorganic Chemistry, Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Lodz, Poland
dSigma Life Science, 9186 Six Pines, The Woodlands, Texas 77380, USA
First published on 20th November 2014
Abstract
Small interfering RNAs (siRNAs) with phosphorodithioate modifications (PS2-RNA) possess favourable properties for use as RNAi therapeutics. Beneficial here is the combining of PS2 and 2′-O-methyl modifications (MePS2). SiRNAs with MePS2 moieties in the sense strand show promising efficacies in vitro and in vivo. Crystal structures of PS2- and MePS2- modified RNAs reveal subtle changes in geometry and hydration compared with natural RNA. A model of an MePS2-RNA–PAZ domain complex points to a hydrophobic effect as the source of the higher affinity of MePS2-RNA for Ago2.
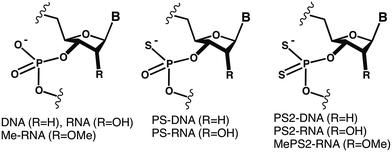
Small interfering RNAs (siRNAs) designed against a wide range of targets, of various formulations, and in many cases comprising chemical modifications are now being evaluated in the clinic.1–4 Chemical modification of the antisense (guide) and/or sense (passenger) siRNA strand modulates RNA affinity, nuclease resistance, immune stimulation, uptake and biodistribution, and is widely expected to be required for efficacy in vivo.5–9 SiRNAs containing phosphorodithioate modifications (PS2-RNA; Fig. 1) were recently shown to exhibit favorable properties for therapeutic applications.10 Thus, the PS2 moiety, despite destabilizing the duplex slightly relative to the native phosphate (PO2), appears not to distort the A-form geometry, improves serum stability, and is tolerated in the central part of the antisense strand as well as in most of the positions tested in the sense strand.
Unlike the phosphorothioate moiety (PS; Fig. 1), PS2 is achiral, thus precluding potential drawbacks that may arise from chiral P-substituted oligonucleotides, owing to variable biophysical and biochemical properties of individual (e.g. PS-) diastereo-isomers.11,12 In addition, PS2-modified siRNAs display increased protection against degradation by nucleases compared with PS-modified oligonucleotides.10,13,14 Combining the PS2 with the 2′-O-methyl ribose modification (MePS2-RNA; Fig. 1) afforded increased loading of modified siRNA duplexes into the RNA-induced silencing complex (RISC) as well as enhanced anti-tumor activity.15 Interestingly, the former property can be attributed to MePS2 modification at two residues adjacent to the 3′-TT overhang in the sense siRNA strand (ESI Fig. S1†). This conclusion is supported by higher association (based on in vitro pull-down assays), and tighter intracellular binding between MePS2-siRNA and Ago2 protein, relative to the corresponding PO2, PS2-, or methoxy-modified RNAs (RNA, PS2-RNA, or Me-RNA, resp.; Fig. 1). MePS2-siRNA showed significantly enhanced silencing of EphA2 involved in taxane resistance compared to all other RNA chemistries (Fig. 1) in both the SKOV3ipl and HeyA8 epithelial ovarian cancer cell lines that highly express target protein.15 Other proteins with a key role in chemo-resistance besides EphA2, such as GRAMD1B, were subsequently targeted by systemic administration of MePS2-siRNA, leading to a re-sensitization of chemo-resistant ovarian tumors to taxane therapy.15
To gain insight into the conformational consequences of PS2 modification and potential changes as a result of combining it with the ribose 2′-O-methyl substitution, we determined crystal structures of the RNA duplexes (CGCPS2GAAUUAGCG)2 (CPS2G-RNA), (CGCGAMePS2AUUAGCG)2 (AMePS2A-RNA), and (CGCGAAUUAMePS2GCG)2 (AMePS2G-RNA) at resolutions of 1.19, 1.18 and 1.13 Å, respectively.‡ The structures were phased by molecular replacement, using the previously determined structure of the native RNA16 as search model [ESI (Table S1†)].
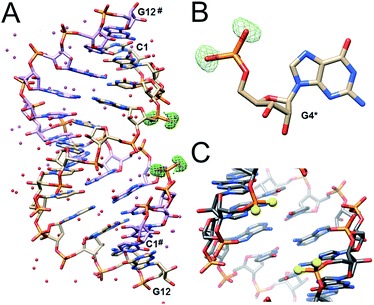
The structures of the PS2-modified RNA duplexes are isomorphous with that of the native RNA.16 Thus, the duplex is located on a crystallographic dyad and a single dodecamer strand constitutes the asymmetric unit. Residues are numbered 1–12 and those in the symmetry-related strand are numbered 1#–12# (Fig. 2). Superimposition of the CPS2G and native RNA duplexes demonstrates their virtually identical conformations (Fig. S2†). In particular PS2 modification does not trigger any changes in the ribose conformation and the backbone torsion angles. The most obvious deviation concerns the P–S bond lengths in the phosphorodithioate moiety (ca. 1.94 Å) compared to the P–O bonds in the native phosphate (ca. 1.60 Å) (Fig. S2†).
