AFM analysis of changes in nucleosome wrapping induced by DNA epigenetic modification†
Seiichiro
Kizaki
a,
Yuki
Suzuki
a,
Tomohiro
Takenaka
a,
Masayuki
Endo
b and
Hiroshi
Sugiyama
*ab
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: hs@kuchem.kyoto-u.ac.jp
bInstitute for Integrated Cell-Material Sciences (WPI-iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto, 606-8501, Japan
First published on 30th June 2014
Abstract
The wrapping and unwrapping of the nucleosome, which is a fundamental packing unit of chromatin, are tied to the regulation of gene expression. The accessibility of DNA within nucleosomes is controlled not only by chromatin-remodeling molecules, but also by chemical modifications of histones and DNA. Understanding the structural changes of a nucleosome during epigenetic modifications is a key to unravel the mechanisms of gene regulation. Here, we reconstituted nucleosomes using methylcytosine- or hydroxymethylcytosine-substituted DNA, and analyzed their morphological features by atomic force microscopy (AFM). Our results indicate that cytosine methylation induces overwrapping of the DNA around the histone octamer, whereas cytosine hydroxymethylation has a lesser effect on the overwrapping of the DNA. These results suggest that two types of DNA modification yield different wrapping states of nucleosomes, which may contribute to the compaction and relaxation of the chromatin structure.
Introduction
Eukaryotic genomic DNA interacts with various proteins and is folded into chromatin fibers. The most fundamental unit of chromatin is the nucleosome, which is composed of ∼147 bp DNA that wraps in ∼1.75 turns around a histone octamer that contains two copies of H2A, H2B, H3, and H4.1,2 The nucleosome represents a significant barrier for DNA-binding regulatory proteins that control the process of gene expression. Therefore, changes in the nucleosome structure are closely related to gene regulation.The key mechanisms that modulate nucleosome properties are enzyme-mediated modifications of histones and DNA. Among these epigenetic modifications, the methylation of cytosine in DNA is essential in genomic imprinting, retrotransposon silencing, and X-chromosome inactivation.3–5 In mammalian cells, DNA methyltransferase (DNMT) transfers the methyl group of S-adenosylmethionine (SAM) to cytosine in CpG dinucleotides. In comparison with other epigenetic modifications, the methylation of cytosine is a relatively stable modification that is usually maintained throughout the cell cycle.6,7 However, in specific developmental stages, such as developing primordial germ cells (PGCs), methylation of cytosine is rapidly removed.8,9 Although the molecular mechanism underlying this active demethylation remains unclear, recent studies revealed that Tet-family proteins convert methylcytosine (mC) to hydroxymethylcytosine (hmC), and further to formylcytosine (fC) and carboxylcytosine (caC) (Fig. 1).10–12 These oxidized derivatives of mC are now considered intermediates of the active demethylation pathway.13
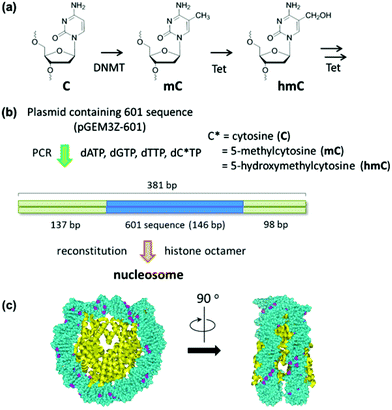 | ||
| Fig. 1 Study on the effect of cytosine modification on nucleosome formation. (a) Cytosine modification for the epigenetic gene regulation. (b) Experimental scheme for the nucleosome reconstitution using a PCR amplified reconstitution sequence containing modified cytosine. The location of the 601 sequence along a 381 bp fragment is shown. (c) The cytosine-methylated nucleosome model was constructed using the crystal structure of the Xenopus laevis nucleosome core particle (Protein Data Bank code: 3LZ0). Methyl groups of all cytosines are colored in purple. | ||
To understand the structural changes of nucleosomes that occur upon DNA methylation and demethylation, it will be necessary to image the nucleosome on modified DNA strands. The capability of atomic force microscopy (AFM) to provide images in solution has made it possible to study structural properties of nucleosomes under physiologically relevant conditions.14–16 In the studies communicated here, we applied this method to investigate the structure of nucleosomes that were reconstituted on unmethylated, methylated, and hydroxymethylated DNA. We showed that DNA modification changes the extent of wrapping of DNA around the histone octamer.
