DOI:
10.1039/D3NJ00426K
(Paper)
New J. Chem., 2023,
47, 10016-10024
AIE active cyanostilbenes for live-cell imaging of lipid droplets†
Received
6th February 2023
, Accepted 3rd April 2023
First published on 4th April 2023
Abstract
Lipid droplets (LDs), owing to their involvement in various physiological processes and diseases such as fatty liver disease, diabetes, and cancer, have attracted considerable attention. Live cell imaging of lipid droplets allows researchers to visualize and track the movement of these organelles within living cells in real time. In this study, we synthesized trifluoromethyl-substituted cyanostilbene fluorophores containing naphthalene or julolidine groups to visualize lipid droplets in live cells and investigate the changes in the size and number of LDs upon oleic acid stimulation. The fluorophores with donor (D) and acceptor (A) substituents show strong solvatochromism due to intramolecular charge transfer (ICT). In addition, these fluorophores show enhanced aggregation-induced emission (AIE) in water, and the lipophilic trifluoromethyl substitution aids the formation of stable organogels. The morphology and size of the aggregates obtained were characterized through scanning electron microscopy and dynamic light scattering experiments. Rheology and differential scanning calorimetry were used to further characterize the organogels. Our findings suggest that these D–π–A-substituted cyanostilbenes are valuable probes for understanding live-cell LD dynamics.
Introduction
Cyanostilbene derivatives that show characteristic aggregation-induced emission have been investigated extensively as functional materials for various applications in sensing, displays, and bioimaging.1–7 The restrictions on molecular rotation imposed by the presence of the nitrile group on the double bond and additional substituents can yield significant changes in either the emission intensity or wavelength. Considering the uniqueness of the scaffold, many cyanostilbene derivatives with several donor or acceptor substituents have been developed. One substituent that has attracted attention is the trifluoromethyl (CF3) group. Trifluoromethyl groups are important in various areas of chemistry and biology and are used as bioisosteres in medicinal chemistry to create potent substrates with improved bioavailability and specificity.8 These groups are known for their strong electron-withdrawing properties, high polarity, high dipole moment and high electric permittivity, and can therefore affect the steric and electronic properties, making them useful in drug design and organic synthesis.9 Considering the unique properties that fluorine substitution can offer, Park and co-workers have developed organogels with CF3 substitution in place of traditional gelator moieties such as hydrocarbon chain, glycol or amide units, along with their aggregation induced emission properties.10–13 Our previous work has also explored the utility of the CF3 group for the formation of various low molecular weight organogels using various aromatic scaffolds such as pyrene, anthracene and triphenylamine.14–17 In addition, trifluoromethyl groups can increase the lipophilicity of molecules, which can improve their ability to penetrate cell membranes and target certain proteins. Thus, we intended (i) to utilize the unique aggregation-induced emission behaviour of cyanostilbene for its utility in biological imaging and (ii) demonstrate the formation of stable organogels. Previously, several derivatives of α-cyanostilbenes have been utilized for cellular imaging. For example, a folic acid conjugate of an unsymmetrical cyanostilbene was utilized for its internalization in a cancer cell line,18 and α-cyanostilbenes bearing alkylammonium pendants were used for hydroxyl radical bioimaging.19 By incorporating a mitochondrial-targeting pyridinium group in the cyanostilbene scaffold, mitochondrial uptake and imaging were demonstrated.20–22 Cellular internalization was also demonstrated by pyrene- or triphenylamine-containing cyanostilbenes with distribution throughout the cytoplasmic region,23 monitoring the morphological changes of the plasma membrane,24 and binding to the mitochondria in zebrafish eggs.25 A pyridine-containing acrylonitrile derivative with coumarin substitution was also shown for the imaging the lipid droplets.26 Recently, Niu and co-workers synthesized various acrylonitrile derivatives to image LDs.24,27 As demonstrated by their results, the lipophilic properties of the CF3-containing cyanostilbenes can be well-suited to the imaging of lipid droplets. LDs are small, spherical droplets of fats and other lipids that are found in cells, and they are composed of a core of neutral lipids, such as triacylglycerols and cholesteryl esters and surrounded by a phospholipid monolayer. LDs play an important role in energy storage and metabolism and are also involved in the regulation of gene expression and cell signaling.28 Therefore, various research groups have explored the imaging of lipid droplets and underlying phenomena using small-molecule fluorescent probes.26,29–35 Utilizing the lipophilic properties of the trifluoromethyl group, and through the incorporation of strong electron-donating groups such as dimethylamino naphthalene or julolidine, we investigated the photophysical properties, organogel formation and utility of cyanostilbene fluorophores for the imaging of lipid-rich organelles. We detail our results below.
Materials and methods
All the necessary chemicals required for the synthesis of the probes were purchased from Aldrich, BLD Pharm, TCI chemicals, Avra synthesis, or SD Fine-chem. The synthesized samples were characterized using 1H and 13C NMR (Bruker Advance III-500 MHz) in chloroform-D or dimethyl sulphoxide (D6) solvents. Absorption studies were carried out using a Shimadzu UV-Vis 3600i instrument, and steady-state and fluorescence studies were carried out using a Horiba Jobin Yvon Fluorolog-3 spectrofluorimeter. The sample concentrations used for the absorption and fluorescence studies were of the order of 10 μM. The excitation wavelength for recording the emission spectra was typically kept at the absorption maxima of the samples under investigation. Mass spectral data were recorded using a Waters Synapt G2S ESI Q-ToF mass spectrometer. DLS measurements were performed using a Malvern Zetasizer Ultra instrument. The samples were prepared in water and filtered through a micro-filter; air bubbles were also removed before measurements. The organogel was lyophilized and vacuum dried before SEM measurements. Drop cast (10 μM of the sample) SEM images of the aggregate were obtained using a silicon wafer. SEM measurements were performed using an advanced analytical FE-SEM instrument (JEOL JSM-7900F). The rheology measurements were carried out using an Anton Paar (MCR 702) rheometer. TGA-DLS thermal analysis was performed using a Jupiter thermal analyser.
