Open Access Article
Open Access ArticleChemical approaches to probe and engineer AAV vectors
Quan
Pham
 ,
Jake
Glicksman
and
Abhishek
Chatterjee
,
Jake
Glicksman
and
Abhishek
Chatterjee
 *
*
Department of Chemistry, Boston College, 2609 Beacon Street, Chestnut Hill, MA 02467, USA. E-mail: abhishek.chatterjee@bc.edu
First published on 18th June 2024
Abstract
Adeno-associated virus (AAV) has emerged as the most promising vector for in vivo human gene therapy, with several therapeutic approvals in the last few years and countless more under development. Underlying this remarkable success are several attractive features that AAV offers, including lack of pathogenicity, low immunogenicity, long-term gene expression without genomic integration, the ability to infect both dividing and non-dividing cells, etc. However, the commonly used wild-type AAV capsids in therapeutic development present significant challenges, including inadequate tissue specificity and the need for large doses to attain therapeutic effectiveness, raising safety concerns. Additionally, significant preexisting adaptive immunity against most natural capsids, and the development of such anti-capsid immunity after the first treatment, represent major challenges. Strategies to engineer the AAV capsid are critically needed to address these challenges and unlock the full promise of AAV gene therapy. Chemical modification of the AAV capsid has recently emerged as a powerful new approach to engineer its properties. Unlike genetic strategies, which can be more disruptive to the delicate capsid assembly and packaging processes, “late-stage” chemical modification of the assembled capsid—whether at natural amino acid residues or site-specifically installed noncanonical amino acid residues—often enables a versatile approach to introducing new properties to the capsid. This review summarizes the significant recent progress in AAV capsid engineering strategies, with a particular focus on chemical modifications in advancing the next generation of AAV-based gene therapies.
Introduction
Adeno-associated virus (AAV) is a small, non-enveloped virus belonging to the parvoviridae family, which has exhibited tremendous potential as a vector for the development of human gene therapies.1–7 With 12 distinct human serotypes, none of which are linked to any human disease, AAV efficiently transduces a broad spectrum of both dividing and non-dividing cells, allowing for the prolonged expression of transgenes. Furthermore, AAV triggers minimal innate immune responses and is inherently replication deficient. Thanks to these attractive features, AAV stands out as a highly favorable vector for gene delivery in both in vitro and in vivo settings. To date, the FDA has approved five AAV-based drugs for gene therapy that address various genetic diseases:8–13 Luxturna, an AAV2 drug for inherited retinal disease; Zolgensma, an AAV9 drug for spinal muscular dystrophy; Hemgenix, an AAV5 drug for hemophilia B; Roctavian, an AAV5 drug for hemophilia A; and Elevidys, an AAVrh74 drug to treat Duchenne muscular dystrophy. Numerous other clinical trials are underway, showing promising results.3AAV possesses a compact genome, approximately 4.7 kb of single-stranded DNA flanked by two inverted terminal repeats (ITR).6,14 The native genome features two main open reading frames —rep and cap—each of which produces multiple non-structured and structured proteins through alternative splicing and the use of alternative start codons. The capsid comprises three proteins, VP1, VP2, and VP3, approximately in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio, forming an icosahedral particle with extensive interactions related to two-, three-, and five-fold symmetry.14 The initial step in AAV's infectious pathway involves binding to a primary cell-surface receptor, followed by interaction with a secondary receptor.15–17 This secondary interaction initiates diverse internalization pathways, resulting in trafficking to endosomal and Golgi compartments. Subsequently, AAV particles escape into the cytoplasm, accumulating in the perinuclear space before ultimately entering the nucleus for genome release and replication. These processes entail complex changes in capsid conformation and intricate interactions between the capsid and cellular factors, many of which remain poorly characterized. One of the main reasons behind this knowledge gap is the lack of appropriate tools to probe the biology underlying the entry of this virus. Many established tools used to probe protein localization and interaction networks involve tagging the target with other proteins (e.g., proteins with intrinsic fluorescence or proximity labeling capabilities), which are typically incompatible with the delicate and complex AAV capsid. Consequently, new tools are needed to elucidate the biology of AAV, which are associated with minimal perturbation to the target. In addition, recent advances in mass-spectrometry-based platforms have also revealed the presence of many novel post-translational modifications on the AAV capsid residues across multiple serotypes.18,19 However, the distribution of these modifications is quite heterogeneous and, except for a few well-documented cases such as phosphorylation mediated capsid degradation,20–22 and their physiological significance remains poorly understood.
10 ratio, forming an icosahedral particle with extensive interactions related to two-, three-, and five-fold symmetry.14 The initial step in AAV's infectious pathway involves binding to a primary cell-surface receptor, followed by interaction with a secondary receptor.15–17 This secondary interaction initiates diverse internalization pathways, resulting in trafficking to endosomal and Golgi compartments. Subsequently, AAV particles escape into the cytoplasm, accumulating in the perinuclear space before ultimately entering the nucleus for genome release and replication. These processes entail complex changes in capsid conformation and intricate interactions between the capsid and cellular factors, many of which remain poorly characterized. One of the main reasons behind this knowledge gap is the lack of appropriate tools to probe the biology underlying the entry of this virus. Many established tools used to probe protein localization and interaction networks involve tagging the target with other proteins (e.g., proteins with intrinsic fluorescence or proximity labeling capabilities), which are typically incompatible with the delicate and complex AAV capsid. Consequently, new tools are needed to elucidate the biology of AAV, which are associated with minimal perturbation to the target. In addition, recent advances in mass-spectrometry-based platforms have also revealed the presence of many novel post-translational modifications on the AAV capsid residues across multiple serotypes.18,19 However, the distribution of these modifications is quite heterogeneous and, except for a few well-documented cases such as phosphorylation mediated capsid degradation,20–22 and their physiological significance remains poorly understood.
Despite its remarkable success over the last decade, the wild-type AAV capsids that the industry and most clinical studies continue to rely on face several considerable disadvantages. One such key drawback is the limited control over the tissue tropism of native AAV serotypes. The broad tropism of AAV, affecting various tissues such as lung, liver, spleen, muscle, brain, kidney, etc., makes it challenging to deliver a transgene selectively to a target tissue. The liver, a common target for many AAV serotypes, acts as a sink, consuming the majority of delivered particles. Consequently, high doses of viral vectors are typically required to achieve sufficient transgene expression at the desired tissues, leading to potential toxicity in the liver and possibly other organs. Additionally, a large percentage of the human population have AAV-neutralizing antibodies due to prior exposure to this virus, excluding them from AAV vector-based treatments. Furthermore, treatment of eligible patients with a particular therapeutic AAV vector typically triggers significant adaptive immune response, and the resulting neutralizing antibodies prevent subsequent redosing. To overcome these limitations, various strategies, including genetic engineering and chemical modification, have been employed to engineer the viral capsid of AAV.1,2,6,23–29 In this review, we summarize these strategies, with a particular focus on the chemical approaches that have been utilized to both probe and engineer the biology of AAV.
