Research highlights
Cole A.
DeForest
a,
Huaibin
Zhang
b,
Adnan
Memic
c,
Mehmet
Dokmeci
def and
Ali
Khademhosseini
*defg
aDivision of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California 91125, USA
bDepartment of Chemistry, Tufts University, Medford, 02155, USA
cCenter of Nanotechnology, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
dCenter for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Cambridge, Massachusetts 02139, USA. E-mail: alik@rics.bwh.harvard.edu
eHarvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
fWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts 02115, USA
gWorld Premier International-Advanced Institute for Materials Research (WPI-AIMR), Tohoku University, Sendai, 980-8577, Japan
First published on 16th August 2012
Sugar-templated hydrogels for vascular tissue engineering
Mass transport within native human tissue enables delivery of nutrients to and removal of waste from cells embedded in a dense 3D matrix material. Blood flow through cardiovascular networks (e.g., veins and arteries) provides nutrients that can diffuse into the tissue. The ability to deliver nutrients deeper into the tissues is a critical issue in the development of large-scale multicellular organisms. As researchers attempt to regrow and recreate tissue-like structures, they face a major challenge in engineering an efficient vasculature. The desired vasculature is expected to promote cell survival within thick tissue constructs, where bulk diffusion is insufficient in supporting basic function.1Recently, Chen and colleagues have introduced an elegant and effective solution that utilizes simple sugars to create desired vasculature within synthetic tissues.2 In their work, carbohydrate glass was extruded with a custom-modified 3D printer to generate open solid lattice networks. This self-supporting structure was then encapsulated inside cell-laden hydrogels, which mimicked the native extracellular matrix (ECM). As the carbohydrate glass rapidly dissolved in water and cell media, liberating only fully cytocompatible sugar monomers (e.g., sucrose, glucose), the user was left with a scaffold with hollow channels that is akin to the native vasculature (Fig. 1). These microscale channels could then be perfused with nutrients to promote the survival and function of the encapsulated cells.
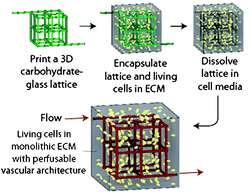 | ||
| Fig. 1 Schematic overview of vascular network generation by carbohydrate network 3D printing, encapsulation within cell-containing hydrogels, and subsequent lattice dissolution. Figure reprinted with permission from the Nature Publishing Group from Miller et al.2 | ||
Utilizing carbohydrate glass as a sacrificial material has many advantages from a fabrication standpoint.2,3 First, the degradation products of the carbohydrate glass are cytocompatible. Next, the diameter of the printed filaments can be readily controlled by varying the translational velocity of the extrusion nozzle. This yields structures with diameters similar to those of human blood vessels. Additionally, the sugar mixture is capable of physically supporting its own weight, allowing for complex interconnected structures to be printed via standard 3D printing techniques. Finally, the material is optically clear and transparent to light wavelengths that enable cellular imaging, fluorescence microscopy, and photopolymerization.
Upon infusing human umbilical vein endothelial cells (HUVECs) into the hollow microchannels of the hydrogel, uniform cell seeding along the channel walls and generation of key vascular components was observed. HUVECs lined the cylindrical walls, resulting in a lumen structure required for nutrient perfusion. Spontaneous sprouting of new vessels, one of the holy grails of vascular tissue engineering, was observed from the vein-like structures and into the bulk hydrogel. Furthermore, encapsulated cells uniformly distributed throughout the synthetic ECM exhibited increased viability and function within hydrogel constructs containing perfusion channels, owing to the increased nutrient availability deep within the material construct.
By combining emerging 3D printing techniques with established tissue engineering approaches, Miller et al. have made a major stride in the creation of synthetic cardiovascular structures. The proposed approach is highly versatile, compatible with a variety of cell types and synthetic ECM mimics, and should prove successful in the engineering of thick tissues for applications in regenerative medicine.