Cell culture and live-cell imaging
The COS-7 cells (monkey kidney fibroblast; ATCC) were grown in cell media containing DMEM (Dulbecco's Modified Eagle Medium; Gibco) supplemented with 10% FBS (fetal bovine serum; Gibco). The cells were maintained in a humidified environment at 37 °C with 5% CO2. For live-cell imaging, a cell density of 1.5 × 105 cells was seeded on a 35 mm glass-bottom dish 24 h before the dye treatment. The colocalization experiment was performed with the addition of 2 μM of the dye (N-1, N-2, J-1, and J-2, respectively) in FluoroBrite™ DMEM, followed by the addition of 50 nM of Nile red to stain the lipid droplets of the COS-7 cells, with subsequent incubation for a period of 10 minutes. The cells were then imaged using a 63× oil immersion objective lens (1.40 numerical aperture) of a Leica laser scanning confocal microscope (TCS SP8) in a live cell chamber at 37 °C with 97% humidity and 5% CO2. The Nile red dye was excited using a 561 nm laser, and the emission signal was collected using a HyD detector. For the synthesized dyes, excitation was via a 488 nm laser, and again the HyD detector was used to the record emission signal.
LD accumulation experiments
COS-7 cells were seeded on a 35 mm glass-bottom dish for 24 h, and then treated with 100 μM oleic acid for 4 h. After incubation, the medium was removed, and the cells were washed three times with PBS. The LD accumulation experiment was performed with the addition of J-2 (2 μM) in FluoroBrite™ DMEM. For the analysis of changes in the number and size of the lipid droplets upon oleic acid treatment, 15 cells with and without oleic acid treatment were considered for these calculations.
Cytotoxicity measurements
The MTT assay was used for checking the cytotoxicity level of the dyes. A cell population of 5 × 103 cells per well was seeded in a 96-well plate in Dulbecco's modified Eagle medium (DMEM) complemented with 10% fetal bovine serum (FBS) with 5% CO2 in an incubator at 37 °C for 24 h. The cells were then treated with different concentrations of each synthesized dye, i.e., 0.25, 0.5, 1, 2 and 4 μM. The dye was incubated with the cells for 4 h. After incubation, the medium was removed from the wells, and 100 μl of MTT solution (0.5 mg mL−1) was added to each well and again incubated for 4 h, leading to the formation of formazan crystals due to the reduction of MTT by viable cells. Using a multichannel pipette, the MTT solution was then removed from the corners of the well without disturbing the purple-colored formazan crystals. DMSO (100 μL) was added to the wells to dissolve the formazan crystals. The plate was shaken for 10 min at RT to dissolve the crystals completely. The absorbance values corresponding to different concentrations were then recorded at 570 nm using a PerkinElmer multimode plate reader. Triplets of each concentration of the probe were prepared for MTT assay. The experiment was repeated twice, and the mean ± standard deviation of the percentage cell viability was plotted against the different concentrations. The percentage cell viability of the dye was calculated with respect to the negative control (cells in DMEM and 10% FBS medium).
Data analysis
ImageJ software (nih.gov) was used to analyze and quantify the live-cell images. For the calculation of the Pearson correlation coefficient (PCC) in the colocalization experiment, the coloc2 plugin of ImageJ was used. Origin 9.1 software was used to plot the data for the cell viability assay.
Results and discussion
Synthesis
The series of LD-CF3 probes containing trifluoromethyl substituents using the Knoevenagel condensation reaction between the corresponding aldehydes and trifluoromethyl phenylacetonitrile derivatives (Fig. 1). The dimethylamino naphthalene carboxaldehyde was synthesized as per earlier literature.36 The synthetic procedures with characterization and spectral details are provided in the ESI.†
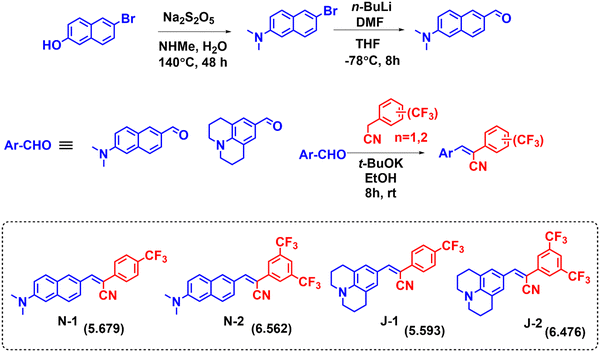 |
| | Fig. 1 Synthetic scheme for preparation of the LD-CF3 probes. (The c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values are given in parentheses.) P values are given in parentheses.) | |
Absorption and emission
The compounds were characterized by their strong electron-donating amino groups (dimethylamine for N1 and N2, and julolidine for J1 and J2) and electron-accepting nitrile and trifluoromethyl groups, which promote strong charge transfer. In addition, the nitrile group on the double bond is expected to aid in the unique aggregation induced emission (AIE) behaviour by increasing the rigidity of the molecule and reducing the degree of intramolecular rotation.2 Compound N1 absorbs at 423 nm in dioxane and N2 with an additional CF3 group absorbs slightly higher at 430 nm. Similarly, compound J1 with one CF3 group shows an absorption maximum at 434 nm, and J2 with two CF3 groups yields a moderate bathochromic shift. These bathochromic absorption shifts are attributed to the electron accepting nature of the CF3 group. The fused ring nature of the julolidine substituents (J1 and J2) also plays a role in electron donation to yield the bathochromic shifts in absorption. Compound J2 shows the highest absorption maximum, which is attributed to the increased planarity. Despite the presence of extended conjugation in N1 and N2, the julolidine-containing compounds J1 and J2 show a higher absorption, indicating their greater electron-donating nature. The order of absorption wavelength maxima for all the compounds is N1 < N2 < J1 < J2 (Table 1 and Fig. 2). In water, all the compounds show a broad absorption spectrum due to their weaker solubility and propensity to form aggregates. The solvent polarity has a weaker effect on the absorption maxima (Fig. 2).