An overview of the genetic approaches to engineer AAV
In this section we seek to provide a brief overview of the genetic approaches employed to engineer the properties of the AAV capsid, representative example of which are also summarized in Table 1. These examples can be broadly divided into two categories, those relying on rational engineering and on directed evolution. Rational design offers a more direct approach, and relies on prior knowledge of the structure and the function of the capsid to engineer the capsid proteins in a hypothesis-dependent manner. Such approaches typically involve rational mutation of residues known to negatively impact AAV (e.g., phosphodegrons),20–22 or the insertion of a peptide or a protein domain into permissive sites of the capsid proteins to introduce novel properties such as tissue specificity. Insertion of novel peptide sequences, such as those identified from in vitro selection experiments, into permissive loops of the AAV capsid proteins is straightforward, and has been used with some success for targeting the virus to specific cell-types.30–35 However, permissive sites in the capsid that withstand such insertions are limited, and only small peptides are typically tolerated. Fusion of larger proteins – such as antibody fragments and DARPINs – has also been used, but these are largely restricted to the N-terminus, or into the variable domain IV of the minor capsid protein through extensive linker optimization.36–43 Such strategies have also been used to improve the selectivity of AAV vectors. However, fusion of a foreign protein to the N-terminus of the minor capsid protein VP2 disrupts the natural capsid architecture, since VP2 N-terminus is normally tucked inside the capsid. Similarly, the disruptive effect of internal fusion on the capsid has also significantly limited the size and complexity of proteins that can be amenable to such fusion. Although there are numerous examples of using peptide and protein fusion strategies to engineer AAV, potential disruption to the complex and delicate capsid architecture represents a potential limitation of this strategy. Another notable rational approach involves the use of bispecific antibodies, which can mediate the interaction between an AAV capsid and a target receptor.44,45 Insertion of peptide tags into the capsid, which can be subsequently covalently (e.g., formylglycine, HUH etc.), or noncovalently (e.g., asymmetric leucine zippers) labeled with function-altering entities with high specificity, has also been employed to engineer the properties of the capsid.46–50| Strategies | Selected methods | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| Directed evolution | Error-prone PCR | Enables diversification across the whole capsid. | Low mutation rate in library generation | 51–53 |
| Can find unexpected mutations which improve function | Limited coverage of the possible sequence space | |||
| Serotype shuffling | Combines the benefits of multiple serotypes. | Unlikely to acquire novel and unique functions | 54–57 | |
| Enables protein fusions at logical break points | ||||
| AAV display | Allows in vivo selection of cell-binding peptides incorporated in the capsid | Selections are technically demanding, outcome is unpredictable | 58–66 | |
| Structure-guided approach | Can be used on any serotype to retarget virus to tissue of interest | Requires relevant structural information. | 71 and 72 | |
| Evolved variants can gain altered tissue tropism and immune evasion | ||||
| Rational design | Point mutation to ablate phospho-degrons on the capsid surface | Improves both in vitro and in vivo transduction abilities via point mutations (such as tyrosine residues) | Limited scope | 20–22 |
| Peptide loop insertion | Can be well tolerated at specific sites | May destabilize the capsid in most sites, limited to short peptides | 30–35 | |
| Straightforward approach | ||||
| N-terminal fusion and loop insertion to minor capsid proteins | Can be used to attach larger, folded protein domains (such as affibodies and DARPins) | Can disrupt capsid, and thus infectivity and viral titer | 36–43 | |
| Non-covalent modification using biochemical tags | Can be used to attach small to large, folded proteins (antibodies, protein vault nanoparticles) | Some bacteria-derived tags can be immunogenic in vivo Non-covalent conjugation can cause loss of the attached molecules. | 46–50 | |
| In silico design | Computational reconstruction of ancestral AAVs | Allows discovery of ancestral capsids that are not recognized by the immune system | Unpredictable tropism. | 69 and 70 |
| Acquires adaptive immune response after first dose | ||||
| Machine-learning assisted capsid evolution | Diverse, synthetic capsid variants can be generated with higher throughput than rational design and random mutagenesis approaches | Requires necessary machine learning models and algorithms | 67 and 68 | |
| Require library cloning capable of covering the vast amount of generated variants |
Directed evolution approaches have been extensively utilized to create AAV variants with enhanced capabilities such as selective transduction of specific tissues and evading neutralizing antibodies. Several approaches have been used to generate the libraries of capsid variants for such directed evolution experiments, including error-prone mutagenesis of the cap gene,51–53 DNA shuffling of the cap gene from various existing AAV serotypes,54–57 as well as the insertion of randomized peptide libraries into a permissive site of the capsid (e.g., at the variable domains IV and VIII of the capsid).58–66 Facilitated by the available structural information of the AAV capsid, and the rapid development of DNA synthesis and next-generation sequencing capabilities, numerous AAV directed evolution studies have been reported in both cell cultures and animal models. This process has successfully generated variants capable of efficiently transducing challenging targets such as the brain, muscles, and T cells while exhibiting lower immunogenicity, which are excellent potential candidates for gene therapy. Despite these success stories, directed evolution of AAV capsid can be a time- and labor-intensive process, which is not guaranteed to yield desirable results. Moreover, mutant capsids with novel properties selected using a specific setup (in cell culture or non-human animal models) may not always translate to humans, due to subtle differences in receptor structures and other factors in vivo.
Finally, in silico engineering of the AAV capsid has recently emerged as a promising alternative approach. For example, machine learning strategies, that capitalize large sequence-function correlation datasets, such deep-mutation profiling data of the AAV capsid gene, are being used to design novel capsid mutants.67,68 Computational approaches have also been used to resurrect extinct ancestral AAV capsid sequences, which may have limited preexisting immunity.69,70
Chemical modification of the AAV capsid
Genetic approaches to engineer AAV are typically associated with significant sequence alterations (mutation or insertions) which may have unanticipated impacts on the complex structure and biology of AAV. These concerns are further amplified by our limited understanding of AAV biology, which compromises our ability to fully map the associated impacts of such sequence changes. Chemical modification of native AAV capsids, or those harboring site-specifically incorporated bioorthogonal conjugation handles, has emerged as an alternative approach for introducing function-altering entities on them to engineer their properties. Chemical modification has also been used to introduce small biophysical probes on the capsid to study the biology of AAV. Since the modification is introduced on packaged capsids, this strategy largely maintains the native capsid architecture and packaging behavior of AAV. The degree of capsid modification can also be carefully controlled by modulating reaction conditions to avoid introducing perturbation to its structure and function. Furthermore, this approach allows the introduction of entities with practically unlimited chemical diversity on the AAV capsid, enabling the creation of diverse AAV conjugates with novel properties. This includes user-defined synthetic molecules, fluorescent dyes, quantum dots, DNA, RNA, peptides, and even proteins. The resulting conjugates have many potential applications such as: (1) aiding the virus in evading the immune system, (2) retargeting the virus to tissues of interest with high efficiency and selectivity, (3) studying the biology of the virus through real-time imaging and controlling its behavior with light, and (4) using the virus as a tool for the directed evolution of biomolecules. Below, we discuss the advances in this area, focusing first on the modification of native AAV capsids at canonical amino acid residues, and then bioorthogonal site-specific modification of AAV capsids harboring precisely incorporated noncanonical amino acid residues.Modification of the AAV capsid at canonical amino acid residues
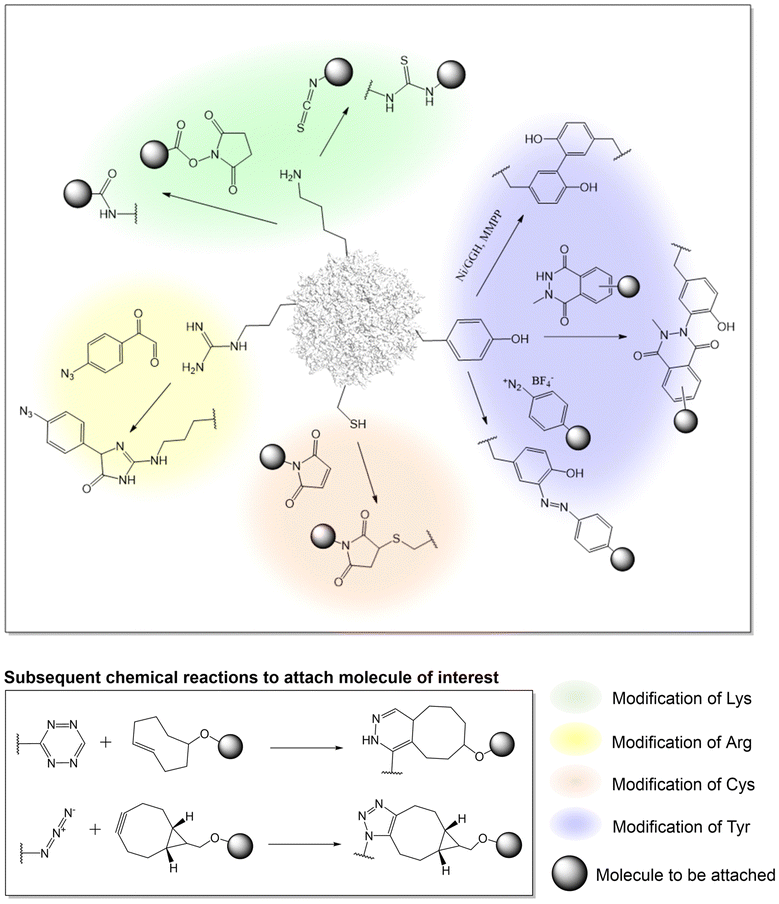
Advances in protein-labeling chemistries provide numerous ways to modify the capsid proteins at various canonical amino acid residues with high chemoselectivity, including lysine, arginine, cysteine, and tyrosine residues with no genetic manipulation needed (Fig. 1, Table 2).| Amino acid to modify | AAV serotype | Reagent and chemistry to modify | Molecule to be attached | Application | Ref. |
|---|---|---|---|---|---|
| Lys | AAV2 | N-Hydroxy-succinimide (NHS) | Fluorescence dyes | FACS analysis, real time imaging, real-time single particle tracking during infection | 73–76 |
| AAV3 | |||||
| AAV9 | |||||
| AAVrh.10 | |||||
| Quantum dot | Long-term live-cell imaging | 78 | |||
| Radioactive isotope | Imaging in animal models | 79–81 | |||
