Multivalent DNA recognition by self-assembled clusters: deciphering structural effects by fragments screening and evaluation as siRNA vectors†
Abstract
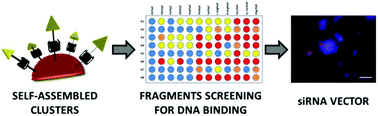
The identification of low-molecular-weight clusters that effectively complex oligonucleotides of therapeutic interest is of great importance for applications in gene delivery. We recently reported the use of self-assembly processes based on chemoselective ligation in order to generate biomolecular clusters for the multivalent recognition of DNA. Herein, we exploit the modularity of this methodology to perform a one-pot fragments screening of scaffolds and binding groups. Structural parameters affecting DNA binding were observed and hits have been identified by fluorescence displacement and gel electrophoresis assays. Finally, we evaluated the potential of these systems for siRNA transfection. One biomolecular cluster was found to effectively complex and transport a 21-mer siRNA inside MCF7 human breast cancer cells, resulting in a significant knockdown of the target gene.
- This article is part of the themed collection: Multivalent Biomolecular Recognition

 Please wait while we load your content...
Please wait while we load your content...