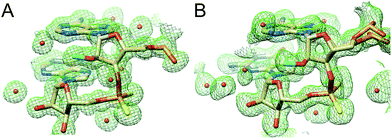
In the AMePS2A- and AMePS2G-RNA duplexes, 2′-O-methyl groups are directed into the minor groove, a result of the familiar ap orientation around the C2′–O2′ bond (Fig. 3).17,18 Therefore, neither the PS2 modification alone nor the combination of the PS2 and 2′-O-methyl modifications appear to have a notable effect on the conformation of A-form RNA judging from the crystallographic results.
However, the 2′-O-methyl modification can offset the slight thermodynamic destabilization caused by the PS2 moiety compared with the native RNA duplex.10 We determined the melting temperatures (Tm) of siRNAs containing two to six MePS2 modifications and found that they exhibit thermal stabilities that are comparable to that of the unmodified RNA (PO2) (Table 1).
| RNA | Sequence | T m [°C] |
|---|---|---|
| siRNA-1 | 5′ AUA CAG GCA GCA GUA ACU UU TT 3′ | 73.7 ± 0.8 |
| 3′ TT UAU GUC CGU CGU CAU UGA AA 5′ | ||
| MePS2-1 | 5′ AUA CAG GCA GCA GUA ACU UMePS2UMePS2TT 3′ | 74.0 ± 0.2 |
| 3′ TT UAU GUC CGU CGU CAU UGA A A 5′ | ||
| MePS2-3 | 5′ AUA CAMePS2GGCAGMePS2CAGUAMePS2ACU UUMePS2TT 3′ | 73.7 ± 0.5 |
| 3′ TT UAU GU C CGUC GUCAU UGA AA 5′ | ||
| MePS2-4 | 5′ AUA CAG GCA GCA GUA AC U UU TT 3′ | 74.2 ± 0.8 |
| 3′ TT UAU GUC CGU CGU CAU UGMePS2AMePS2AA 5′ | ||
| MePS2-9 | 5′ AUAMePS2CAG GCA GCAMePS2GUA ACU UU TT 3′ | 74.4 ± 0.5 |
| 3′ TT UAU GUC CGU CGUMePS2CAU UGA AA 5′ | ||
| MePS2-11 | 5′ AUA CAMePS2GGCAGMePS2CAGUAMePS2ACU UUMePS2TT 3′ | 73.2 ± 0.2 |
| 3′ TT UAU GU CCGUMePS2CMePS2GUCAU UGA AA 5′ |
To assess potential conformational consequences of the MePS2 modification in solution, we measured the circular dichroism (CD) spectra of the siRNA duplexes listed in Table 1. All spectra were closely similar to the spectrum of the native duplex and are consistent with the typical A-type structure (a maximum of the positive Cotton effect at 268 nm and a crossover point at 240 nm; Fig. S3†).
The lack of an obvious change in both the conformation and stability of MePS2-RNA relative to native RNA points to another cause of the favorable therapeutic properties of this modification.15 To better understand the increased Ago2 affinity of siRNA containing MePS2-modified sense strands (Fig. S1†), we built models of PAZ–siRNA complexes† based on the crystal structure of a short RNA fragment featuring a 3′ two-residue overhang bound to the human Ago2 PAZ domain.19 This domain is most likely the only region of Ago2 affected by MePS2 modification near the siRNA 3′-end (Fig. S4†). Comparison of the complexes with siRNA and MePS2-siRNA reveals formation of a hydrophobic patch in the latter, that involves both the 2′-O-methyl and PS2 moieties as well as methionine and cysteine side chains from the PAZ domain (Fig. 4). The hypothesis that increased hydrophobicity conferred by the MePS2 modification plays a key role in the stronger binding to Ago2 and its enhanced efficacy is supported by the relative Ago2 affinities of siRNAs containing no modification, PS, MePS, PS2 or MePS2 (Table 2).