| Polyethylene glycol | Vector protection against neutralizing antibodies | 82 | |||
| Paclitaxel | Delivery of taxol | 94 | |||
| Biotin | Noncovalently conjugate with streptavidin – EGF fusion protein, or with T4 phage | 95 and 96 | |||
| Streptavidin | Noncovalently conjugate with biotin tagged antibody for retargeting | 86 | |||
| AAV8 | NHS followed by maleimide | Zwitterionic phosphoserine | Suppress preexisting immunity by evading anti-AAV antibodies | 83 | |
| AAV8 | 3,3′-Dithiobis(sulfo-succinimidyl propionate) (DTSSP) | L-Fucose, cross-linked AAV | Targeted gene delivery in pancreatic cancer cells | 97 | |
| AAV2, AAV8 | Isothiocyanates | FITC, GalNac, Mannose | Live-cell imaging, targeted delivery to hepatocytes | 85 | |
| Eosin to trigger photopolymerization | Encapsulate AAV in an acid-degradable polymeric shell embedded with siRNA for (1) gene and siRNA co-delivery and (2) immune-evasion | 84 | |||
| AAV2, AAV8, AAV9 | N-Ethyl Maleimide (NEM) | Retargeting to murine bone marrow | 98 | ||
| Arg | AAV2 | Methylglyoxal | Hydroimidazol-ones | - Retarget from liver to skeletal and cardiac muscle | 87 |
| - Immune evasion | |||||
| AAV2 | 4-Azidophenyl glyoxal, followed by BCN (via SPAAC) | Anti V-CAM1 scFv | Retargeting to endothelial cells | 88 | |
| Tyr | AAV2 | MMPP | Cross-linked VP protein | Investigate the impact of inter-subunit protein dynamics on externalization of VP N-terminus | 89 |
| AAV2 | Diazonium salts | GalNac, Mannose | Targeted delivery to hepatocytes and retina | 92 | |
| AAV2 | N-Methylluminol | GalNac, Mannose | Improved transduction of hepatocyte carcinoma | 91 | |
| Cys | AAV9 | TCEP, followed by maleimide | Fluorescence dye, biotin | In vivo imaging | 93 |
| Characterization of the capsid interactome |
Utilizing lysine residues
Earlier work on chemical modification of AAV has centered around the surface exposed lysine residues on the viral capsid.33–37,73–77 Due to the nucleophilicity of the lysine side-chain, it is possible to acylate the amino groups with either N-hydroxysuccinimide (NHS) esters or isothiocyanates in buffered aqueous solutions near physiological pH. In an effort to understand how AAV invades the cell, early research used lysine-selective NHS esters to introduce fluorescent cyanine dyes on the capsid. These probe-labeled virus particles allowed the real time imaging of viral entry into live cells.73–76 Such experiments provided initial knowledge about the kinetics of AAV entry and trafficking. This strategy has also been used to introduce other probes such as quantum dots, which are less susceptible to photobleaching, as well as radioactive isotopes (which was attached to AAV by NHS-tetrazine, followed by TCO ligation), enabling other ways to probe AAV entry and trafficking.78–81Additionally, lysine-targeted NHS ester chemistry was also used by the Schaffer group to attach polyethylene glycol (PEG) to AAV to shield the virus from the immune system.82 Various PEG lengths were appended to uncover the optimal shielding effect against neutralizing antibodies in serum. By testing multiple virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG ratios, the study underscored the importance of regulating capsid modification levels to achieve optimal shielding without compromising viral infectivity. Similarly, the Jiang group tethered an immunosuppressive zwitterionic phosphoserine (PS)-containing polypeptide as an immunomodulatory signal to the viral capsid.83 The resulting conjugates were observed to significantly diminish anti-AAV immune responses without substantially altering viral transduction and tropism. Kwon et al. conjugated AAV with eosin (a photo-initiator) at surface-exposed lysine residues using isothiocyanate,84 which was subsequently used to initiate a polymerization reaction to create an acid-degradable polyketal shell around the virus, which was further embedded with siRNAs. This polymer-coated virus evaded immune response while co-delivering both the virus and the siRNA into cells. In a different approach, Mével et al. employed isothiocyanate chemistry to attach carbohydrates as hepatocyte-targeting moiteties.85
PEG ratios, the study underscored the importance of regulating capsid modification levels to achieve optimal shielding without compromising viral infectivity. Similarly, the Jiang group tethered an immunosuppressive zwitterionic phosphoserine (PS)-containing polypeptide as an immunomodulatory signal to the viral capsid.83 The resulting conjugates were observed to significantly diminish anti-AAV immune responses without substantially altering viral transduction and tropism. Kwon et al. conjugated AAV with eosin (a photo-initiator) at surface-exposed lysine residues using isothiocyanate,84 which was subsequently used to initiate a polymerization reaction to create an acid-degradable polyketal shell around the virus, which was further embedded with siRNAs. This polymer-coated virus evaded immune response while co-delivering both the virus and the siRNA into cells. In a different approach, Mével et al. employed isothiocyanate chemistry to attach carbohydrates as hepatocyte-targeting moiteties.85
To develop a more universal platform, streptavidin or biotin was conjugated to the AAV capsid using NHS reagents. Taking advantage of the rapid and near-irreversible association between biotin and avidin, the platform allowed the attachment of various biomolecules of interest to the AAV capsids which was chemically conjugated with either of the two. For example, Lee and Ahn demonstrated that AAV2 modified with streptavidin-NHS ester can be noncovalently linked to biotin-tagged antibodies.86 The anti-EpCAM-AAV2 conjugates exhibited remarkable specificity in targeting and slowing tumor progression in mouse models by transporting EGFR shRNA. In an alternative approach, the Castillas, Jr. team engineered AAV2 by introducing biotin-NHS, then coupling it with EGF-Streptavidin fusion, resulting in over a 100-fold increase in transduction in SK-OV-3 cells. Similarly, Rao et al. employed biotin-labeled AAV along with T4 phage, connected through a streptavidin bridge, yielding a hybrid vector capable of carrying unprecedented payloads of up to 170 kb DNA genome and up to 1025 protein molecules. Although this approach is valuable, a downside of this strategy includes the poor control over attachment site and stoichiometry. Potential immunogenicity associated with bacteria-derived avidin is also a concern.
Utilizing arginine residues
Many serotypes of AAV utilize positively charged arginine residues to bind heparan sulfate proteoglycan for cellular entry. Alterations to these arginine residues can ‘detarget’ the virus from its primary receptor for reengineering AAV tropism. Asokan et al. demonstrated the feasibility of masking surface-exposed arginines of AAV2 using methylglyoxal, generating charge-neutral hydroimidazolones and thereby disrupting viral binding to heparan sulfate.87 This led to the observed ability to evade neutralizing antibodies, along with introducing novel tissue tropism towards cardiac and skeletal muscle. Similarly, Pearce et al. glycated arginine residues with 4-azidophenyl glyoxal, introducing a click chemistry handle to the AAV6 capsid.88 The azide group reacts with BCN through strain-promoted azide-alkyne cycloaddition (SPAAC), facilitating the crosslinking of anti-VCAM1-scFv to the virus as a retargeting agent. Similar to lysine residues, targeting arginine residues also faces intrinsic challenges in controlling the site of attachment and the right degree of modification to optimize AAV conjugate. Disruption of the endogenous net charge of AAV, as well as the cross-reactivity of methylglyoxal with other nucleophilic residues can also be problematic.Utilizing tyrosine residues
In addition to modifying lysine and arginine, tyrosine-selective conjugation reactions have also been used to modify AAV. Targeting tyrosine residues is advantageous because, unlike lysine and arginine-selective reactions, tyrosine modification is typically not associated with a change in charge, minimizing the chances of perturbing virus structure and function. In addition, tyrosine residues are less abundant than lysine and arginine on the capsid, affording a lower degree of heterogeneity when chemically modified. Leveraging the unique redox properties of the tyrosine phenol, Asokan and colleagues used a mild metal-catalyzed oxidation reaction to crosslink a pair of neighboring tyrosine resides bridging two capsid proteins together.89 This selective crosslinking reaction was used to probe the role of interfacial dynamics at the AAV capsid's two-fold symmetry axis, in the externalization of N-terminal domain of a capsid protein during infection. Recently, a novel tyrosine-selective electrochemical conjugation strategy was developed by Gouin et al. using N-methylluminol derivatives.90 The tyrosine residues of AAV2 were used to attach carbohydrates such as acetylgalactosamine (GalNAc) or mannose (Man) as retargeting moieties.91 Alternatively, Leray et al. have utilized the azo-coupling reaction to label tyrosine residues on the AAV capsid to attach carbohydrates for selective transduction of liver and retinal targets.92Utilizing cysteine residues