Table 1 Absorption and emission properties of all the molecules in different solvents
| Solvent |
N-1
|
N-2
|
J-1
|
J-2
|
|
λ
abs (nm) |
λ
em (nm) |
ε* |
λ
abs (nm) |
λ
em (nm) |
ε*
|
λ
abs (nm) |
λ
em (nm) |
ε*
|
λ
abs (nm) |
λ
em (nm) |
ε*
|
| Dioxane |
423 |
528 |
6.1 |
430 |
546 |
5.3 |
434 |
493 |
5.3 |
441 |
508 |
5.5 |
| THF |
425 |
556 |
6.0 |
434 |
577 |
5.2 |
438 |
551 |
5.1 |
448 |
523 |
4.9 |
| CH3CN |
413 |
579 |
5.2 |
422 |
595 |
4.4 |
435 |
526 |
4.0 |
444 |
536 |
4.3 |
| DMF |
419 |
587 |
4.6 |
429 |
602 |
4.2 |
441 |
529 |
4.1 |
451 |
540 |
4.2 |
| DMSO |
420 |
593 |
4.9 |
427 |
609 |
2.8 |
445 |
532 |
3.7 |
452 |
543 |
3.6 |
| Ethanol |
411 |
564 |
5.0 |
422 |
587 |
4.3 |
428 |
510 |
3.8 |
446 |
524 |
4.4 |
| Methanol |
411 |
568 |
5.2 |
420 |
591 |
4.2 |
430 |
513 |
3.8 |
444 |
526 |
4.2 |
| Water |
413 |
591 |
3.6 |
426 |
614 |
3.2 |
428 |
566 |
2.4 |
441 |
593 |
3.0 |
| Solid state (λem) |
540 |
648 |
544 |
588 |
|
ε* = value × 104 mol−1 cm−1. |
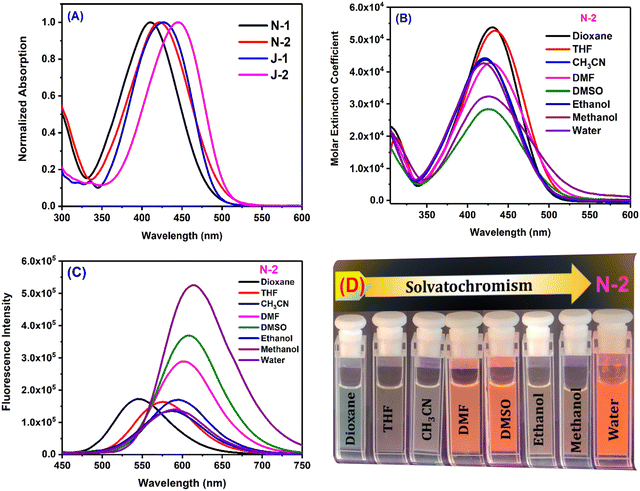 |
| | Fig. 2 (A) Absorption of all compounds in ethanol; (B) Absorption of N2 in all solvents; (C) Emission of N2 in all solvents; (D) Pictures of solutions for the emission of N2 in different solvents under UV lamp irradiation of 365 nm. [Concentration = 10 μM.]. (Data for the other compounds are given in Fig. S3, ESI.†) | |
However, the emission of the compounds show a strong solvatochromic shift with the solvent polarity. With a change in the solvent polarity from dioxane to DMSO, N1 shows a bathochromic shift in emission of 65 nm, N2 of 63 nm, J1 of 39 nm, and J2 of 35 nm. The additional CF3 moiety shows no major influence on the emission maxima. The naphthalene-substituted compounds N1 and N2 show a greater solvatochromic variation than the julolidine derivatives and also exhibit an intense color demarcation (Fig. 2 and Fig. S2, ESI†). The spectroscopic purity of all the compounds is shown by the perfect overlap between the absorption and excitation spectra (Fig. S1, ESI†). As can be seen in Table 1, all the compounds show significant emission wavelength shifts in water attributed to the aggregation induced emission2,37 coupled with solvent polarity effects. The emission spectra were also recorded in a dioxane–water binary mixture to learn about the emission behavior through the systematic variation of the solvent polarity. Dioxane is a good solvent in which molecules are readily soluble, whereas water is a poor solvent.
As the percentage of water in dioxane was increased, we noted prominent changes in both the emission wavelength and emission intensity for all the compounds at a water fraction of >60% (Fig. 3 and Fig. S4, ESI†). Along with the wavelength shifts, a concomitant intensity enhancement was also noted: N1 and J1 yield an intensity enhancement of ∼6–8-fold, and N2 and J2 (with the additional CF3 substituent) show an intensity enhancement of ∼16 fold and ∼20 fold, respectively, with distinct colors seen with different water fractions (inset of Fig. 3B and Fig. S4A–C, ESI†). The enhanced emission is attributed to the formation of the aggregates, and the aggregate morphology was studied via drop-casting of the aqueous solution on a Si wafer (Fig. 4A, B and Fig. S5a, ESI†). Irregular, spheroid, and rectangular shaped aggregates were obtained for all the compounds in aqueous medium (Fig. 4 and Fig. S5a, ESI†). The aggregate particle size was measuring using dynamic light scattering (DLS), which showed a mean hydrodynamic diameter of 397 nm for N1, 171.8 nm for N2, 152.4 nm for J1 and 143.1 nm for J2 (Fig. 4C, D and Fig. S5b, ESI†).