| siRNA ID | Sequence | K D |
|---|---|---|
| a The PS2- and MePS2-siRNA affinities based on bio-layer interferometry are similar, although those from cellular Ago2 association or pull-down assays consistently show tighter binding by MePS2-modified RNAs.15 | ||
| AF163A | 5′-UGA CAU GCC GAU CUA CAUMePS2GMePS2dTdT-3′ | 21.1 pMa |
| 3′-Biotin dTdT ACU GUA CGG CUA GAU GUA C-5′ | ||
| AF163B | 5′-UGA CAU GCC GAU CUA CAU GdTdT-3′ | 0.6 μM |
| 3′-Biotin dTdT ACU GUA CGG CUA GAU GUA C-5′ | ||
| AF163C | 5′-UGA CAU GCC GAU CUA CAUMePSGMePSdTdT-3′ | 41.1 nM |
| 3′-Biotin dTdT ACU GUA CGG CUA GAU GUA C-5′ | ||
| AF163D | 5′-UGA CAU GCC GAU CUA CAUPS2GPS2dTdT-3′ | 12.4 pMa |
| 3′-Biotin dTdT ACU GUA CGG CUA GAU GUA C-5′ | ||
| AF163F | 5′-UGA CAU GCC GAU CUA CAUPSGPSdTdT-3′ | 14.5 nM |
| 3′-Biotin dTdT ACU GUA CGG CUA GAU GUA C-5′ | ||
Conclusions
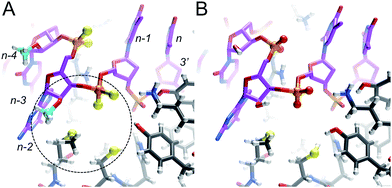
The MePS2 modification in the backbone region adjacent to the 3′-dTdT overhang of the siRNA sense strand triggers a strongly enhanced affinity to the RISC Ago2 slicer and silencing of proteins that confer chemoresistance in ovarian cancer cell lines and tumors.15 This finding is surprising given the fact that the antisense siRNA strand mediates cleavage of the target RNA at the Ago2 active site and indicates that chemical modification of the sense strand can play a key role in siRNA loading and efficacy.We demonstrate here that MePS2 modification does not affect RNA conformation and stability. Instead the increased Ago2 affinity is likely the result of favourable hydrophobic interactions with the PAZ domain mediated by both the 2′-O-methyl and PS2 moieties. This conclusion is consistent with the inability of PS2 sulphur atoms to form H-bonds with water molecules lining the RNA backbone in the crystal structure (Fig. 5).
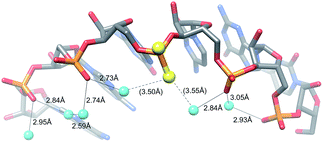 | ||
| Fig. 5 The PS2 moiety disrupts H-bonding interactions in the water chain bridging adjacent O2P atoms in the major groove. | ||
Both steric and electronic factors can contribute to the increased nuclease resistance afforded by a modification.18,20,21 The bulkier sulphur atoms in the PS2 moiety along with the increased hydrophobicity relative to PO2 might exclude nonthiophilic metal ions (e.g. Mg2+) from an exonuclease active site or result in suboptimal positioning of the water nucleophile for attack at the phosphate group.
Acknowledgements
Supported by US NIH grant R44 GM086937 (to X.Y. and M.E.).Notes and references
- M. E. Davis, J. E. Zuckerman, C. H. J. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
- C. Pecot, G. Calin, R. Coleman, G. Lopez-Berestein and A. Sood, Nat. Rev. Cancer, 2011, 11, 59–67 CrossRef CAS PubMed.
- J. C. Burnett, J. J. Rossi and K. Tiemann, Biotechnol. J., 2011, 6, 1130–1146 CrossRef CAS PubMed.
- J. Tabernero, G. I. Shapiro, P. M. LoRusso, A. Cervantes, G. K. Schwartz, G. J. Weiss, L. Paz-Ares, D. C. Cho, J. R. Infante, M. Alsina, M. M. Gounder, R. Falzone, J. Harrop, A. C. White, I. Toudjarska, D. Bumcrot, R. E. Meyers, G. Hinkle, N. Svrzikapa, R. M. Hutabarat, V. A. Clausen, J. Cehelsky, S. V. Nochur, C. Gamba-Vitalo, A. K. Vaishnaw, D. W. Sah, J. A. Gollob and H. A. Burris III, Cancer Discovery, 2013, 3, 406–417 CrossRef CAS PubMed.
- S. Choung, Y. Kim, S. Kim, H. Park and Y. Choi, Biochem. Biophys. Res. Commun., 2006, 342, 919–927 CrossRef CAS PubMed.
- D. Bumcrot, M. Manoharan, V. Koteliansky and D. W. Y. Sah, Nat. Chem. Biol., 2006, 2, 711–719 CrossRef CAS PubMed.