A key limitation of targeting canonical amino acids for protein labeling is the limited control over site and stoichiometry. Due to the relative scarcity of cysteines in proteins, coupled with its unique reactivity as a potent nucleophile, engineered cysteine residues have been frequently used for labeling proteins with improved site-specificity. Given the lack of exposed cysteine residues in AAV capsids, the introduction of engineered cysteine residues represents an attractive strategy for site-specific and stoichiometry control capsid modification. However, use of this strategy has remained relatively rare for AAV capsid engineering, possibly due to associated technical challenges such as oxidation of the exposed cysteines, as well as potential capsid aggregation. The Azzouz lab incorporated a 12-amino acid tetracysteine motif into the interface of VP1/VP2 proteins of AAV9.93 After packaging, the disulfide bonded cysteine residues were reduced with TCEP, yielding nucleophilic thiols, followed by labeling with maleimide-containing molecules. Using this strategy, fluorescent dyes were attached to the capsid, enabling real-time in vivo imaging of viral particles in the mouse brain. Additionally, by conjugating the virus with biotin to enable streptavidin enrichment, it was possible to identify potential host interaction partners, including actin/cytoskeletal proteins, proteins involved in RNA splicing/processing, chromatin modification, intracellular trafficking, etc.Chemical modification of AAV capsid on non-canonical amino acids
Although labeling canonical amino acid residues has been used extensively, this approach provides limited control over the sites and stoichiometry of capsid modification, and typically results in heterogeneous mixture of conjugates. The lack of site-specificity also may be problematic due to modification of functionally important residues. The genetic code expansion technology (GCE) provides an exciting solution to these challenges by enabling site-specific incorporation of a noncanonical amino acid (ncAA) into the virus capsid with a bioorthogonal conjugation handle, which can be subsequently labeled with high specificity (Table 3).| Serotype | ncAA | Tested sites | Reagents and chemistry to modify | Application | Ref. |
|---|---|---|---|---|---|
| AAV2 | AzK | S261, S264, A266, Q325, Q325 + 1, D327, N381, Y444, R447,T450-R459, S452 + 1, G453 + 1, S492, Y500, F534, E548, E548 + 1, T573, S578, R585 + 1, N587, N587 + 1, S662, T454 (VP1/VP2 only or VP1 + VP2) | DBCO (via SPAAC) | - Attach cRGD peptides for retargeting to ovarian cancer cell lines | 103, 104 and 106 |
| - Biotin attachment for VADER selection in mammalian cells | 113–116 | ||||
| DIBO (via SPAAC) | - Attach cRGD peptides to improve transduction of glioblastoma cells | 105 | |||
| - Optimizing PEG linker size and site of incorporation to reduce immunogenicity | 109 | ||||
| - Attachment of fluorescent probes for imaging | 112 | ||||
| NBK | R585, R588 | Irradiation with 365 nm decage | - Perturbation of HSPG binding to control AAV infection with light | 111 | |
| AAV5 | AzK | D374, E381, T444, G455, V481, S485, S518, T539, S576, T577 | DBCO (via SPAAC) | -Attachment of fluorescence dyes | 108 |
| - Improve transduction of many cell lines (HEK293, HUH7, C2C12, A549) and tissues (lung, liver, heart, TA muscle) | |||||
| AAV-DJ | AzK | R447, S578, N587, S662 | Alkyne (via Cu-click) | -Attachment of fluorescence dye for imaging | 110 |
| - Attachment of oligonucleotide to coat AAV with lipofectamine, shielding AAV from neutralizing serum | |||||
| T456, D555, A587, N589N589 (VP2/VP3 only) | DBCO (via SPAAC) | - Attachment of biotin for streptavidin blot | 107 | ||
| - Attachment of folic acid or aptamer for retargeting | |||||
| AAV8/AAVLKO3 | AzK | AAV8 (E330, T457, T499, N590, N590 + 1), AAV-LK03 (N588, T455, T456) | DBCO- (via SPAAC) | - Attach folic acid for improved transduction of HeLa cells | 107 |
Production of AAV incorporating genetically encoded ncAAs
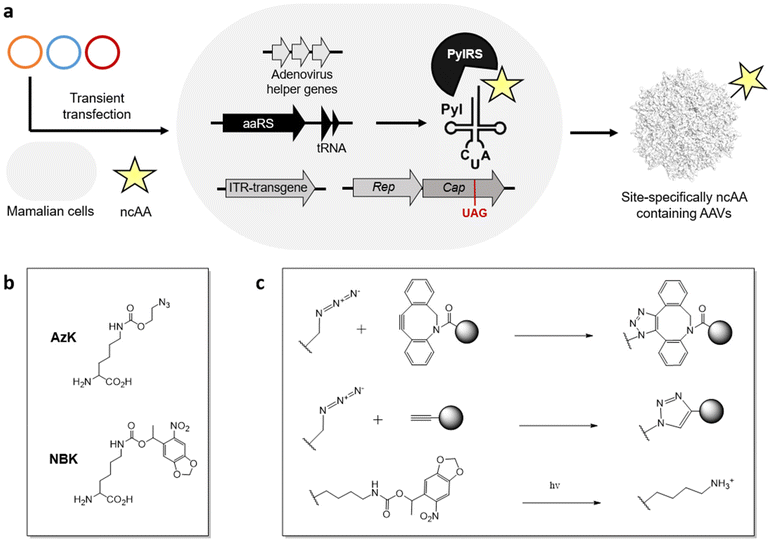
Using the genetic code expansion (GCE) technology, numerous ncAAs have been genetically encoded in various domains of life.99–101 This technology uses an engineered aminoacyl-tRNA synthetase (aaRS)/tRNA pair to incorporate a ncAA in response to a repurposed nonsense codon (typically the amber stop codon UAG). With access to >200 ncAAs in mammalian cells, including those suitable for diverse applications such as bioorthogonal conjugation and photo-crosslinking. Their small structural footprint minimizes potential perturbation to the virus capsid, while providing powerful new capabilities to probe and engineer its properties.Using the GCE technology, our lab and others have generated AAVs site-specifically decorated with ncAAs, through transient transfection of AAV2 genes, adenovirus helper genes, and a suitable aaRS/tRNA pair in mammalian cells (Figure 2a).102–105 To incorporate ncAA into the capsid protein, a stop codon is strategically placed at a surface-exposed site, which is suppressed by the engineered aaRS/tRNA pair. Since all capsid proteins VP1, VP2, VP3 share the same coding sequence with different splicing and alternative starts, introducing a stop codon at the common VP3 region results in the ncAA incorporation at all 60 capsid proteins. Using the pyrrolysyl synthetase (PylRS)/tRNA pair, it has been possible to incorporate azido-lysine (AzK) at various sites on the AAV capsid, without perturbing its properties. The azide-containing side chain of this ncAA enables subsequent bioorthogonal modification of the resulting capsids.
Recently, we developed a split-cap system to selectively incorporate the ncAA into a chosen subset of the capsid proteins (VP1, VP2, and VP3), providing further control over the number of ncAA handles present per capsid; capsids with 5 or 10 or 60 ncAAs can be generated by selectively incorporating them into capsid proteins VP1/VP2, VP1 + VP2 or all VP1 + VP2 + VP3.106 Additionally, the incorporation of ncAAs at minor capsid proteins was shown to provide wild-type-like titer of the recombinant virus, while also showing a greater tolerance to chemical modification with more defined conjugates.
Retargeting AAVs using site-specific capsid modification
It has been possible to retarget AAV to distinct cell surface receptors by attaching targeting ligands at ncAA residues on the capsid. For example, bioorthogonal attachment cyclic-RGD peptides onto AzK-modified AAV2 capsids using strain-promoted azide-alkyne cycloaddition has been used to selectively redirect them to cancer cell lines overexpressing integrin receptors (Figure 2b, c).102–106 A key advantage of this strategy is the ability to systematically modulate the site and stoichiometry of ligand attachment to fine-tune the properties of the resulting conjugate. We have shown that the site of attachment indeed has a significant impact on the efficacy of the resulting capsid conjugates. The number of retargeting ligands attached per capsid is also critically important. For example, cyclic-RGD mediated retargeting was found to be optimal at a labeling density of approximately 12 ligands/capsid.106 Lower stoichiometry was insufficient for strong retargeting, likely due to inefficient binding, while excessive modification with cRGD was detrimental for infectivity. Kay et al. has also functionalized various AAV serotypes at site-specifically incorporated AzK residues with aptamers as retargeting ligands, and demonstrated significant improvement in transduction in cancer cell lines in vitro.107 However, the efficacy of the aptamer-AAV conjugates was modest in vivo in animal models, perhaps due to the aptamers’ instability in blood. There is thus a need to further explore the possibility of attaching retargeting moieties with high receptor binding affinity and stability. Additionally, Wang et al. demonstrated that incorporation AzK into specific sites of the AAV5 capsid alone enhanced their lung-specific transduction.108 This work highlights how the incorporation of diverse chemistries represented in the GCE toolbox – even without further modification – can impact the properties of AAV.Lower the immunogenicity of AAV
GCE technology has significantly enhanced the control over the site and stoichiometry of capsid modification. Using this strategy, PEG molecules with varying molecular weights have been attached onto different sites on the AzK-containing virus particle.109 The resulting viruses exhibited an approximately 50% reduction in anti-AAV antibody generation upon intravenous administration. In a different approach, the Mali et al. incorporated AzK into the AAV-DJ capsid for further conjugation with oligonucleotides using copper click chemistry (Figure 2c).110 By complexing the resulting oligonucleotide-labeled capsids with lipofectamine (a cationic lipid based transfection agent), they showed that the resulting lipofectamine-coated capsids were effectively shielded from neutralizing serum.Probing the biology of viruses
Using GCE, we have incorporated photo-activatable ncAAs into the capsid of AAV2. Incorporating a photocaged lysine (NBK), replacing key arginine residues (R588 or R585) crucial for binding the primary receptor heparan sulfate proteoglycan (HSPG) resulted in non-infective mutants which were unable to bind HSPG (Figure 2b, c).111 However, a brief irradiation fully uncaged the positively charged lysine residue, restoring the HSPG receptor binding and nearly full-infectivity of these mutants. Using this strategy, it should be possible to reversibly perturb distinct molecular interactions between the virus particle and various host factors to probe their roles during viral entry. Additionally, AzK-labeled AAV capsids have also been bioorthogonally labeled with fluorescent dyes, enabling real-time imaging of virus particles during cellular entry and intracellular trafficking.112Despite its remarkable potential, functionalizing AAVs with ncAAs also has some drawbacks. This approach is technically demanding, and requires significant manipulation of the standard genetic machinery needed for AAV production. Incorporating ncAAs using nonsense suppression can also lead to a reduction in viral yield, particularly when the ncAA is incorporated across all 60 capsid proteins. In addition, despite their limited footprint, not all ncAAs are tolerated at all exposed sites on the AAV capsid. In fact, only AzK and its analogs have been successfully integrated into AAV to date using the pyrrolysyl pair. Expanding the variety of ncAAs that can be incorporated into AAVs will significantly broaden the scope of applications achievable through modifying ncAAs on AAVs.