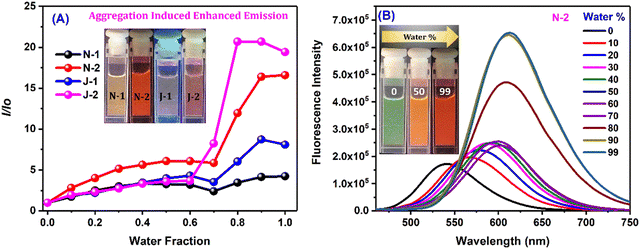 |
| | Fig. 3 (A) I/Io for all the derivatives corresponding to λmax of water vs. water fraction; and (B) emission spectra of N2 in the dioxane/water binary solvent system. [Concentration = 10 μM.] | |
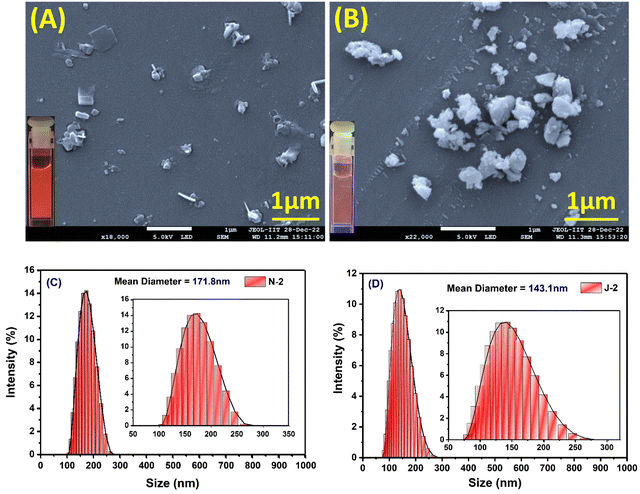 |
| | Fig. 4 Scanning electron microscope (SEM) images of 10 μM drop-cast solutions of (A) N2 and (B) J2. The samples were coated with gold for 60 seconds. [The FESEM images of drop-cast solutions of N1 and J1 are shown in Fig. S5, ESI.†] (C and D) MA-DLS spectrum depicting the size/mean hydrodynamic diameter of the aggregates of N2 and J2. [DLS spectra of N1 and J2 are shown in Fig. S4, ESI.†] [For each sample, a concentration of 10 μM was used.] | |
Low molecular weight organogels
Small molecule organogels are three-dimensional networks of self-assembled molecules that form a gel-like structure in organic solvents. Organogels based on small molecules have unique properties, such as high stability, ease of preparation, and tunable properties, making them useful for a variety of applications such as drug delivery, tissue engineering, imaging, tracking and sensing.38–40 It has been well established that CF3 groups and long-chain alkyl groups help to form organogels with higher stability through various non-covalent interactions, packing and bonding interactions.10,41 We obtained the formation of a stable organogel with compounds bearing two CF3 groups (N2 and J2) in ethanol upon heating, followed by sonication and cooling. The organogels were found to be stable for 24 h and converted to the sol state upon heating at 100 °C. The melting and thermal degradation temperatures for the xerogels were obtained via differential scanning calorimetry and were found to be 190–200 °C for both J2 and N2 (Fig. S7, ESI†), indicating their good stability at higher temperatures. The emission of the organogels shows a red shift compared with their solution state, with J2 having a higher shift (+69 nm) than N2 (+46 nm), with the order of solution state < AIE < gel state for N-2 and solution state < gel phase ≤ AIE phase for J2 (Fig. 5A and B). The viscoelastic properties of the organogels were also investigated using a rheometer. In the rheology experiments, G′ and G′′ refer to the storage and loss moduli, respectively. The storage modulus (G′) represents the elastic component of a material's response to an applied stress, while the loss modulus (G′′) represents the viscous component. The ratio of G′′ to G′ is known as the loss tangent (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ) and can be used to determine the degree of viscoelasticity of a material. The observed storage modulus for J2 and N2 was dominant over their respective viscosity modulus or loss modulus. This indicates that the material can return to its original shape more easily when the applied stress is removed. The moduli are recorded with respect to the angular frequency ω (rad s−1) as well as time (s), and the storage modulus for both was found to be dominant when monitored till ω = 100 rad s−1 and t = 1400 s, which indicates greater gel stability with respect to ω and time (Fig. 5C, D and Fig. S6, ESI†). After a time sweep of 800 s, N2 was found to lose its elasticity (Fig. S6B, ESI†) although J2 exhibited an increased elasticity owing to the additional stabilization offered due to the presence of the fused ring system of julolidine. The SEM images of xerogels of the compounds show fibrous and network-like structures of varying thickness, indicating the gel-formation (Fig. 6).
δ) and can be used to determine the degree of viscoelasticity of a material. The observed storage modulus for J2 and N2 was dominant over their respective viscosity modulus or loss modulus. This indicates that the material can return to its original shape more easily when the applied stress is removed. The moduli are recorded with respect to the angular frequency ω (rad s−1) as well as time (s), and the storage modulus for both was found to be dominant when monitored till ω = 100 rad s−1 and t = 1400 s, which indicates greater gel stability with respect to ω and time (Fig. 5C, D and Fig. S6, ESI†). After a time sweep of 800 s, N2 was found to lose its elasticity (Fig. S6B, ESI†) although J2 exhibited an increased elasticity owing to the additional stabilization offered due to the presence of the fused ring system of julolidine. The SEM images of xerogels of the compounds show fibrous and network-like structures of varying thickness, indicating the gel-formation (Fig. 6).
