Differential accumulation of host mRNAs on polyribosomes during obligate pathogen-plant interactions†
Abstract
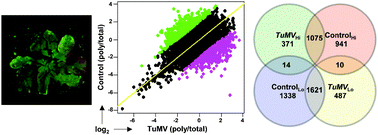
Plant pathogens elicit dramatic changes in the expression of host genes during both compatible and incompatible interactions. Gene expression profiling studies of plant-pathogen interactions have only considered

 Please wait while we load your content...
Please wait while we load your content...