Fluorescent 7-azaindole N-linked 1,2,3-triazole: synthesis and study of antimicrobial, molecular docking, ADME and DFT properties†
Kanika
Sharma
a,
Ram Kumar
Tittal
 *a,
Kashmiri
Lal
*a,
Kashmiri
Lal
 b,
Ramling S.
Mathpati
a and
Ghule Vikas
D.
b,
Ramling S.
Mathpati
a and
Ghule Vikas
D.
 a
a
aDepartment of Chemistry, National Institute of Technology Kurukshetra, Haryana 136119, India. E-mail: rktittaliitd@nitkkr.ac.in
bDepartment of Chemistry, GJUS&T Hisar, Haryana 12500, India
First published on 4th April 2023
Abstract
Two libraries of biologically essential and fluorescent 7-azaindole N-linked benzyl 1,2,3-triazole (4a–4i) and 7-azaindole N-linked phenyl 1,2,3-triazole (4j–4q) hybrids have been designed and synthesized from N-propargyl 7-azaindole, i.e., 1-prop-2-ynyl-1H-pyrrolo[2,3-b]pyridine (2) with benzyl azides (3a–3i) and phenyl azides (3j–3q), respectively, via a Cu(I)-catalyzed alkyne azide cycloaddition (CuAAC) click reaction. The structures of the synthesized hybrids were confirmed with the help of the IR, 1H-NMR, 13C-NMR, and HRMS data. However, the structure of one of the solid crystalline compounds, 4f, was finally supported by its single-crystal X-ray data. In vitro, antimicrobial activity results revealed that most compounds exhibited superior efficacy towards Aspergillus compared to fluconazole. Compounds 4e, 4f, and 4g showed high potency against A. niger with MIC values of 0.0193, 0.0170, and 0.0187 μmol mL−1, respectively. Compound 4f also showed increased activity against P. aeruginosa and E. coli (MIC: 0.0170 μmol mL−1). The most promising compounds, 4d, 4f, and 4n, showed excellent binding energies of −8.81, −8.76, and −8.79 kcal mol−1 with a dynamic DNA site 14α-demethylase lanosterol enzyme (PDB ID:1EA1). Chemical properties such as chemical potential (μ), chemical hardness (η), and electrophilicity index (ω) were studied by DFT at the B3LYP level with a 6-311G(d,p) basis set. All compounds excellently follow Lipinski's rule of five.
Introduction
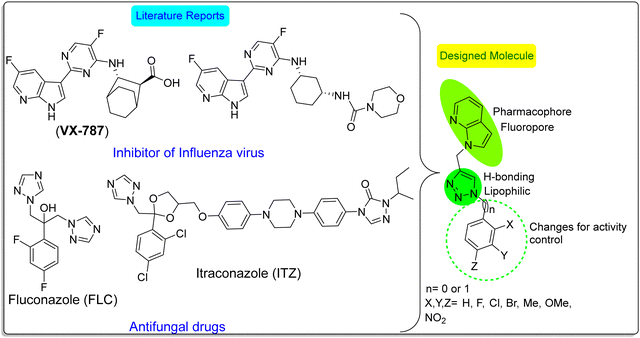
Post-corona (COVID-19) effects have significantly threatened immune-compromised people and corona-recovered patients. Different complications are observed in COVID-19 patients, such as severe respiratory syndromes due to invasive pulmonary aspergillosis, also called COVID-19-associated pulmonary aspergillosis (CAPA), and other Aspergillus genus-based infections. The rapid growth of Aspergillus infections leads to critical conditions in infected patients with respiratory disorders. Unfortunately, in many cases, diseases due to the Aspergillus genus are not diagnosed at the early stage due to its rapid growth, which leads to severe respiratory impairment.1 Sometimes, prolonged use of steroids and other heavy antibiotic drugs during the treatment of COVID-19 and other diseases causes loss of the immune system. It weakens the defense mechanism of the body to fight pathogenic attacks, especially fungal attacks, which increases the risk for similar and other severe fungal infections.2 The most commonly reported fungal infections in patients with COVID-19 include aspergillosis, candidiasis, and mucormycosis, also called “black fungus”.3 The intensity of various infectious pathogenic diseases, especially viral and fungal diseases, is increasing daily. It was observed that these microbial infections are asymptomatic, which slowly progress to active disease. The situation becomes more complicated when some microbial strains resist any drug(s). It alarms an immediate concern for developing some new class of antimicrobial drugs to eliminate resistant microbial strains with a different mode of action.4 Our lab employed a molecular hybridization approach for the development of new chemicals with improved properties.5 Molecular assimilation involves a combination of two or more biologically significant compounds to develop a new hybrid molecule with augmented properties compared to the parent fragments.67-Azaindole (7-AI) is well-known for its chromophoric properties and