PredCSO: an ensemble method for the prediction of S-sulfenylation sites in proteins†
Abstract
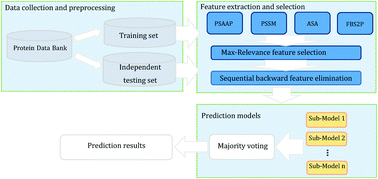
Protein S-sulfenylation is a type of reversible post-translational modification (PTM) through which cysteine (CYS) thiols of proteins are reversibly oxidized to cysteine sulfenic acids (CSO). Recent studies have shown that this event plays an essential role in cell signaling, transcriptional regulation and protein functions. Therefore, the identification of S-sulfenylation sites is important to understand the functions of S-sulfenylated proteins. In this study, we proposed PredCSO, a computational method for predicting S-sulfenylation sites in proteins. PredCSO is built on four kinds of features, including position-specific scoring matrix, position-specific amino acid propensity, the absolute solvent accessibility and four-body statistical pseudo-potential. In particular, 21 crucial features were refined out using a two-step feature selection procedure consisting of a max-relevance algorithm and a sequential backward elimination algorithm. To overcome the problem of imbalanced sample sizes, we adopt an ensemble method, which combines bootstrap resampling, gradient tree boosting and majority voting. Our performance evaluation shows that PredCSO achieves state-of-the-art performance in identifying S-sulfenylation sites in proteins.


 Please wait while we load your content...
Please wait while we load your content...