Experimental
DNAs and histones
The 381 bp DNA fragment containing the 601 positioning sequence was amplified by PCR using forward 5′-TAATACGACTCACTATAGG-3′ and reverse 5′-ATTTAGGTGACACTATAGAATAC-3′ primers from pGEM3Z-601.17 After the reaction, the amplified DNA was purified using the GenElute PCR Clean-Up Kit (Sigma-Aldrich, St. Louis, MO, USA). Methylcytosine- and hydroxymethylcytosine-substituted DNAs were prepared by conducting the PCR with the 5-methylcytosine dNTP mix (Zymo Research, Irvine, CA, USA) and the 5-hydroxymethylcytosine dNTP mix (Zymo Research), respectively. The HeLa core histone was purchased from Active Motif (Carlsbad, CA, USA).Nucleosome reconstitution
Nucleosomes were reconstituted as described previously.18,19 Briefly, equal amounts (0.5 μg) of the purified DNA and the histone octamer were mixed in Hi-buffer [10 mM Tris–HCl (pH 7.5), 2 M NaCl, 1 mM EDTA, 0.05% NP-40, and 5 mM β-mercaptoethanol], and placed in a dialysis tube (total volume 50 mL). The dialysis was started in 150 mL of Hi-buffer with stirring at 4 °C. Lo-buffer [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.05% NP-40, and 5 mM β-mercaptoethanol] was added to the dialysis buffer at the rate of 0.46 mL min−1, and the dialysis buffer was pumped out at the same rate with a peristaltic pump so that the final dialysis buffer contained 50 mM NaCl after 20 h. The sample was collected from the dialysis tube and stored at 4 °C until use.AFM imaging
The reconstituted nucleosome was diluted to a concentration of 0.5 ng μL−1 in a buffer containing 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2, and 1 mM EDTA, and 3 μL of the sample was immediately deposited onto freshly cleaved mica discs (φ 1.5 mm) pretreated with 0.1% (3-aminopropyl)triethoxysilane (APTES). After 1 min incubation, the sample was rinsed with 2 × 10 μL of the buffer and then imaged in the same buffer without the drying step. The AFM experiments were performed using a high-speed AFM (Nano Live Vision, RIBM, Tsukuba, Japan). The sample was imaged in buffer solution at ambient temperature with a small cantilever of dimensions L × W × H = 10 × 2 × 0.1 μm3 (BL-AC10EGS, Olympus, Tokyo, Japan). These cantilevers had a spring constant of 0.1–0.2 N m−1 with a resonant frequency in water of 400–1000 kHz and 320 × 240 pixel images were obtained at the scan rate of 0.2 fps. Individual images were imported into ImageJ (http://rsb.info.nih.gov/ij/) and analyzed. The length of wrapped DNA was calculated by subtracting the sum of the measured lengths of both DNA arms from the measured length of free DNA (Fig. S1†). Each nanometer-to-base-pair conversion factor (nm/bp) for unmethylated, methylated, and hydroxymethylated DNA was also obtained by dividing the measured lengths of free DNA in nm (Fig. S1†) by 381 bp.Results and discussion
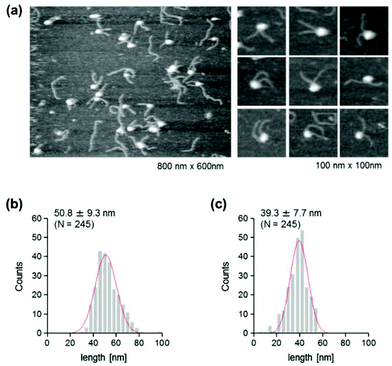