is a basic 7-azatryptophan (7-ATph) unit. 7-ATph is a non-natural amino acid. It has been used for the development of synthetic peptides and bacterial proteins.7 7-AI is a better substitute for tryptophan (Tph) and is used as an optical probe for protein structure over most of the physiological pH range.8 Although the charge transfer (π–π*) excited state of the 7-AI dimer has traditionally been considered as a model for base pairing in DNA9 molecules, the double proton transfer properties of 7-AI in water and other protic solvents such as methanol made it a suitable candidate for biological applications.10 Surprisingly, 7-AI moiety has not been given proper attention for developing advanced antimicrobial drugs except for one report for synthesizing only some derivatives with 4-Cl or 4-Br substituted 7-AI, e.g., 1-((1-(4-substituted benzyl)-1H-1,2,3-triazl-4-yl)methyl)-4-halo-1H-pyrolo[2,3-b]pyridine.11 However, Michael P. Clark and his group reported the development of a novel AI-based compound, VX-787 (Fig. 1), which was a first-in-class, orally bioavailable inhibitor of human influenza virus replication. It offers treatment for seasonal and pandemic influenza with essential advantages in care treatments, such as potency and efficacy.12–15 Benhamou et al. have utilized the fluorescence properties of azoles for constructing a molecular tool as a fluorescent probe to access detailed information about the site of drug action via real-time imaging. For the last three decades, azole-based antifungal drugs, especially fluconazole (FLC), as shown in Fig. 1, have extensively been explored as a first-line treatment drug for fatal fungal diseases. However, resistance to azole drugs, especially for Candida and Aspergillus species, has become a significant clinical concern.16 The much interesting and fascinating properties of the 7-AI moiety motivated us to utilize this moiety to construct some hybrid lead molecules.
The pharmacophoric properties of the five-membered 1,2,3-triazole, such as its lipophilic character, aromatic stability, fair dipole moment, and resistance to both acid and base hydrolysis, made it a suitable pharmacophoric candidate for the development of medicinally important compounds.17–19 Also, the lipophilicity and H-bonding features of triazoles help them associate and bind with biological targets such as DNA to improve solubility and metabolic stability under physiological conditions.20 The property of azoles to inhibit the antifungal target lanosterol 14α-demethylase16 and their broad spectrum of applications in medicine and materials science further add new dimensions to their importance.21–24 The important biological applications of 1,4-disubstituted 1,2,3-triazoles include their antifungal,25 antibacterial,26,27 anticancer,28,29 antiviral,30 anti-tubercular,22 anti-HIV,31,32 antidiabetic,33 anti-inflammatory,22,34 antimalarial,35 antiallergic,36 and antioxidant properties.37,38 The regioselectivity compromised thermal synthesis of 1,2,3-triazole initially proposed by Huisgen39,40 was effortlessly modified and improved by Sharpless and Meldal using a Cu(I)-catalyst under the name of “click chemistry,” which is also called the copper-catalyzed alkyne-azide cycloaddition (CuAAC) reaction, and it got tremendous recognition.41–45 The wide use of CuAAC led to its use up to the biological level under “Bioorthogonal Chemistry” by Carolyn Bertozzi, who recently shared a Nobel Prize in the field.46–48
In the current scenario, using fluorescent bioactive molecules for fluorescent labeling methods has become a vital tool in medical and biological applications.16 The abovementioned observations and our group's research interest49–51 motivated us to synthesize 7-AI linked 1,2,3-triazole hybrid molecules as fluorescent and antimicrobial agents. Herewith, we report the synthesis of two libraries of biologically significant 7-azaindole N-linked benzyl/phenyl appended 1,2,3-triazole hybrids via a CuAAC-based click reaction as potential fluorescent antifungal agents.