Virus as a tool for directed evolution
Beyond exploring the biology and engineering of AAV for enhanced gene therapy, we recently showed that AAV can be used to perform directed evolution of tRNAs (and potentially other biomolecules) in mammalian cells.113,114 In this process, the activity of an AAV-encoded library of UAG-suppressing tRNA variants is coupled to the expression of a UAG-encoding AAV Cap gene. Consequently, tRNA variants with higher activity enable the production of ncAA-labeled Cap protein, facilitating the packaging of the progeny virus displaying the bioorthogonal ncAA handle on its capsid. These capsids can be further enriched through bioorthogonal biotinylation, followed by avidin enrichment. This chemical modification/enrichment strategy to improve the stringency of the selection was crucial for its success. This strategy has already been used for successful engineering of pyrrolysyl113,115 and bacterial leucyl tRNAs,116 and it has tremendous potential to facilitate directed evolution of additional biomolecules in mammalian cells. Gratifyingly, the engineered tRNAs generated by this method significantly improved the efficiency of ncAA incorporation into the capsid of AAV, providing wild-type-like yields (Fig. 2).Conclusion
In this review, we have outlined various exciting strategies for modifying the capsid of AAV to understand and engineer its biology, especially for developing next-generation gene therapy vectors. Among these methods, we've particularly emphasized on the chemical approaches to modify the AAV capsid, either at canonical amino acid residues, or at site-specifically incorporated ncAA residues using the GCE technology. This approach allows the capsid to be decorated with diverse entities, including biophysical probes, peptides, proteins, sugars, oligonucleotides, carbohydrates, etc. to probe and manipulate AAV biology. Modification of the canonical amino acid residues provides a technically simple, yet enabling approach to modify the AAV capsid. However, this strategy is typically associated with a lack of control over the site of modification. Although technically more demanding, the site-specific ncAA incorporation technology offers a complementary strategy with an exquisite level of control over the site and stoichiometry of capsid modification. Recent results have underscored how such controlled modification is crucial for producing AAV conjugates with optimal properties. Further development of these strategies has the potential to overcome the existing hurdles in AAV gene therapy, including retargeting the virus to specific tissues and protecting it from the immune system, thereby fully harnessing the remarkable potential of this exciting new therapeutic modality.Data availability
No relevant data is associated with this minireview.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
A. C. acknowledges support from National Science Foundation (MCB-1817893) and NIH (R35GM136437).References
- D. Wang, P. W. L. Tai and G. Gao, Adeno-associated virus vector as a platform for gene therapy delivery, Nat. Rev. Drug Discovery, 2019, 18, 358–378 CrossRef CAS PubMed.
- C. Li and R. J. Samulski, Engineering adeno-associated virus vectors for gene therapy, Nat. Rev. Genet., 2020, 21, 255–272 CrossRef CAS PubMed.
- D. A. Kuzmin, M. V. Shutova, N. R. Johnston, O. P. Smith, V. V. Fedorin, Y. S. Kukushkin, J. C. M. van der Loo and E. C. Johnstone, The clinical landscape for AAV gene therapies, Nat. Rev. Drug Discovery, 2021, 20, 173–174 CrossRef CAS PubMed.
- R. J. Samulski and N. Muzyczka, AAV-Mediated Gene Therapy for Research and Therapeutic Purposes, Annu. Rev. Virol., 2014, 1, 427–451 CrossRef PubMed.
- M. F. Naso, B. Tomkowicz, W. L. Perry III and W. R. Strohl, Adeno-associated virus (AAV) as a vector for gene therapy, BioDrugs, 2017, 31, 317–334 CrossRef CAS PubMed.
- A. Pupo, A. Fernández, S. H. Low, A. François, L. Suárez-Amarán and R. J. Samulski, AAV vectors: The Rubik's cube of human gene therapy, Mol. Ther., 2022, 30, 3515–3541 CrossRef CAS PubMed.
- F. Mingozzi and K. A. High, Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges, Nat. Rev. Genet., 2011, 12, 341–355 CrossRef CAS PubMed.
- A. M. Keeler and T. R. Flotte, Recombinant Adeno-Associated Virus Gene Therapy in Light of Luxturna (and Zolgensma and Glybera): Where Are We, and How Did We Get Here?, Annu. Rev. Virol., 2019, 6, 601–621 CrossRef CAS PubMed.
- J. Gao, R. M. Hussain and C. Y. Weng, Voretigene Neparvovec in Retinal Diseases: A Review of the Current Clinical Evidence, Clin. Ophthalmol., 2020, 14, 3855–3869 CrossRef CAS PubMed.
- H. A. Blair, Onasemnogene Abeparvovec: A review in spinal muscular atrophy, CNS Drugs, 2022, 36, 995–1005 CrossRef PubMed.
- Y.-A. Heo, Etranacogene dezaparvovec: first approval, Drugs, 2023, 83, 347–352 CrossRef CAS PubMed.
- A. Philippidis, BioMarin's ROCTAVIAN wins food and drug administration approval as first gene therapy for severe hemophilia A, Hum. Gene Ther., 2023, 34, 665–668 CrossRef CAS PubMed.
- S. M. Hoy, Delandistrogene Moxeparvovec: first approval, Drugs, 2023, 83, 1323–1329 CrossRef CAS PubMed.
- M. Agbandje-McKenna and J. Kleinschmidt, AAV capsid structure and cell interactions, Methods Mol. Biol., 2011, 807, 47–92 CrossRef CAS PubMed.
- A. M. Dudek, S. Pillay, A. S. Puschnik, C. M. Nagamine, F. Cheng, J. Qiu, J. E. Carette and L. H. Vandenberghe, An Alternate Route for Adeno-associated Virus (AAV), Entry Independent of AAV Receptor, J. Virol., 2018, 92, e02213–17 Search PubMed.
- B. P. Dhungel, C. G. Bailey and J. E. J. Rasko, Journey to the Center of the Cell: Tracing the Path of AAV Transduction, Trends Mol. Med., 2021, 27, 172–184 CrossRef CAS PubMed.
- S. Pillay, N. L. Meyer, A. S. Puschnik, O. Davulcu, J. Diep, Y. Ishikawa, L. T. Jae, J. E. Wosen, C. M. Nagamine and M. S. Chapman, et al., An essential receptor for adeno-associated virus infection, Nature, 2016, 530, 108–112 CrossRef CAS PubMed.
- B. Mary, S. Maurya, S. Arumugam, V. Kumar and G. R. Jayandharan, Post-translational modifications in capsid proteins of recombinant adeno-associated virus (AAV) 1-rh10 serotypes, FEBS J., 2019, 286, 4964–4981 CrossRef CAS PubMed.
- F. Guapo, L. Strasser, S. Millán-Martín, I. Anderson and J. Bones, Fast and efficient digestion of adeno associated virus (AAV) capsid proteins for liquid chromatography mass spectrometry (LC-MS) based peptide mapping and post translational modification analysis (PTMs), J. Pharm. Biomed. Anal., 2022, 207, 114427 CrossRef CAS PubMed.
- R. Nakahama, A. Saito, S. Nobe, K. Togashi, I. K. Suzuki, A. Uematsu and K. Emoto, The tyrosine capsid mutations on retrograde adeno-associated virus accelerates gene transduction efficiency, Mol. Brain, 2022, 15, 70 CrossRef CAS PubMed.
- L. Zhong, B. Li, C. S. Mah, L. Govindasamy, M. Agbandje-McKenna, M. Cooper, R. W. Herzog, I. Zolotukhin, K. H. Warrington Jr and K. A. Weigel-Van Aken, et al., Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 7827–7832 CrossRef CAS PubMed.
- M. Whitehead, A. Sage, T. Burgoyne, A. Osborne, P. Yu-Wai-Man and K. R. Martin, Immunobiology of a rationally-designed AAV2 capsid following intravitreal delivery in mice, Gene Ther., 2023, 30, 723–735 CrossRef CAS PubMed.
- A. Asokan, D. V. Schaffer and R. J. Samulski, The AAV vector toolkit: poised at the clinical crossroads, Mol. Ther., 2012, 20, 699–708 CrossRef CAS PubMed.
- H. Büning and A. Srivastava, Capsid Modifications for Targeting and Improving the Efficacy of AAV Vectors, Mol. Ther.–Methods Clin. Dev., 2019, 12, 248–265 CrossRef PubMed.
- E. J. Lee, C. M. Guenther and J. Suh, Adeno-Associated Virus (AAV) Vectors: Rational Design Strategies for Capsid Engineering, Curr. Opin. Biomed. Eng., 2018, 7, 58–63 CrossRef PubMed.
- M. A. Kotterman and D. V. Schaffer, Engineering adeno-associated viruses for clinical gene therapy, Nat. Rev. Genet., 2014, 15, 445–451 CrossRef CAS PubMed.
