Denaturation mapping of Saccharomyces cerevisiae
Abstract
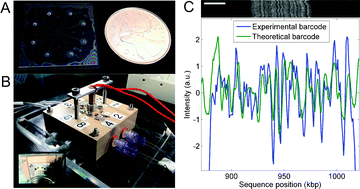
Optical mapping of DNA provides large-scale genomic information that can be used to assemble contigs from next-generation sequencing, and to detect rearrangements between single cells. A recent optical mapping technique called denaturation mapping has the advantage of using physical principles rather than the action of enzymes to probe genomic structure. Denaturation mapping uses

 Please wait while we load your content...
Please wait while we load your content...