- S. Shukla, C. S. Sumaria and P. I. Pradeepkumar, ChemMedChem, 2009, 5, 328–349 CrossRef PubMed.
- M. Manoharan, A. Akinc, R. K. Pandey, J. Qin, P. Hadwiger, M. John, K. Mills, K. Charisse, M. A. Maier, L. Nechev, E. M. Greene, P. S. Pallan, E. Rozners, K. G. Rajeev and M. Egli, Angew. Chem., Int. Ed., 2011, 50, 2284–2288 CrossRef CAS PubMed.
- D. M. Kenski, G. Butora, A. T. Willingham, A. J. Cooper, W. Fu, N. Qi, F. Soriano, I. W. Davies and W. M. Flanagan, Mol. Ther.–Nucleic Acids, 2012, 1, e5 CrossRef PubMed.
- X. Yang, M. Sierant, M. Janicka, L. Peczek, C. Martinez, T. Hassell, N. Li, X. Li, T. Wang and B. Nawrot, ACS Chem. Biol., 2012, 7, 1214–1220 CrossRef CAS PubMed.
- A. V. Lebedev and E. Wickstrom, Perspect. Drug Discovery Des., 1996, 4, 17–40 CrossRef CAS.
- W. S. Marshall and M. H. Caruthers, Science, 1993, 259, 1564–1570 CAS.
- A. Detzer, M. Overhoff, A. Mescalchin, M. Rompf and G. Sczakiel, Curr. Pharm. Des., 2008, 14, 3666–3673 CrossRef CAS.
- D. A. Braasch, Z. Paroo, A. Constantinescu, G. Ren, O. K. Öz, R. P. Mason and D. R. Corey, Bioorg. Med. Chem. Lett., 2004, 14, 1139–1143 CrossRef CAS PubMed.
- S. Y. Wu, X. Yang, K. Gharpure, H. Hatakeyama, M. Egli, M. McGuire, A. S. Nagaraja, T. Miyake, R. Rupaimoole, C. V. Pecot, M. Taylor, S. Pradeep, M. Sierant, C. Rodriguez-Aguayo, H. J. Choi, R. A. Previs, G. N. Armaiz-Pena, L. Huang, C. Martinez, T. Hassell, C. Ivan, V. Sehgal, R. Singhania, H.-D. Han, C. Su, J.-H. Kim, H. Dalton, C. Kowali, K. Keyomarsi, N. A. J. McMillan, W. W. Overwijk, J. Liu, J.-S. Lee, K. Baggerly, G. Lopez-Berestein, P. Ram, B. Nawrot and A. K. Sood, Nat. Commun., 2014, 5, 3459 Search PubMed.
- F. Li, P. S. Pallan, M. A. Maier, K. G. Rajeev, S. L. Mathieu, C. Kreutz, Y. Fan, J. Sanghvi, R. Micura, E. Rozners, M. Manoharan and M. Egli, Nucleic Acids Res., 2007, 35, 6424–6438 CrossRef CAS PubMed.
- P. Lubini, W. Zürcher and M. Egli, Chem. Biol., 1994, 1, 39–45 CrossRef CAS.
- M. Egli, G. Minasov, V. Tereshko, P. S. Pallan, M. Teplova, G. B. Inamati, E. A. Lesnik, S. R. Owens, B. S. Ross, T. P. Prakash and M. Manoharan, Biochemistry, 2005, 44, 9045–9057 CrossRef CAS PubMed.
- J.-B. Ma, K. Ye and D. J. Patel, Nature, 2004, 429, 318–322 CrossRef CAS PubMed.
- M. Teplova, S. T. Wallace, G. Minasov, V. Tereshko, A. Symons, P. D. Cook, M. Manoharan and M. Egli, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14240–14245 CrossRef CAS.
- C. A. Brautigam, S. Sun, J. A. Piccirilli and T. A. Steitz, Biochemistry, 1999, 38, 696–704 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: General methods, including crystallographic experiments (Table S1) and model building, and illustrations of EphA2 siRNA (Fig. S1), superimposition of PS2-RNA and RNA duplexes (Fig. S2), CD spectra (Fig. S3), and comparisons between crystal structures of Ago2–siRNA and PAZ–siRNA complexes (Fig. S4). See DOI: 10.1039/c4ra10986d |
| ‡ Data deposition: final coordinates and structure factor files have been deposited in the Brookhaven Protein Data Bank (http://www.rcsb.org). The entry codes are 4RBY (CPS2G-RNA), 4RBZ (AMePS2A-RNA), and 4RC0 (AMePS2G-RNA). |
| This journal is © The Royal Society of Chemistry 2014 |