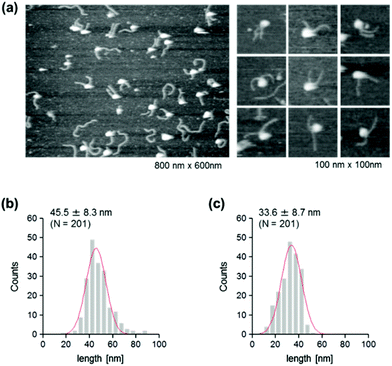
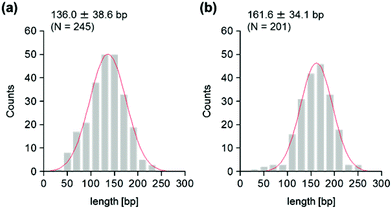
The use of reconstituted nucleosomal systems that contain a set of nucleosome-positioning signals has been a powerful approach to the analysis of the structure–function relationship of nucleosomes.20,21 We used the fragment of 381 bp carrying the 146 bp nucleosome-positioning 601 sequence flanked with two DNA arms of different lengths, 137 and 98 bp (Fig. 1b). AFM images in a liquid of the reconstituted nucleosomes depict a typical morphology for the mononucleosome, i.e. a bright particle with the two DNA arms at both sides (Fig. 2a). Note that the prepared sample was not treated with any crosslinkers, such as glutaraldehyde. The mean lengths of the longer and shorter arms (±SD) were 50.8 ± 9.3 nm and 39.3 ± 7.7 nm, respectively (Fig. 2b and c). Although the mean values obtained were comparable to the expected lengths of 50 nm for the longer arm and 36 nm for the shorter arm based on a nanometer-to-base-pair conversion factor of 0.366 nm bp−1, the ranges of the both distributions were broad. This may reflect the dynamic feature of nucleosomes,22–25 thus allowing varying extents of DNA wrapping, ranging from ∼1 to ∼1.75 turns.14We were interested in determining whether the structure of the nucleosome changes after the methylation of the template DNA (Fig. 3a). To assess this, we reconstituted nucleosomes using an mC-substituted DNA template and compared their features with those of unmethylated nucleosomes. Intriguingly, in AFM images (Fig. 3b), the assembled methylated nucleosomes were more compact than were the unmethylated ones. The analysis of the contour length of the two DNA arms provided quantitative information about this observation. As shown in Fig. 3c and d, the frequency distribution of the longer and shorter arms of the methylated nucleosome showed a peak at 45.5 ± 8.3 nm and 33.6 ± 8.7 nm, respectively. These values were smaller than the values obtained for the unmethylated nucleosomes (Fig. 2b), which suggests that cytosine methylation induces overwrapping of DNA around the histone octamer in a nucleosome. Note that nearly no considerable change was observed in the length of free DNA after substitution of cytosine with methylcytosine (Fig. S1†). To assess the degree of nucleosome wrapping, we calculated the length of the wrapped DNA for all nucleosomes, as described in the Methods section. Fig. 4a and b show the histograms of this value for the unmethylated nucleosomes and for the cytosine-methylated nucleosomes, respectively. Compared with unmethylated nucleosomes, an increase of ∼26 bp in the average of wrapped DNA was observed in methylated nucleosomes. These results indicate that cytosine methylation changes the nucleosome structure and is accompanied by overwrapping of DNA around the histone octamer.