- J. Becker, J. Fakhiri and D. Grimm, Fantastic AAV Gene Therapy Vectors and How to Find Them-Random Diversification, Rational Design and Machine Learning, Pathogens, 2022, 11, 756 CrossRef CAS PubMed.
- H. J. Wagner, W. Weber and M. Fussenegger, Synthetic Biology: Emerging Concepts to Design and Advance Adeno-Associated Viral Vectors for Gene Therapy, Adv. Sci., 2021, 8, 2004018 CrossRef CAS PubMed.
- S. Zolotukhin and L. Vandenberghe, AAV capsid design: A Goldilocks challenge, Trends Mol. Med., 2022, 28, 183–193 CrossRef CAS PubMed.
- H. Büning, M. U. Ried, L. Perabo, F. M. Gerner, N. A. Huttner, J. Enssle and M. Hallek, Receptor targeting of adeno-associated virus vectors, Gene Ther., 2003, 10, 1142–1151 CrossRef PubMed.
- A. Girod, M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deléage and M. Hallek, Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2, Nat. Med., 1999, 5, 1438 CrossRef CAS PubMed.
- P. Wu, W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte and N. Muzyczka, Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism, J. Virol., 2000, 74, 8635–8647 CrossRef CAS PubMed.
- M. Grifman, M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini and M. D. Weitzman, Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids, Mol. Ther., 2001, 3, 964–975 CrossRef CAS PubMed.
- S. A. Nicklin, H. Buening, K. L. Dishart, M. de Alwis, A. Girod, U. Hacker, A. J. Thrasher, R. R. Ali, M. Hallek and A. H. Baker, Efficient and selective AAV2-mediated gene transfer directed to human vascular endothelial cells, Mol. Ther., 2001, 4, 174–181 CrossRef CAS PubMed.
- M. U. Ried, A. Girod, K. Leike, H. Büning and M. Hallek, Adeno-associated virus capsids displaying immunoglobulin-binding domains permit antibody-mediated vector retargeting to specific cell surface receptors, J. Virol., 2002, 76, 4559–4566 CrossRef CAS PubMed.
- A. M. Eichhoff, K. Börner, B. Albrecht, W. Schäfer, N. Baum, F. Haag, J. Körbelin, M. Trepel, I. Braren and D. Grimm, et al., Nanobody-Enhanced Targeting of AAV Gene Therapy Vectors, Mol. Ther.–Methods Clin. Dev., 2019, 15, 211–220 CrossRef CAS PubMed.
- R. C. Münch, H. Janicki, I. Völker, A. Rasbach, M. Hallek, H. Büning and C. J. Buchholz, Displaying High-affinity Ligands on Adeno-associated Viral Vectors Enables Tumor Cell-specific and Safe Gene Transfer, Mol. Ther., 2013, 21, 109–118 CrossRef PubMed.
- R. C. Münch, A. Muth, A. Muik, T. Friedel, J. Schmatz, B. Dreier, A. Trkola, A. Plückthun, H. Büning and C. J. Buchholz, Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors, Nat. Commun., 2015, 6, 6246 CrossRef PubMed.
- Q. Yang, M. Mamounas, G. Yu, S. Kennedy, B. Leaker, J. Merson, F. Wong-Staal, M. Yu and J. R. Barber, Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy, Hum. Gene Ther., 1998, 9, 1929–1937 CrossRef CAS PubMed.
- J. Hartmann, R. C. Münch, R. T. Freiling, I. C. Schneider, B. Dreier, W. Samukange, J. Koch, M. A. Seeger, A. Plückthun and C. J. Buchholz, A Library-Based Screening Strategy for the Identification of DARPins as Ligands for Receptor-Targeted AAV and Lentiviral Vectors, Mol. Ther.–Methods Clin. Dev., 2018, 10, 128–143 CrossRef CAS PubMed.
- S. A. Theuerkauf, E. Herrera-Carrillo, F. John, L. J. Zinser, M. A. Molina, V. Riechert, F. B. Thalheimer, K. Börner, D. Grimm and P. Chlanda, et al., AAV vectors displaying bispecific DARPins enable dual-control targeted gene delivery, Biomaterials, 2023, 303, 122399 CrossRef CAS PubMed.
- M. V. Hamann, N. Beschorner, X. K. Vu, I. Hauber, U. C. Lange, B. Traenkle, P. D. Kaiser, D. Foth, C. Schneider and H. Büning, et al., Improved targeting of human CD4+ T cells by nanobody-modified AAV2 gene therapy vectors, PLoS One, 2021, 16, e0261269 CrossRef CAS PubMed.
- A. Michels, A. M. Frank, D. M. Günther, M. Mataei, K. Börner, D. Grimm, J. Hartmann and C. J. Buchholz, Lentiviral and adeno-associated vectors efficiently transduce mouse T lymphocytes when targeted to murine CD8, Mol. Ther.–Methods Clin. Dev., 2021, 23, 334–347 CrossRef CAS PubMed.
- J. S. Bartlett, J. Kleinschmidt, R. C. Boucher and R. J. Samulski, Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab'gamma)2 antibody, Nat. Biotechnol., 1999, 17, 181–186 CrossRef CAS PubMed.
- J. Kuklik, S. Michelfelder, F. Schiele, S. Kreuz, T. Lamla, P. Müller and J. E. Park, Development of a Bispecific Antibody-Based Platform for Retargeting of Capsid Modified AAV Vectors, Int. J. Mol. Sci., 2021, 22, 8355 CrossRef CAS PubMed.
- A. C. Zdechlik, Y. He, E. J. Aird, W. R. Gordon and D. Schmidt, Programmable Assembly of Adeno-Associated Virus-Antibody Composites for Receptor-Mediated Gene Delivery, Bioconjugate Chem., 2020, 31, 1093–1106 CrossRef CAS PubMed.
- N. N. Thadani, J. Yang, B. Moyo, C. M. Lee, M. Y. Chen, G. Bao and J. Suh, Site-Specific Post-translational Surface Modification of Adeno-Associated Virus Vectors Using Leucine Zippers, ACS Synth. Biol., 2020, 9, 461–467 CrossRef CAS PubMed.
- A. Muik, J. Reul, T. Friedel, A. Muth, K. P. Hartmann, I. C. Schneider, R. C. Münch and C. J. Buchholz, Covalent coupling of high-affinity ligands to the surface of viral vector particles by protein trans-splicing mediates cell type-specific gene transfer, Biomaterials, 2017, 144, 84–94 CrossRef CAS PubMed.
- M. D. Stachler, I. Chen, A. Y. Ting and J. S. Bartlett, Site-specific Modification of AAV Vector Particles With Biophysical Probes and Targeting Ligands Using Biotin Ligase, Mol. Ther., 2008, 16, 1467–1473 CrossRef CAS PubMed.
- Y. Liu, Y. Fang, Y. Zhou, E. Zandi, C. L. Lee, K. I. Joo and P. Wang, Site-specific modification of adeno-associated viruses via a genetically engineered aldehyde tag, Small, 2013, 9, 421–429 CrossRef CAS PubMed.
- N. Maheshri, J. T. Koerber, B. K. Kaspar and D. V. Schaffer, Directed evolution of adeno-associated virus yields enhanced gene delivery vectors, Nat. Biotechnol., 2006, 24, 198–204 CrossRef CAS PubMed.
- R. Qian, B. Xiao, J. Li and X. Xiao, Directed Evolution of AAV Serotype 5 for Increased Hepatocyte Transduction and Retained Low Humoral Seroreactivity, Mol. Ther.–Methods Clin. Dev., 2021, 20, 122–132 CrossRef CAS PubMed.
- J. T. Koerber, N. Maheshri, B. K. Kaspar and D. V. Schaffer, Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles, Nat. Protoc., 2006, 1, 701–706 CrossRef CAS PubMed.
- W. Li, A. Asokan, Z. Wu, T. Van Dyke, N. DiPrimio, J. S. Johnson, L. Govindaswamy, M. Agbandje-McKenna, S. Leichtle and D. E. Redmond Jr, et al., Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles, Mol. Ther., 2008, 16, 1252–1260 CrossRef CAS PubMed.
- Y. B. Liu, B. C. Xu, Y. T. Chen, X. Yuan, J. Y. Liu, T. Liu, G. Z. Du, W. Jiang, Y. Yang and Y. Zhu, et al., Directed evolution of AAV accounting for long-term and enhanced transduction of cardiovascular endothelial cells in vivo, Mol. Ther.–Methods Clin. Dev., 2021, 22, 148–161 CrossRef CAS PubMed.
- D. Grimm, J. S. Lee, L. Wang, T. Desai, B. Akache, T. A. Storm and M. A. Kay, In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses, J. Virol., 2008, 82, 5887–5911 CrossRef CAS PubMed.
- A. K. Herrmann, C. Bender, E. Kienle, S. Grosse, J. El Andari, J. Botta, N. Schürmann, E. Wiedtke, D. Niopek and D. Grimm, A Robust and All-Inclusive Pipeline for Shuffling of Adeno-Associated Viruses, ACS Synth. Biol., 2019, 8, 194–206 CrossRef CAS PubMed.