3D culturing of captured circulating tumor cells
Circulating tumor cells (CTCs) are epithelial cells released from a primary tumor into the blood stream. The main advantage of microfluidics in the analysis of CTCs lies in the improved capture efficiency of target cells through an increased surface area to volume ratio.4,5 To capture the cells, blood is injected into microfluidic chips containing microfabricated posts that are functionalized with epithelial specific antibodies. Out of millions of blood cells in a sample only CTCs bind to these antibodies, such that they are captured on-chip and can be analysed later on. The number of captured CTCs is proportional to the size of the tumor, and is also related to the metastasis potential of the disease. Some of the remaining challenges in the field include separation of cells from the posts following capture and their subsequent analysis.In a recent study, Lutolf and colleagues have made a major advance in this area by developing a method to directly measure the metastasis potential of CTCs by culturing the captured cells. A key aspect of their work was accessing the captured cells after capture, and also forming 3D tumor spheroids. In this work, Bichsel et al. fabricated a reversibly bonded channel system such that the device was taken apart after use and the captured cells were readily extracted.6 To protect cells during this process, a biodegradable hydrogel matrix was formed in the channel for temporary cell encapsulation.
Microfluidic channels with pillars were formed by standard soft lithography using poly(dimethylsiloxane) (PDMS). The PDMS channel surface was thiolated by silane, and coated with maleimide-linked Protein G. The surface was then functionalized with antibodies specific to epithelial cells (EpCAM and CD49f), via the immunoglobin-binding Protein G intermediate. The functionalized PDMS channel was covered with another piece of PDMS and the setup was sandwiched inside a mechanical scaffold to form a closed microscale device. Prostate cancer cells expressing mCherry fluorescent protein (PC3-mCherry) were spiked into the cell culture medium or whole blood at a concentration of 1000 cells mL−1 and were flowed through the device. For cells spiked in the cell culture medium, the capture efficiencies for chips functionalized with EpCAM and CD49f were significantly higher than for those functionalized with a control antibody (∼40% versus 8%). The capture efficiency observed using EpCAM, however, was lower than that of a similar system that uses biotin–avidin linkage to couple antibodies onto silicon chips (>60%).4 For cells spiked in whole blood, the difference between the capture efficiency for EpCAM, CD49f and control functionalized flow cells was not statistically significant.
To culture the captured cells, a pre-polymer solution of a commercially available QGel matrix was injected into the channel. After polymerization, the device was disassembled and the PDMS channel with encapsulated cells was immersed into a cell culture medium (Fig. 2). About half of the captured cells were alive after this procedure. Among the remaining cells, 17% survived until day 6 of culture. At this time, colonies of spheroids were observed, which were used as an in vitro model for tumors.
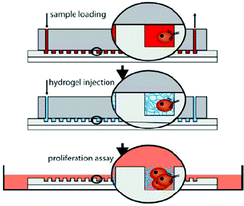 | ||
| Fig. 2 Schematic of the experimental procedure. The sample is first flushed through the chip and the CTCs are captured, then the chip is washed and the hydrogel is injected, and finally the gel-encapsulated cells are transferred to cell culture conditions. Figure reprinted with permission from the Royal Society of Chemistry from Bichsel et al.6 | ||
This work stands out due to the simplicity of the method for transferring contents obtained by on-chip modules (captured CTCs) to off-chip applications (cell culturing). The biodegradable hydrogel matrix serves as a gentle and effective carrier for this purpose. To make this approach truly useful for cancer diagnostics, it is important to demonstrate the viability of this method in whole blood samples with higher capture efficiency. This could potentially be improved by optimizing the attachment of antibodies to the PDMS surface and the placement of the functionalized PDMS pillars.
Weaving hydrogels into mosaic art
Advanced biomaterials may be useful in creating tissues with biomimetic tissue structure. However, so far approaches that have aimed to emulate such hierarchical architectures on meaningful tissue scales have been limited, requiring multiple steps and complex production processes. Emerging approaches7 aim to generate complex materials through automated, continuous methods that would be amenable to biological systems.A recent communication following such an approach is reported by Günther and coworkers. Leng et al.8 developed a single step, continuous flow fabrication of scalable mosaic hydrogel sheets. They were able to extrude hydrogels with both temporal and spatial resolution, therefore controlling the soft material composition. The advantage of this approach is that it does not require substrate support or moving device components during the tessellation (repetition or patterning of geometric shapes with no overlaps or gaps) and coding of planar soft materials. At the same time this method is highly compatible with a range of biopolymers and different cell types.