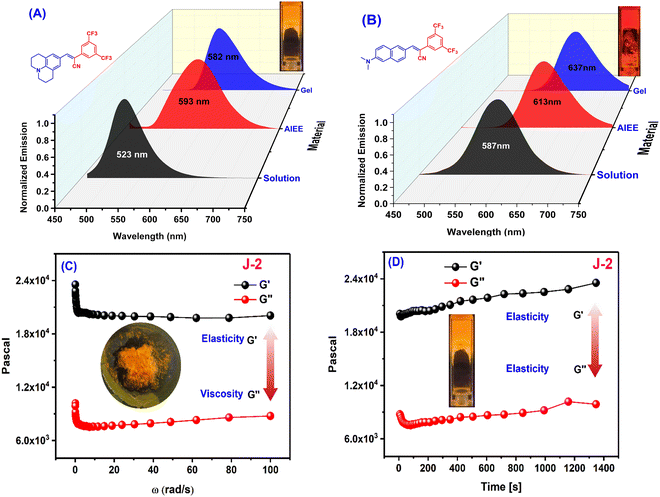 |
| | Fig. 5 Emission spectra of (A) J2 and (B) N2 in ethanol solution (10 μM), water solution (10 μM, AIE) and the gel state. (C and D) Frequency sweep (C) and time sweep (D) measurements for organogel J2. (Insets show gel pictures of J2 kept in the rheometer after applying pressure; a photo of gel in the cuvette. The frequency sweep and time sweep measurements for organogel N2 are given in Fig. S6, ESI.†) | |
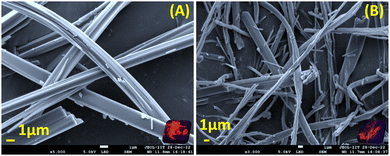 |
| | Fig. 6 Scanning electron microscope images of xerogels of (A) N-2 and (B) J-2 (where the insets show pictures of xerogels under UV lamp irradiation of 365 nm). The xerogels were obtained via lyophilization and vacuum drying. The samples were coated with gold for 60 seconds. | |
Live cell imaging of lipid droplets
With the strong solvatochromic behavior of the compounds, we extended the utility of the molecules for the imaging of lipid droplets. The synthesized compounds exhibit c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values ranging from 5.6 to 6.5, which is a measure of the compound's lipophilicity, i.e., its ability to dissolve in lipids. In the context of lipid droplet imaging, c
P values ranging from 5.6 to 6.5, which is a measure of the compound's lipophilicity, i.e., its ability to dissolve in lipids. In the context of lipid droplet imaging, c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P can be a useful descriptor for predicting a compound's ability to localize to lipid droplets. Compounds with c
P can be a useful descriptor for predicting a compound's ability to localize to lipid droplets. Compounds with c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values greater than ∼3 are considered to be lipophilic and tend to localize to lipid droplets.42 Therefore, given their favorable c
P values greater than ∼3 are considered to be lipophilic and tend to localize to lipid droplets.42 Therefore, given their favorable c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values, we investigated the potential of these fluorescent probes (N1, N2, J1, and J2) for the labelling of lipid droplets (LDs) in COS-7 cells. We found that when these cells were incubated with the molecules, they quickly entered the cell membrane and produced a distinct, bright, and uniform punctate like structures throughout the cytoplasm. The images of the cells both in the bright field and the confocal microscope are given in Fig. 7 and Fig. S8 (ESI†) and show structures that resemble lipid droplets. To confirm that these structures were indeed lipid droplets, we also stained the cells with Nile red,43 a known marker for lipid droplets, and found a significant overlap between the two labelling methods, indicating greater selectivity for the lipid droplets.
P values, we investigated the potential of these fluorescent probes (N1, N2, J1, and J2) for the labelling of lipid droplets (LDs) in COS-7 cells. We found that when these cells were incubated with the molecules, they quickly entered the cell membrane and produced a distinct, bright, and uniform punctate like structures throughout the cytoplasm. The images of the cells both in the bright field and the confocal microscope are given in Fig. 7 and Fig. S8 (ESI†) and show structures that resemble lipid droplets. To confirm that these structures were indeed lipid droplets, we also stained the cells with Nile red,43 a known marker for lipid droplets, and found a significant overlap between the two labelling methods, indicating greater selectivity for the lipid droplets.
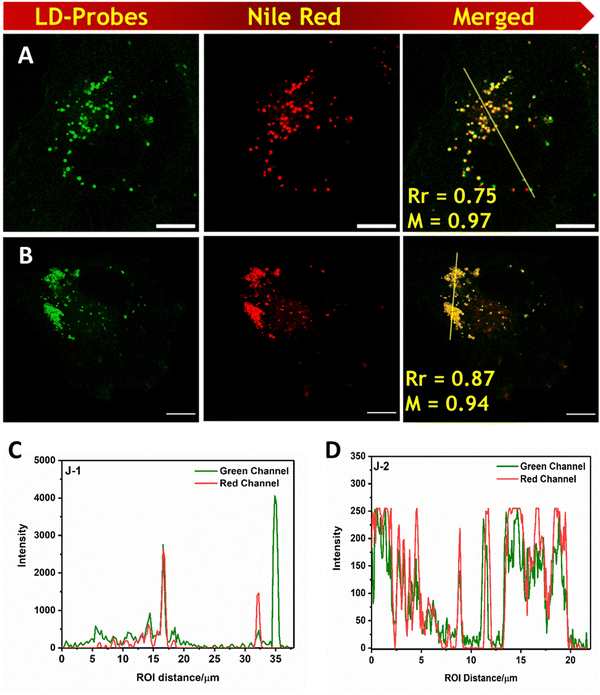 |
| | Fig. 7 A and B are cells stained with J1 and J2 respectively J1 and J2 in COS-7 cells (Column-1). Nile red was used as a lipid droplet marker and merged images are shown in column-3. Scale bar = 10 μm. C and D Intensity profile overlaps of for J1 and J2 with Nile red respectively. Bright-field images are shown in the ESI.† (λex = 488 nm, λem = 505–560 nm for all the molecules; and λex = 561 nm, λem = 570–679 nm for Nile red.) | |
Among the molecules investigated, J2 shows the highest PCC value indicating strong selectivity compared with the other fluorophores. We also examined the cytotoxicity via MTT assay of the probes at different concentrations for 4 h, and we did not find any adverse cellular behavior (Fig. S9, ESI†).