- L. Perabo, H. Büning, D. M. Kofler, M. U. Ried, A. Girod, C. M. Wendtner, J. Enssle and M. Hallek, In vitro selection of viral vectors with modified tropism: the adeno-associated virus display, Mol. Ther., 2003, 8, 151–157 CrossRef CAS PubMed.
- K. Börner, E. Kienle, L. Y. Huang, J. Weinmann, A. Sacher, P. Bayer, C. Stüllein, J. Fakhiri, L. Zimmermann and A. Westhaus, et al., Pre-arrayed Pan-AAV Peptide Display Libraries for Rapid Single-Round Screening, Mol. Ther., 2020, 28, 1016–1032 CrossRef PubMed.
- J. Körbelin, G. Dogbevia, S. Michelfelder, D. A. Ridder, A. Hunger, J. Wenzel, H. Seismann, M. Lampe, J. Bannach and M. Pasparakis, et al., A brain microvasculature endothelial cell-specific viral vector with the potential to treat neurovascular and neurological diseases, EMBO Mol. Med., 2016, 8, 609–625 CrossRef PubMed.
- K. Varadi, S. Michelfelder, T. Korff, M. Hecker, M. Trepel, H. A. Katus, J. A. Kleinschmidt and O. J. Müller, Novel random peptide libraries displayed on AAV serotype 9 for selection of endothelial cell-directed gene transfer vectors, Gene Ther., 2012, 19, 800–809 CrossRef CAS PubMed.
- B. E. Deverman, P. L. Pravdo, B. P. Simpson, S. R. Kumar, K. Y. Chan, A. Banerjee, W. L. Wu, B. Yang, N. Huber and S. P. Pasca, et al., Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain, Nat. Biotechnol., 2016, 34, 204–209 CrossRef CAS PubMed.
- R. Lin, Y. Zhou, T. Yan, R. Wang, H. Li, Z. Wu, X. Zhang, X. Zhou, F. Zhao and L. Zhang, et al., Directed evolution of adeno-associated virus for efficient gene delivery to microglia, Nat. Methods, 2022, 19, 976–985 CrossRef CAS PubMed.
- S. R. Kumar, T. F. Miles, X. Chen, D. Brown, T. Dobreva, Q. Huang, X. Ding, Y. Luo, P. H. Einarsson and A. Greenbaum, et al., Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types, Nat. Methods, 2020, 17, 541–550 CrossRef PubMed.
- M. Tabebordbar, K. A. Lagerborg, A. Stanton, E. M. King, S. Ye, L. Tellez, A. Krunnfusz, S. Tavakoli, J. J. Widrick and K. A. Messemer, et al., Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species, Cell, 2021, 184, 4919–4938 CrossRef CAS PubMed.
- M. Davidsson, G. Wang, P. Aldrin-Kirk, T. Cardoso, S. Nolbrant, M. Hartnor, J. Mudannayake, M. Parmar and T. Björklund, A systematic capsid evolution approach performed in vivo for the design of AAV vectors with tailored properties and tropism, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 27053–27062 CrossRef CAS PubMed.
- P. J. Ogden, E. D. Kelsic, S. Sinai and G. M. Church, Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design, Science, 2019, 366, 1139–1143 CrossRef CAS PubMed.
- D. H. Bryant, A. Bashir, S. Sinai, N. K. Jain, P. J. Ogden, P. F. Riley, G. M. Church, L. J. Colwell and E. D. Kelsic, Deep diversification of an AAV capsid protein by machine learning, Nat. Biotechnol., 2021, 39, 691–696 CrossRef CAS PubMed.
- E. Zinn, S. Pacouret, V. Khaychuk, H. T. Turunen, L. S. Carvalho, E. Andres-Mateos, S. Shah, R. Shelke, A. C. Maurer and E. Plovie, et al., In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector, Cell Rep., 2015, 12, 1056–1068 CrossRef CAS PubMed.
- E. Zinn, C. Unzu, P. F. Schmit, H. T. Turunen, N. Zabaleta, J. Sanmiguel, A. Fieldsend, U. Bhatt, C. Diop and E. Merkel, et al., Ancestral library identifies conserved reprogrammable liver motif on AAV capsid, Cell Rep. Med., 2022, 3, 100803 CrossRef CAS PubMed.
- L. V. Tse, K. A. Klinc, V. J. Madigan, R. M. Castellanos Rivera, L. F. Wells, L. P. Havlik, J. K. Smith, M. Agbandje-McKenna and A. Asokan, Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E4812–E4821 CrossRef CAS PubMed.
- W. A. Nyberg, J. Ark, A. To, S. Clouden, G. Reeder, J. J. Muldoon, J. Y. Chung, W. H. Xie, V. Allain and Z. Steinhart, et al., An evolved AAV variant enables efficient genetic engineering of murine T cells, Cell, 2023, 186, 446–460 CrossRef CAS PubMed.
- J. S. Bartlett and R. J. Samulski, Fluorescent viral vectors: a new technique for the pharmacological analysis of gene therapy, Nat. Med., 1998, 4, 635–637 CrossRef CAS PubMed.
- J. S. Bartlett, R. J. Samulski and T. J. McCown, Selective and rapid uptake of adeno-associated virus type 2 in brain, Hum. Gene Ther., 1998, 9, 1181–1186 CrossRef CAS PubMed.
- S. Sanlioglu, P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds and J. F. Engelhardt, Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation, J. Virol., 2000, 74, 9184–9196 CrossRef CAS PubMed.
- G. Seisenberger, M. U. Ried, T. Endress, H. Büning, M. Hallek and C. Bräuchle, Real-time single-molecule imaging of the infection pathway of an adeno-associated virus, Science, 2001, 294, 1929–1932 CrossRef CAS PubMed.
- W. Xiao, K. H. Warrington Jr, P. Hearing, J. Hughes and N. Muzyczka, Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2, J. Virol., 2002, 76, 11505–11517 CrossRef CAS PubMed.
- K. I. Joo, Y. Fang, Y. Liu, L. Xiao, Z. Gu, A. Tai, C. L. Lee, Y. Tang and P. Wang, Enhanced real-time monitoring of adeno-associated virus trafficking by virus-quantum dot conjugates, ACS Nano, 2011, 5, 3523–3535 CrossRef CAS PubMed.
- P. Kothari, B. P. De, B. He, A. Chen, M. J. Chiuchiolo, D. Kim, A. Nikolopoulou, A. Amor-Coarasa, J. P. Dyke and H. U. Voss, et al., Radioiodinated Capsids Facilitate In Vivo Non-Invasive Tracking of Adeno-Associated Gene Transfer Vectors, Sci. Rep., 2017, 7, 39594 CrossRef CAS PubMed.
- J. W. Seo, E. S. Ingham, L. Mahakian, S. Tumbale, B. Wu, S. Aghevlian, S. Shams, M. Baikoghli, P. Jain and X. Ding, et al., Positron emission tomography imaging of novel AAV capsids maps rapid brain accumulation, Nat. Commun., 2020, 11, 2102 CrossRef CAS PubMed.
- J. Zengel, Y. X. Wang, J. W. Seo, K. Ning, J. N. Hamilton, B. Wu, M. Raie, C. Holbrook, S. Su and D. R. Clements, et al., Hardwiring tissue-specific AAV transduction in mice through engineered receptor expression, Nat. Methods, 2023, 20, 1070–1081 CrossRef CAS PubMed.
- G. K. Lee, N. Maheshri, B. Kaspar and D. V. Schaffer, PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization, Biotechnol. Bioeng., 2005, 92, 24–34 CrossRef CAS PubMed.
- Z. Yuan, B. Li, W. Gu, S. Luozhong, R. Li and S. Jiang, Mitigating the Immunogenicity of AAV-Mediated Gene Therapy with an Immunosuppressive Phosphoserine-Containing Zwitterionic Peptide, J. Am. Chem. Soc., 2022, 144, 20507–20513 CrossRef CAS PubMed.
- C. A. Hong, S. K. Cho, J. A. Edson, J. Kim, D. Ingato, B. Pham, A. Chuang, D. A. Fruman and Y. J. Kwon, Viral/Nonviral Chimeric Nanoparticles To Synergistically Suppress Leukemia Proliferation via Simultaneous Gene Transduction and Silencing, ACS Nano, 2016, 10, 8705–8714 CrossRef CAS PubMed.
- M. Mével, M. Bouzelha, A. Leray, S. Pacouret, M. Guilbaud, M. Penaud-Budloo, D. Alvarez-Dorta, L. Dubreil, S. G. Gouin and J. P. Combal, et al., Chemical modification of the adeno-associated virus capsid to improve gene delivery, Chem. Sci., 2019, 11, 1122–1131 RSC.
- S. Lee and H. J. Ahn, Anti-EpCAM-conjugated adeno-associated virus serotype 2 for systemic delivery of EGFR shRNA: Its retargeting and antitumor effects on OVCAR3 ovarian cancer in vivo, Acta Biomater., 2019, 91, 258–269 CrossRef CAS PubMed.
- E. D. Horowitz, M. S. Weinberg and A. Asokan, Glycated AAV vectors: chemical redirection of viral tissue tropism, Bioconjugate Chem., 2011, 22, 529–532 CrossRef CAS PubMed.