Results and discussion
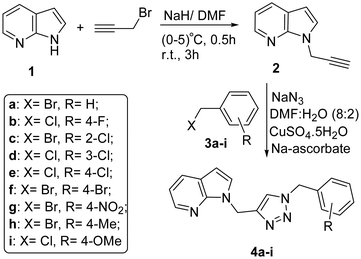
For the synthesis of 7-azaindole N-linked benzyl 1,2,3-triazole (4a–4i) hybrids as presented in Scheme 1, commercially available 7-azaindole (1) was subjected to N-propargylation as per the reported procedure,52 with the help of propargyl bromide in the presence of sodium hydride in dimethylformamide. The as-obtained N-propargyl 7-azaindole, i.e., 1-prop-2-ynyl-1H-pyrrolo[2,3-b]pyridine (2), was purified by column chromatography using an ethylacetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (1
n-hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 v/v) solution for eluting the pure compound in 80% yield. Compound 2 was used as a dipolarophile to synthesize the 7-azaindole N-linked 1,2,3-triazole hybrids (4a–4q).
9 v/v) solution for eluting the pure compound in 80% yield. Compound 2 was used as a dipolarophile to synthesize the 7-azaindole N-linked 1,2,3-triazole hybrids (4a–4q).
First, 7-azaindole N-linked benzyl 1,2,3-triazole (4a–4i) derivatives were prepared using the CuAAC mediated click reactions from in situ generated organic azides with N-propargyl 7-azaindole (2) using CuSO4·5H2O (5 mol%) and sodium ascorbate (10 mol%) as a catalyst at room temperature. Organic azides were prepared as per the reported method from benzyl halides (3a–3i).53 The desired representative 1,2,3-triazole compound 4a was isolated in 97% yield through a short silica gel band of column chromatography using an ethyl acetate and n-hexane (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 v/v) solvent system for elution. The synthesized compound 4a was thoroughly characterized with FTIR, 1H-NMR, 13C-NMR, and ESI-MS data. The formation of 1,2,3-triazole was ascertained by the disappearance of the alkyne
7 v/v) solvent system for elution. The synthesized compound 4a was thoroughly characterized with FTIR, 1H-NMR, 13C-NMR, and ESI-MS data. The formation of 1,2,3-triazole was ascertained by the disappearance of the alkyne ![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–H and C
C–H and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C stretching bands at 3292 cm−1 and 2121 cm−1, respectively, and the appearance of a new characteristic stretching band of 1,2,3-triazole due to
C stretching bands at 3292 cm−1 and 2121 cm−1, respectively, and the appearance of a new characteristic stretching band of 1,2,3-triazole due to ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H stretching at νmax 3119 cm−1 in the FTIR spectrum of compound 4a. The absorption bands at 1508 and 1306 cm−1 due to the stretching vibrations of –C
C–H stretching at νmax 3119 cm−1 in the FTIR spectrum of compound 4a. The absorption bands at 1508 and 1306 cm−1 due to the stretching vibrations of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and –C–N of the 7-azaindole moiety were retained in compound 4a, which appeared initially in 7-azaindole linked alkyne 2 at the exact wavenumber, i.e., 1508 and 1306 cm−1.
N and –C–N of the 7-azaindole moiety were retained in compound 4a, which appeared initially in 7-azaindole linked alkyne 2 at the exact wavenumber, i.e., 1508 and 1306 cm−1.
The 1H-NMR spectrum of 4a showed all the proton signals for eleven C–H signals of extended aromatic protons from δ 6.45 to 8.30 ppm and the rest of four aliphatic proton signals as singlets at δ 5.43 along with δ 5.57 ppm for two CH2 protons. A sharp singlet signal at δ 7.41 ppm, which could be attributed to the triazolyl proton, showed confirmation of the triazole ring in the compound. The 13C-NMR spectrum of 4a showed all the required seventeen carbon signals at δ 147.40, 144.58, 143.05, 136.58, 129.41, 129.31, 129.18, 128.70, 128.53, 124.00, 120.60, 116.38, 100.22, 53.33, and 39.41 including two of the overlapped phenyl carbon signals at δ 129.319 (2C) and 128.533 (2C) which were counted for two carbons.
The ESI mass spectrum of compound 4a exhibited a peak at m/z 290.14 (M + H)+, which is precisely similar to the calculated value of (M + H)+ of 290.14 and thus showed good agreement with the molecular formula (C17H15N5H+).