LD accumulation
Oleic acid (OA) stimulates the formation of lipid droplets in cells by activating certain signalling pathways, such as the FPAR pathway.44 To investigate lipid droplet accumulation, cells were treated with oleic acid (100 μM) for 4 h in cell culture media. Visualization of the cells revealed a significant increase in the number and size of lipid droplets. Fig. 8 clearly illustrates the increase in the number of lipid droplets as well as their size after the addition of OA. The mean diameter (size) of the lipid droplets increased from 261.93 nm to 581 nm upon treatment with oleic acid, and the number of lipid droplets per cell increased from n = 119 to n = 251. These results indicate that the probes can effectively label and visualize lipid droplets, even at low concentrations (2 μM) and with shorter incubation times (15 minutes).
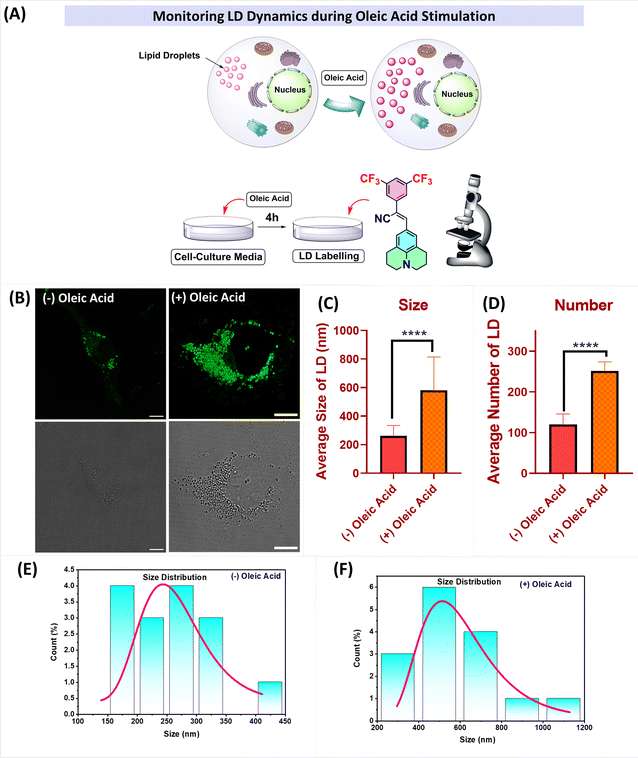 |
| | Fig. 8 (A) Pictorial representation of the changes observed after oleic acid stimulation; (B) (left) CLSM images of cells incubated with J-2 (2 μM), and (right) the images of oleic acid-treated COS-7 cells followed by incubation with J-2 (2 μM). (C and D) Plots representing the changes in LD dynamics with and without oleic acid stimulation: (C) size of LDs, and (D) number of LDs. (E and F) Size distribution histograms showing the sizes of the lipid droplets without (left) and with (right) oleic acid stimulation. | |
Conclusions
In summary, we have designed and synthesized four trifluoromethyl-substituted cyanostilbene fluorophores containing naphthalene or julolidine donor groups for effective visualization of lipid droplets in COS-7 cells. These molecules have proven to be good candidates for visualizing changes in lipid droplet number and size upon stimulation with oleic acid, due to their efficient colocalizing ability for lipid droplets. The dynamic visualization of these changes using these potential probes opens opportunities in areas such as detecting accumulated fats in tissues, fatty liver syndrome, and other lipid droplet-associated diseases. Furthermore, these fluorophores, which bear both donor and acceptor substituents, exhibit strong solvatochromism and excellent aggregation-induced emission in aqueous media. Additionally, we demonstrated the formation of organogels stabilized by intermolecular interactions using fluorophores bearing two trifluoromethyl substituents.
Author contributions
RD performed the overall investigation and methodology, including the synthesis, fluorescence studies, organogelation studies, analysis of the cellular images and developing the first draft of the manuscript. DS and DR performed cell biology experiments, including colocalization of the probes and cell viability assay. VS and SK supervised, planned, and conceived the idea and were involved in manuscript editing and writing.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors acknowledge SERB (CRG/2018/004020) for the financial support and IIT Gandhinagar for general research support. VS acknowledges the funding from DBT (Grant No. BT/PR15214/BRB/10/1449/2015 and BT/RLF/Re-entry/45/2015) and DST-SERB (Grant No. ECR/2016/000913).
References
- S. Lin, K. G. Gutierrez-Cuevas, X. Zhang, J. Guo and Q. Li, Adv. Funct. Mater., 2021, 31, 2007957 CrossRef CAS.
- P. Mahalingavelar and S. Kanvah, Phys. Chem. Chem. Phys., 2022, 24, 23049–23075 RSC.
- C. Hang, H.-W. Wu and L.-L. Zhu, Chin. Chem. Lett., 2016, 27, 1155–1165 CrossRef CAS.
- M. Martinez-Abadia, R. Gimenez and M. B. Ros, Adv. Mater., 2018, 30, 1704161 CrossRef PubMed.
- L. Zhu and Y. Zhao, J. Mater. Chem. C, 2013, 1, 1059–1065 RSC.
- M. Paramasivam and S. Kanvah, J. Phys. Chem. C, 2016, 120, 10757–10769 CrossRef CAS.