The authors accomplished this by developing a novel microfluidic device capable of dynamically incorporating single or multiple secondary biopolymers woven into the flowing base polymer, thereby generating a mosaic structure. These secondary biopolymers consisted of either a chemically different composition or were carrying a molecular, cellular or colloidal payload. The group was able to achieve tessellations with defined x,y planar secondary biopolymer incorporation. These varying compositions were retained following the cross-linking step, forming the mosaic hydrogel.
Specifically, they fabricated the microfluidic device by vertically stacking and bonding individual PDMS layers. The completed device could withstand pressures of up to 600 kPa and therefore could sustain high flow rates for an extended period of time. Each of the three PDMS sections was molded via soft lithography from a unique master and had additional features at the device exit. The focusing fluid flowing through the bottom and top layers contained the cross-linking ions, which induced gelation of the base polymer. The template polymer fluid stacked between the two focusing flows was connected to 7 parallel channels allowing encoding with secondary biopolymers. By adjusting the flow rate of the focusing fluid, they were able to obtain planar soft materials with a range of thicknesses (170–700 μm). The thickness of the layers could also be controlled by manually attaching the hydrogel sheet to a collection drum 50 mm downstream of the device exit. The ability to vary the speed of the collection drum offered an additional parameter for adjustment of the focusing fluid flow and resulted in improved control during the formation and image-based inspection of collected mosaic hydrogels. For temporal resolution, the microfluidic device was outfitted with computer-actuated solenoid valves. These valves initiated the outflow from the on-chip secondary biopolymer reservoirs by raising the pressure. This approach prevented cross-talk between reservoirs while incorporating the secondary biopolymers within the mosaic hydrogel sheet with high spatiotemporal resolution.
The researchers were able to not only demonstrate predictable, accurate, and dynamic control of soft material composition, but they also illustrated an approach that is high throughput and combines continuous, mask-free patterning and formation of soft materials. They characterized the effect of tessellations both locally and in the bulk material, and they also quantified the bulk elastic modulus of mosaic sheets. In addition, the proposed method is also compatible with patterning dense cell populations within soft material sheets, which was demonstrated using primary rat cardiomyocytes.
References
- G. Vunjak-Novakovic and D. T. Scadden, Cell Stem Cell, 2011, 8, 252–261 CrossRef CAS.
- J. S. Miller, K. R. Stevens, M. T. Yang, B. M. Baker, D. H. Nguyen, D. M. Cohen, E. Toro, A. A. Chen, P. A. Galie, X. Yu, R. Chaturvedi, S. N. Bhatia and C. S. Chen, Nat. Mater., 2012 DOI:10.1038/nmat3357.
- J. Lee, J. Paek and J. Kim, Lab Chip, 2012, 12, 2638–2642 RSC.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U. J. Balis, R. G. Tompkins, D. A. Haber and M. Toner, Nature, 2007, 450, 1235–1239 CrossRef CAS.
- S. L. Stott, C.-H. Hsu, D. I. Tsukrov, M. Yu, D. T. Miyamoto, B. A. Waltman, S. M. Rothenberg, A. M. Shah, M. E. Smas, G. K. Korir, F. P. Floyd, A. J. Gilman, J. B. Lord, D. Winokur, S. Springer, D. Irimia, S. Nagrath, L. V. Sequist, K. J. Isselbacher, S. Maheswaran, D. A. Haber and M. Toner, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18392–18397 CrossRef CAS.
- C. A. Bichsel, S. Gobaa, S. Kobel, C. Secondini, G. N. Thalmann, M. G. Cecchini and M. P. Lutolf, Lab Chip, 2012, 12, 2313–2316 RSC.
- E. Kang, G. S. Jeong, Y. Y. Choi, K. H. Lee, A. Khademhosseini and S.-H. Lee, Nat. Mater., 2011, 10, 877–883 CrossRef CAS.
- L. Leng, A. McAllister, B. Zhang, M. Radisic and A. Günther, Adv. Mater., 2012, 24, 3650–3658 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2012 |