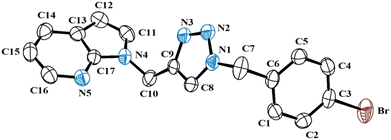
The final confirmation about the formation of 7-azaindole N-linked aryl 1,2,3-triazole derivatives was authenticated after revealing the correct structural parameters of the anticipated structure about one of the solid crystalline compounds with the help of X-ray crystallographic data of 4f (recrystallized from a solution of ethanol and chloroform). The crystal structure of compound 4f and other details are given in Fig. 2 and Table SI-1 (ESI†). The X-ray crystallographic structure of compound 4f (CCDC no. 2192737) crystallizes in a triclinic crystal system in the P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (2) space group. It was observed that two crystallographically independent molecular units were present in an asymmetric unit of compound 4f, as shown in Fig. SI-1 (ESI†).
(2) space group. It was observed that two crystallographically independent molecular units were present in an asymmetric unit of compound 4f, as shown in Fig. SI-1 (ESI†).
Before moving to various derivatization of 7-azaindole N-linked benzyl 1,2,3-triazoles, efforts were made to improve the yield of the reaction products by changing different solvent systems such as t-BuOH, H2O, DMSO, THF, and DMF. The reaction temperature was also optimised, as detailed in Table 1. It was observed that the reaction did not complete in a single solvent such as tBuOH, DMSO, THF, or H2O due to the solubility issue of inorganic catalysts and an organic compound. Thus, a minimum amount of H2O for the catalyst solubility in a suitable solvent such as DMF for the solubility of the organic compound was the best choice for the completion of the reaction. All other derivatives of 7-azaindole N-linked benzyl 1,2,3-triazoles, i.e., 4b–4i, were synthesized and characterized with the help of the same spectral pattern of bands and peaks in the FTIR, 1H-NMR, 13C-NMR, and ESI-MS data. The product reproducibility and generalization of the reaction conditions were tested by extending the scheme for the synthesis of other derivatives of 7-azaindole N-linked benzyl 1,2,3-triazoles (4a–4i) in good to excellent yields of 80–94%, as detailed in Table 2.
| S. no. | Solvent(s) solution (v/v) | Temperature (°C) | Time (h) | Yieldb (%) |
|---|---|---|---|---|
| a Reaction conditions: aryl azide and 7-azaindole N-linked alkyne 2 (1 mmol); CuSO4·5H2O (10 mol%) and Na-ascorbate (20 mol%), rt, and 3–24 h. b Yield refers to purification by recrystallization. c No reaction. | ||||
| 1 |
t-BuOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
35 | 7 | 74 |
| 2 |
t-BuOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8 H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
35 | 5.5 | 78 |
| 3 | t-BuOH | 35 | 24 | |
| 4 | DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
35 | 5 | 82 |
| 5 | DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8 H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
35 | 4 | 86 |
| 6 | DMSO | 35, 60 | 24 | |
| 7 | THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
35 | 6 | 75 |
| 8 | THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8 H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
35 | 4.5 | 79 |
| 9 | THF | 35, 60 | 24 | |
| 10 | DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1 H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
35 | 5 | 88 |
| 11 | DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8 H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
35 | 3 | 97 |
| 12 | DMF | 35 | 24 | 68 |
| 13 | H2O | 35, 60 | 24 | |
| S. no. | Compound | PhCH2/Ph | R | Time (h) | Yieldb (%) |
|---|---|---|---|---|---|
a
2 (1 mmol), 3a–q (1 mmol), sodium azide (1.2 mmol), CuSO4·5H2O (10 mol%), Na-ascorbate (20 mol%), DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8 H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v ratio), rt (35 °C), and 3–7 h.
b Yields refer to purification via column chromatography using ethyl acetate 2 v/v ratio), rt (35 °C), and 3–7 h.
b Yields refer to purification via column chromatography using ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hexane with increased polarity gradient. hexane with increased polarity gradient.