- S. Lin, K. G. Gutierrez-Cuevas, X. Zhang, J. Guo and Q. Li, Adv. Funct. Mater., 2021, 31, 2007957 CrossRef CAS.
- S. Debnath, R. Ghosh, R. R. Nair, D. Pradhan and P. B. Chatterjee, ACS Omega, 2022, 7, 38122–38149 CrossRef CAS PubMed.
- A. Abula, Z. Xu, Z. Zhu, C. Peng, Z. Chen, W. Zhu and H. A. Aisa, J. Chem. Inf. Model., 2020, 60, 6242–6250 CrossRef CAS PubMed.
- B.-K. An, D.-S. Lee, J.-S. Lee, Y.-S. Park, H.-S. Song and S. Y. Park, J. Am. Chem. Soc., 2004, 126, 10232–10233 CrossRef CAS PubMed.
- S. Park, J. E. Kwon, S.-Y. Park, O.-H. Kwon, J. K. Kim, S.-J. Yoon, J. W. Chung, D. R. Whang, S. K. Park, D. K. Lee, D.-J. Jang, J. Gierschner and S. Y. Park, Adv. Opt. Mater., 2017, 5, 1700353 CrossRef.
- J. Seo, J. W. Chung, E.-H. Jo and S. Y. Park, Chem. Commun., 2008, 2794–2796 RSC.
- J. W. Chung, Y. You, H. S. Huh, B.-K. An, S.-J. Yoon, S. H. Kim, S. W. Lee and S. Y. Park, J. Am. Chem. Soc., 2009, 131, 8163–8172 CrossRef CAS PubMed.
- J. Katla, A. Ojha, A. J. M. Nair, K. Rangan and S. Kanvah, New J. Chem., 2018, 42, 18297–18304 RSC.
- J. Katla, A. J. M. Nair, A. Ojha and S. Kanvah, Photochem. Photobiol. Sci., 2018, 17, 395–403 CrossRef CAS PubMed.
- B. Kumari, S. P. Singh, R. Santosh, A. Dutta, S. S. Mallajosyula, S. Ghosal and S. Kanvah, New J. Chem., 2019, 43, 4106–4115 RSC.
- B. Kumari, M. Paramasivam, T. Mukherjee, S. Khandelwal, A. Dutta and S. Kanvah, New J. Chem., 2021, 45, 4683–4693 RSC.
- A. K. Mandal, S. Sreejith, T. He, S. K. Maji, X.-J. Wang, S. L. Ong, J. Joseph, H. Sun and Y. Zhao, ACS Nano, 2015, 9, 4796–4805 CrossRef CAS PubMed.
- N. Y. Lim, J. Ahn, M. Won, W. Choi, J. S. Kim and J. H. Jung, ACS Appl. Bio Mater., 2019, 2, 936–942 CrossRef CAS PubMed.
- T. C. OwYong, S. Ding, N. Wu, T. Fellowes, S. Chen, J. M. White, W. W. H. Wong and Y. Hong, Chem. Commun., 2020, 56, 14853–14856 RSC.
- Y. Zhang, S. Wang, X. Wang, Q. Zan, X. Yu, L. Fan and C. Dong, Anal. Bioanal. Chem., 2021, 413, 3823–3831 CrossRef CAS PubMed.
- C. Y. Y. Yu, W. Zhang, R. T. K. Kwok, C. W. T. Leung, J. W. Y. Lam and B. Z. Tang, J. Mater. Chem. B, 2016, 4, 2614–2619 RSC.
- Z. Huang, F. Tang, F. He, L. Kong, J. Huang, J. Yang and A. Ding, Org. Chem. Front., 2022, 9, 5118–5124 RSC.
- G. Niu, R. Zhang, J. P. C. Kwong, J. W. Y. Lam, C. Chen, J. Wang, Y. Chen, X. Feng, R. T. K. Kwok, H. H. Y. Sung, I. D. Williams, M. R. J. Elsegood, J. Qu, C. Ma, K. S. Wong, X. Yu and B. Z. Tang, Chem. Mater., 2018, 30, 4778–4787 CrossRef CAS.
- A. Gopinath, K. Ramamurthy, M. Subaraja, C. Selvaraju and A. S. Nasar, New J. Chem., 2018, 42, 10243–10253 RSC.
- P. Jana, A. Siva, V. Soppina and S. Kanvah, Org. Biomol. Chem., 2020, 18, 5608–5616 RSC.
- X. Zhu, L. Feng, S. Cao, J. Wang and G. Niu, Org. Lett., 2022, 24, 8305–8309 CrossRef CAS PubMed.
- J. A. Olzmann and P. Carvalho, Nat. Rev. Mol. Cell Biol., 2019, 20, 137–155 CrossRef CAS PubMed.
- Y.-L. Qi, H.-R. Wang, L.-L. Chen, Y.-T. Duan, S.-Y. Yang and H.-L. Zhu, Chem. Soc. Rev., 2022, 51, 7752–7778 RSC.
- L. Wang, X. Chen, X. Ran, H. Tang and D. Cao, Dyes Pigm., 2022, 203, 110332 CrossRef CAS.
- H. Tian, A. C. Sedgwick, H.-H. Han, S. Sen, G.-R. Chen, Y. Zang, J. L. Sessler, T. D. James, J. Li and X.-P. He, Coord. Chem. Rev., 2021, 427, 213577 CrossRef CAS.
- T. K. Fam, A. S. Klymchenko and M. Collot, Materials, 2018, 11, 1768 CrossRef PubMed.
- Y. Zhao, W. Shi, X. Li and H. Ma, Chem. Commun., 2022, 58, 1495–1509 RSC.
- L. Fan, X. Wang, Q. Zan, L. Fan, F. Li, Y. Yang, C. Zhang, S. Shuang and C. Dong, Anal. Chem., 2021, 93, 8019–8026 CrossRef CAS PubMed.