The effect of DNA methylation on the nucleosome structure has been studied extensively by targeting CpG dinucleotides.26,27 Our results from cytosine-methylated nucleosomes were in line with those of previous studies, in which CpG methylation was reported to induce the compaction and stabilization of the nucleosome by causing a tighter wrapping of DNA around the histone octamer.27 The studies of CpG-methylated nucleosome core particles suggest that the changes in the wrapping can be attributed to the reduced twist of DNA upon methylation.27 In our system, almost all cytosines in the 381 bp are methylated, including the two flanking DNA arms. This highly methylated state of DNA might reduce its extent of twisting, causing a drastic increase in the average of wrapped DNA.
In addition to DNA methylation, DNA demethylation is also a fundamental process for epigenetic regulation of gene expression. It has been suggested that mC can be demethylated via stepwise oxidization in vivo.12 Among the oxidized derivatives of mC, hmC has been proposed as a key epigenetic mark per se.28 Considering that the modification of DNA alters its physical properties, the reduction of hydrophobicity upon oxidization from mC to hmC should also affect the structure of nucleosomes.
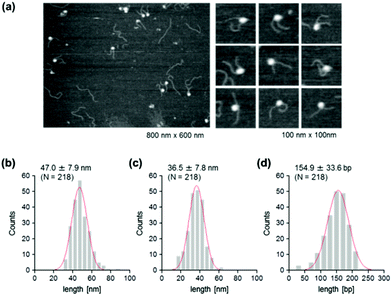
To address this hypothesis, next we performed similar AFM analyses on cytosine-hydroxymethylated nucleosomes. The results obtained are shown in Fig. 5. The mean lengths of the two arms were 47.0 ± 7.9 nm and 36.5 ± 7.8 nm, respectively. These values were larger than the values obtained for the methylated nucleosomes, but smaller than those recorded for the unmethylated ones. The frequency distribution of the wrapped DNA for cytosine-hydroxymethylated nucleosomes had a peak at 154.9 ± 33.6 bp, which represented a value that was between that of unmethylated nucleosomes (136.0 ± 38.6 bp) and that of methylated nucleosomes (161.6 ± 34.1 bp). These findings indicate that the cytosine-hydroxymethylated nucleosomes adopt a structural state that is different from those of both unmethylated and methylated nucleosomes. The introduction of hydroxyl groups at almost all mCs greatly reduces the hydrophobicity of DNA, which may allow the unwrapping and exposure of nucleosomal DNA on the nucleosome.
Conclusions
In the present study, we have shown that DNA methylation causes profound changes in the morphology of nucleosomes, with an increase in the wrapping of DNA around the histone octamer. This overwrapping was mitigated when the mCs in the template DNA were substituted with hmCs. However, the extent of wrapping in cytosine-hydroxymethylated nucleosomes was still larger than that of unmodified nucleosomes. These findings suggest that three different types of DNA modification yield different wrapping states of nucleosomes, probably reflecting the modification-induced changes in the physical properties of DNA strands. Recently, AFM imaging has also been applied to identify the changes in nucleosomes induced by a histone modification.29 Information regarding the interplay between epigenetic modifications and the morphology of nucleosomes obtained from AFM analyses will promote our basic understanding of the mechanisms of gene regulation.Notes and references
- R. D. Kornberg, Science, 1974, 184, 868–871 CAS.
- K. Luger, A. W. Mader, R. K. Richmond, D. F. Sargent and T. J. Richmond, Nature, 1997, 389, 251–260 CrossRef CAS PubMed.
- A. Bird, Genes Dev., 2002, 16, 6–21 CrossRef CAS PubMed.
- D. Takai and P. A. Jones, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 3740–3745 CrossRef CAS PubMed.
- M. Esteller, Nat. Rev. Genet., 2007, 8, 286–298 CrossRef CAS PubMed.
- M. F. Robert, S. Morin, N. Beaulieu, F. Gauthier, I. C. Chute, A. Barsalou and A. R. MacLeod, Nat. Genet., 2003, 33, 61–65 CrossRef CAS PubMed.
- R. J. Klose and A. P. Bird, Trends Biochem. Sci., 2006, 31, 89–97 CrossRef CAS PubMed.
