Small-world networks of residue interactions in the Abl kinase complexes with cancer drugs: topology of allosteric communication pathways can determine drug resistance effects
Abstract
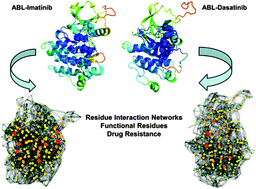
The human protein kinases play a fundamental regulatory role in orchestrating functional processes in complex cellular networks. Understanding how conformational equilibrium between functional kinase states can be modulated by ligand binding or mutations is critical for quantifying molecular basis of allosteric regulation and drug resistance. In this work, molecular dynamics simulations of the Abl kinase complexes with cancer drugs (Imatinib and Dasatinib) were combined with structure-based network modeling to characterize dynamics of the residue interaction networks in these systems. The results have demonstrated that structural architecture of kinase complexes can produce a small-world topology of the interaction networks. Our data have indicated that specific Imatinib binding to a small number of highly connected residues could lead to network-bridging effects and allow for efficient allosteric communication, which is mediated by a dominant pathway sensitive to the unphosphorylated Abl state. In contrast, Dasatinib binding to the active kinase form may activate a broader ensemble of allosteric pathways that are less dependent on the phosphorylation status of Abl and provide a better balance between the efficiency and resilience of signaling routes. Our results have unveiled how differences in the residue interaction networks and allosteric communications of the Abl kinase complexes can be directly related to drug resistance effects. This study offers a plausible perspective on how efficiency and robustness of the residue interaction networks and allosteric pathways in kinase structures may be associated with protein responses to drug binding.

 Please wait while we load your content...
Please wait while we load your content...