- H. A. Pearce, H. Qian, T. U. Connell, D. Huang, C. Gottstein, P. S. Donnelly, K. Peter, P. Gregorevic and C. E. Hagemeyer, Site-Specific Glycation and Chemo-enzymatic Antibody Sortagging for the Retargeting of rAAV6 to Inflamed Endothelium, Mol. Ther.–Methods Clin. Dev., 2019, 14, 261–269 CrossRef CAS PubMed.
- E. D. Horowitz, M. G. Finn and A. Asokan, Tyrosine cross-linking reveals interfacial dynamics in adeno-associated viral capsids during infection, ACS Chem. Biol., 2012, 7, 1059–1066 CrossRef CAS PubMed.
- D. Alvarez-Dorta, C. Thobie-Gautier, M. Croyal, M. Bouzelha, M. Mével, D. Deniaud, M. Boujtita and S. G. Gouin, Electrochemically Promoted Tyrosine-Click-Chemistry for Protein Labeling, J. Am. Chem. Soc., 2018, 140, 17120–17126 CrossRef CAS PubMed.
- S. Depienne, M. Bouzelha, E. Courtois, K. Pavageau, P. A. Lalys, M. Marchand, D. Alvarez-Dorta, S. Nedellec, L. Marín-Fernández and C. Grandjean, et al., Click-electrochemistry for the rapid labeling of virus, bacteria and cell surfaces, Nat. Commun., 2023, 14, 5122 CrossRef CAS PubMed.
- A. Leray, P. A. Lalys, J. Varin, M. Bouzelha, A. Bourdon, D. Alvarez-Dorta, K. Pavageau, S. Depienne, M. Marchand and A. Mellet, et al., Novel chemical tyrosine functionalization of adeno-associated virus improves gene transfer efficiency in liver and retina, Biomed. Pharmacother., 2024, 171, 116148 CrossRef CAS PubMed.
- J. S. Chandran, P. S. Sharp, E. Karyka, J. Aves-Cruzeiro, I. Coldicott, L. Castelli, G. Hautbergue, M. O. Collins and M. Azzouz, Site Specific Modification of Adeno-Associated Virus Enables Both Fluorescent Imaging of Viral Particles and Characterization of the Capsid Interactome, Sci. Rep., 2017, 7, 14766 CrossRef PubMed.
- F. Wei, K. I. McConnell, T. K. Yu and J. Suh, Conjugation of paclitaxel on adeno-associated virus (AAV) nanoparticles for co-delivery of genes and drugs, Eur. J. Pharm. Sci., 2012, 46, 167–172 CrossRef CAS PubMed.
- S. Ponnazhagan, G. Mahendra, S. Kumar, J. A. Thompson and M. Castillas Jr, Conjugate-based targeting of recombinant adeno-associated virus type 2 vectors by using avidin-linked ligands, J. Virol., 2002, 76, 12900–12907 CrossRef CAS PubMed.
- J. Zhu, P. Tao, M. Mahalingam, J. Sha, P. Kilgore, A. K. Chopra and V. Rao, A prokaryotic-eukaryotic hybrid viral vector for delivery of large cargos of genes and proteins into human cells, Sci. Adv., 2019, 5, eaax0064 CrossRef CAS PubMed.
- S. Yoo, B. Kang, S. Oh, Y. Kim and J. H. Jang, A Versatile Adeno-Associated Viral Vector Cross-Linking Platform Capable of Tuning Cellular Tropisms and Simultaneously Inducing Solid-Phase Gene Delivery, ACS Appl. Bio Mater., 2020, 3, 4847–4857 CrossRef CAS PubMed.
- P. L. Mulcrone, A. K. Lam, D. Frabutt, J. Zhang, M. Chrzanowski, R. W. Herzog and W. Xiao, Chemical modification of AAV9 capsid with N-ethyl maleimide alters vector tissue tropism, Sci. Rep., 2023, 13, 8436 CrossRef CAS PubMed.
- J. W. Chin, Expanding and reprogramming the genetic code, Nature, 2017, 550, 53–60 CrossRef CAS PubMed.
- J. S. Italia, Y. Zheng, R. E. Kelemen, S. B. Erickson, P. S. Addy and A. Chatterjee, Expanding the genetic code of mammalian cells, Biochem. Soc. Trans., 2017, 45, 555–562 CrossRef CAS PubMed.
- D. D. Young and P. G. Schultz, Playing with the molecules of life, ACS Chem. Biol., 2018, 13, 854–870 CrossRef CAS PubMed.
- R. E. Kelemen, S. B. Erickson and A. Chatterjee, Synthesis at the interface of virology and genetic code expansion, Curr. Opin. Chem. Biol., 2018, 46, 164–171 CrossRef CAS PubMed.
- R. E. Kelemen, S. B. Erickson and A. Chatterjee, Production and Chemoselective Modification of Adeno-Associated Virus Site-Specifically Incorporating an Unnatural Amino Acid Residue into Its Capsid, Methods Mol. Biol., 2018, 1728, 313–326 CrossRef CAS PubMed.
- R. E. Kelemen, R. Mukherjee, X. Cao, S. B. Erickson, Y. Zheng and A. Chatterjee, A Precise Chemical Strategy To Alter the Receptor Specificity of the Adeno-Associated Virus, Angew. Chem., Int. Ed., 2016, 55, 10645–10649 CrossRef CAS PubMed.
- C. Zhang, T. Yao, Y. Zheng, Z. Li, Q. Zhang, L. Zhang and D. Zhou, Development of next generation adeno-associated viral vectors capable of selective tropism and efficient gene delivery, Biomaterials, 2016, 80, 134–145 CrossRef CAS PubMed.
- S. B. Erickson, Q. Pham, X. Cao, J. Glicksman, R. E. Kelemen, S. S. Shahraeini, S. Bodkin, Z. Kiyam and A. Chatterjee, Precise Manipulation of the Site and Stoichiometry of Capsid Modification Enables Optimization of Functional Adeno-Associated Virus Conjugates, Bioconjugate Chem., 2024, 35, 64–71 CrossRef CAS PubMed.
- F. Puzzo, C. Zhang, B. P. Gray, F. Zhang, B. A. Sullenger and M. A. Kay, Aptamer-programmable adeno-associated viral vectors as a novel platform for cell-specific gene transfer, Mol. Ther.–Nucleic Acids, 2023, 31, 383–397 CrossRef CAS PubMed.
- H. Chang, A. Du, J. Jiang, L. Ren, N. Liu, X. Zhou, J. Liang, G. Gao and D. Wang, Non-canonical amino acid incorporation into AAV5 capsid enhances lung transduction in mice, Mol. Ther.–Methods Clin. Dev., 2023, 31, 101129 CrossRef CAS PubMed.
- T. Yao, X. Zhou, C. Zhang, X. Yu, Z. Tian, L. Zhang and D. Zhou, Site-Specific PEGylated Adeno-Associated Viruses with Increased Serum Stability and Reduced Immunogenicity, Molecules, 2017, 22, 1155 CrossRef PubMed.
- D. Katrekar, A. M. Moreno, G. Chen, A. Worlikar and P. Mali, Oligonucleotide conjugated multi-functional adeno-associated viruses, Sci. Rep., 2018, 8, 3589 CrossRef PubMed.
- S. B. Erickson, R. Mukherjee, R. E. Kelemen, C. J. Wrobel, X. Cao and A. Chatterjee, Precise Photoremovable Perturbation of a Virus-Host Interaction, Angew. Chem., Int. Ed., 2017, 56, 4234–4237 CrossRef CAS PubMed.
- C. Zhang, X. Zhou, T. Yao, Z. Tian and D. Zhou, Precision Fluorescent Labeling of an Adeno-Associated Virus Vector to Monitor the Viral Infection Pathway, Biotechnol. J., 2018, 13, e1700374 CrossRef PubMed.
- D. Jewel, R. E. Kelemen, R. L. Huang, Z. Zhu, B. Sundaresh, X. Cao, K. Malley, Z. Huang, M. Pasha and J. Anthony, et al., Virus-assisted directed evolution of enhanced suppressor tRNAs in mammalian cells, Nat. Methods, 2023, 20, 95–103 CrossRef CAS PubMed.
- D. Jewel, Q. Pham and A. Chatterjee, Virus-assisted directed evolution of biomolecules, Curr. Opin. Chem. Biol., 2023, 76, 102375 CrossRef CAS PubMed.
- D. Jewel, R. E. Kelemen, R. L. Huang, Z. Zhu, B. Sundaresh, K. Malley, Q. Pham, C. Loynd, Z. Huang and T. van Opijnen, et al., Enhanced Directed Evolution in Mammalian Cells Yields a Hyperefficient Pyrrolysyl tRNA for Noncanonical Amino Acid Mutagenesis, Angew. Chem., Int. Ed., 2024, 63, e202316428 CrossRef CAS PubMed.
- R. L. Huang, D. Jewel, R. E. Kelemen, Q. Pham, S. Wang, S. J. S. Roy, Z. Huang, S. D. Levinson, B. Sundaresh, S. E. Miranda, et al., Directed evolution of a bacterial leucyl tRNA in mammalian cells for enhanced noncanonical amino acid mutagenesis, ACS Synth. Biol., 2024, DOI:10.1021/acssynbio.4c00196.
| This journal is © The Royal Society of Chemistry 2024 |