- H. W. Lee, T. K. Pati, I.-J. Lee, J.-M. Lee, B. R. Kim, S. Y. Kwak and H. M. Kim, Anal. Chem., 2022, 94, 15100–15107 CrossRef CAS PubMed.
- B. Kumari, A. Yadav, S. P. Pany, P. I. Pradeepkumar and S. Kanvah, J. Photochem. Photobiol., B, 2019, 190, 128–136 CrossRef CAS PubMed.
- N. Yamamoto, J. Phys. Chem. C, 2018, 122, 12434–12440 CrossRef CAS.
- J. Li, W.-Y. Wong and X.-M. Tao, Nanoscale, 2020, 12, 1281–1306 RSC.
- A. Ajayaghosh and V. K. Praveen, Acc. Chem. Res., 2007, 40, 644–656 CrossRef CAS PubMed.
- C. L. Esposito, P. Kirilov and V. G. Roullin, J. Controlled Release, 2018, 271, 1–20 CrossRef CAS PubMed.
- R. Dahiwadkar, A. Murugan, D. Johnson, R. Chakraborty, V. Thiruvenkatam and S. Kanvah, J. Photochem. Photobiol., A, 2023, 434, 114227 CrossRef CAS.
- R. W. Horobin, F. Rashid-Doubell, J. D. Pediani and G. Milligan, Biotech. Histochem., 2013, 88, 440–460 CrossRef CAS PubMed.
- P. Greenspan, E. P. Mayer and S. D. Fowler, J. Cell Biol., 1985, 100, 965–973 CrossRef CAS PubMed.
- A. Rohwedder, Q. Zhang, S. A. Rudge and M. J. O. Wakelam, J. Cell Sci., 2014, 127, 3104–3115 CAS.
|
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2023 |
 *b and
Sriram
Kanvah
*b and
Sriram
Kanvah
 *a
*a

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values are given in parentheses.)
P values are given in parentheses.)


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ) and can be used to determine the degree of viscoelasticity of a material. The observed storage modulus for J2 and N2 was dominant over their respective viscosity modulus or loss modulus. This indicates that the material can return to its original shape more easily when the applied stress is removed. The moduli are recorded with respect to the angular frequency ω (rad s−1) as well as time (s), and the storage modulus for both was found to be dominant when monitored till ω = 100 rad s−1 and t = 1400 s, which indicates greater gel stability with respect to ω and time (Fig. 5C, D and Fig. S6, ESI†). After a time sweep of 800 s, N2 was found to lose its elasticity (Fig. S6B, ESI†) although J2 exhibited an increased elasticity owing to the additional stabilization offered due to the presence of the fused ring system of julolidine. The SEM images of xerogels of the compounds show fibrous and network-like structures of varying thickness, indicating the gel-formation (Fig. 6).
δ) and can be used to determine the degree of viscoelasticity of a material. The observed storage modulus for J2 and N2 was dominant over their respective viscosity modulus or loss modulus. This indicates that the material can return to its original shape more easily when the applied stress is removed. The moduli are recorded with respect to the angular frequency ω (rad s−1) as well as time (s), and the storage modulus for both was found to be dominant when monitored till ω = 100 rad s−1 and t = 1400 s, which indicates greater gel stability with respect to ω and time (Fig. 5C, D and Fig. S6, ESI†). After a time sweep of 800 s, N2 was found to lose its elasticity (Fig. S6B, ESI†) although J2 exhibited an increased elasticity owing to the additional stabilization offered due to the presence of the fused ring system of julolidine. The SEM images of xerogels of the compounds show fibrous and network-like structures of varying thickness, indicating the gel-formation (Fig. 6).

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values ranging from 5.6 to 6.5, which is a measure of the compound's lipophilicity, i.e., its ability to dissolve in lipids. In the context of lipid droplet imaging, c
P values ranging from 5.6 to 6.5, which is a measure of the compound's lipophilicity, i.e., its ability to dissolve in lipids. In the context of lipid droplet imaging, c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P can be a useful descriptor for predicting a compound's ability to localize to lipid droplets. Compounds with c
P can be a useful descriptor for predicting a compound's ability to localize to lipid droplets. Compounds with c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values greater than ∼3 are considered to be lipophilic and tend to localize to lipid droplets.42 Therefore, given their favorable c
P values greater than ∼3 are considered to be lipophilic and tend to localize to lipid droplets.42 Therefore, given their favorable c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values, we investigated the potential of these fluorescent probes (N1, N2, J1, and J2) for the labelling of lipid droplets (LDs) in COS-7 cells. We found that when these cells were incubated with the molecules, they quickly entered the cell membrane and produced a distinct, bright, and uniform punctate like structures throughout the cytoplasm. The images of the cells both in the bright field and the confocal microscope are given in Fig. 7 and Fig. S8 (ESI†) and show structures that resemble lipid droplets. To confirm that these structures were indeed lipid droplets, we also stained the cells with Nile red,43 a known marker for lipid droplets, and found a significant overlap between the two labelling methods, indicating greater selectivity for the lipid droplets.
P values, we investigated the potential of these fluorescent probes (N1, N2, J1, and J2) for the labelling of lipid droplets (LDs) in COS-7 cells. We found that when these cells were incubated with the molecules, they quickly entered the cell membrane and produced a distinct, bright, and uniform punctate like structures throughout the cytoplasm. The images of the cells both in the bright field and the confocal microscope are given in Fig. 7 and Fig. S8 (ESI†) and show structures that resemble lipid droplets. To confirm that these structures were indeed lipid droplets, we also stained the cells with Nile red,43 a known marker for lipid droplets, and found a significant overlap between the two labelling methods, indicating greater selectivity for the lipid droplets.