- S. C. Wu and Y. Zhang, Nat. Rev. Mol. Cell Biol., 2010, 11, 607–620 CrossRef CAS PubMed.
- Y. Kawasaki, J. Lee, A. Matsuzawa, T. Kohda, T. Kaneko-Ishino and F. Ishino, Sci. Rep., 2014, 4, 3658 Search PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Science, 2009, 324, 930–935 CrossRef CAS PubMed.
- Y. F. He, B. Z. Li, Z. Li, P. Liu, Y. Wang, Q. Tang, J. Ding, Y. Jia, Z. Chen, L. Li, Y. Sun, X. Li, Q. Dai, C. X. Song, K. Zhang, C. He and G. L. Xu, Science, 2011, 333, 1303–1307 CrossRef CAS PubMed.
- S. Ito, L. Shen, Q. Dai, S. C. Wu, L. B. Collins, J. A. Swenberg, C. He and Y. Zhang, Science, 2011, 333, 1300–1303 CrossRef CAS PubMed.
- H. Wu and Y. Zhang, Cell, 2014, 156, 45–68 CrossRef CAS PubMed.
- L. S. Shlyakhtenko, A. Y. Lushnikov and Y. L. Lyubchenko, Biochemistry, 2009, 48, 7842–7848 CrossRef CAS PubMed.
- Y. Suzuki, Y. Higuchi, K. Hizume, M. Yokokawa, S. H. Yoshimura, K. Yoshikawa and K. Takeyasu, Ultramicroscopy, 2010, 110, 682–688 CrossRef CAS PubMed.
- A. Miyagi, T. Ando and Y. L. Lyubchenko, Biochemistry, 2011, 50, 7901–7908 CrossRef CAS PubMed.
- P. T. Lowary and J. Widom, J. Mol. Biol., 1998, 276, 19–42 CrossRef CAS PubMed.
- K. Hizume, S. H. Yoshimura, H. Maruyama, J. Kim, H. Wada and K. Takeyasu, Arch. Histol. Cytol., 2002, 65, 405–413 CrossRef CAS.
- K. Hizume, S. H. Yoshimura and K. Takeyasu, Biochemistry, 2005, 44, 12978–12989 CrossRef CAS PubMed.
- K. Ura, J. J. Hayes and A. P. Wolffe, EMBO J., 1995, 14, 3752–3765 CAS.
- K. Ura and A. P. Wolffe, Methods Enzymol., 1996, 274, 257–271 CAS.
- B. D. Brower-Toland, C. L. Smith, R. C. Yeh, J. T. Lis, C. L. Peterson and M. D. Wang, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1960–1965 CrossRef CAS PubMed.
- S. Mihardja, A. J. Spakowitz, Y. Zhang and C. Bustamante, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15871–15876 CrossRef CAS PubMed.
- G. Li, M. Levitus, C. Bustamante and J. Widom, Nat. Struct. Mol. Biol., 2005, 12, 46–53 CAS.
- H. S. Tims and J. Widom, Methods, 2007, 41, 296–303 CrossRef CAS PubMed.
- J. S. Choy, S. Wei, J. Y. Lee, S. Tan, S. Chu and T. H. Lee, J. Am. Chem. Soc., 2010, 132, 1782–1783 CrossRef CAS PubMed.
- J. Y. Lee and T. H. Lee, J. Am. Chem. Soc., 2012, 134, 173–175 CrossRef CAS PubMed.
- L. Shen and Y. Zhang, Curr. Opin. Cell Biol., 2013, 25, 289–296 CrossRef CAS PubMed.
- N. A. Filenko, C. Kolar, J. T. West, S. A. Smith, Y. I. Hassan, G. E. Borgstahl, J. Zempleni and Y. L. Lyubchenko, PLoS One, 2011, 6, e16299 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4bm00113c |
| This journal is © The Royal Society of Chemistry 2014 |