|
|||||
| 1 | 4a | PhCH2 | H | 3 | 94 |
| 2 | 4b | PhCH2 | 4-F | 5 | 87 |
| 3 | 4c | PhCH2 | 2-Cl | 6 | 83 |
| 4 | 4d | PhCH2 | 3-Cl | 7 | 92 |
| 5 | 4e | PhCH2 | 4-Cl | 5 | 90 |
| 6 | 4f | PhCH2 | 4-Br | 6 | 87 |
| 7 | 4g | PhCH2 | 4-NO2 | 4 | 80 |
| 8 | 4h | PhCH2 | 4-Me | 7 | 92 |
| 9 | 4i | PhCH2 | 4-OMe | 4 | 82 |
| 10 | 4j | Ph | H | 3 | 84 |
| 11 | 4k | Ph | 4-F | 6 | 85 |
| 12 | 4l | Ph | 2-Cl | 5 | 92 |
| 13 | 4m | Ph | 3-Cl | 6 | 85 |
| 14 | 4n | Ph | 4-Cl | 4 | 84 |
| 15 | 4o | Ph | 4-Br | 5 | 84 |
| 16 | 4p | Ph | 4-NO2 | 1 | 83 |
| 17 | 4q | Ph | 4-OMe | 4 | 80 |
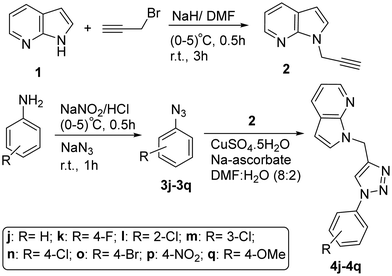
To extend the library of novel 7-azaindole N-linked aryl 1,2,3-triazoles, we thought to synthesize phenyl azides from different aniline derivatives using the reported diazotization and azidation methods.54 Thus prepared phenyl azides (3j–q), as shown in Scheme 2, were subjected to react with N-propargyl 7-azaindole (2) using CuSO4·5H2O (5 mol%) and sodium ascorbate (10 mol%) as a catalyst at room temperature for 1 to 6 h stirring to yield pure 7-azaindole N-linked phenyl 1,2,3-triazole (4j–4q) in 80–92% isolated yield after column chromatography. The library of 7-azaindole N-linked phenyl 1,2,3-triazoles (as detailed in Table 2, entries 10–17) were efficiently synthesized and characterized by following the similar spectral patterns of bands and signals in the FTIR, 1H-NMR, 13C-NMR, and ESI-MS data.
The representative compound of this series, 7-azaindole N-linked phenyl 1,2,3-triazole 4j, obtained in 84% yield, was successfully characterized using the FTIR, 1H-NMR, 13C-NMR, and ESI-MS data. The disappearance of the alkyne ![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C–H and C
C–H and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C stretching bands at 3292 cm−1 and 2121 cm−1, respectively, and the appearance of a new characteristic stretching band of
C stretching bands at 3292 cm−1 and 2121 cm−1, respectively, and the appearance of a new characteristic stretching band of ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H at νmax 3144 cm−1 in the FTIR spectrum confirmed the formation of the 1,2,3-triazole ring. The absorption bands at 1506 and 1300 cm−1 due to the stretching vibrations of –C
C–H at νmax 3144 cm−1 in the FTIR spectrum confirmed the formation of the 1,2,3-triazole ring. The absorption bands at 1506 and 1300 cm−1 due to the stretching vibrations of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and –C–N of the 7-azaindole moiety were observed nearly in the same region as in 7-azaindole linked alkyne (2) at 1508 and 1306 cm−1.
N and –C–N of the 7-azaindole moiety were observed nearly in the same region as in 7-azaindole linked alkyne (2) at 1508 and 1306 cm−1.
The 1H-NMR spectrum of compound 4j confirmed all eleven aromatic protons from δ 6.48 to 8.35 ppm integrating and showing a sharp singlet signal at δ 5.68 ppm integrating two aliphatic protons. A sharp singlet that appeared at δ 7.92 ppm was attributed to the triazolyl proton, which confirmed triazole ring formation.
The 13C-NMR spectrum of 4j showed all the indispensable sixteen carbon signals at δ 147.08, 142.75, 136.87, 129.63, 129.05, 128.74, 128.05, 125.22, 120.81, 120.50, 116.01, 115.59, 100.35 and 39.54 including two of the overlapped phenyl carbon signals at δ 129.638 (2C) and 120.506 (2C).
The ESI mass spectrum of compound 4j exhibited a peak at m/z 276.12 (M + H)+, which is precisely similar to the calculated value of (M + H)+ as per the molecular formula (C16H13N5H+): 276.12. Both theoretical and experimental m/z peaks due to (M + H)+ are in good agreement with each other.
Pharmacological study
Antibacterial activity study
All 7-azaindole N-linked aryl 1,2,3-triazoles 4a–q including the precursor alkyne 2, as shown in Table 3, were screened for in vitro antibacterial activity against gram (+) strains B. subtilis and S. aureus and gram (−) strains E. coli and P. aeruginosa using the standard serial dilution method. In this method, a solution of different triazoles was prepared for stock using DMSO.55 The triazole's minimum inhibitory concentration (MIC) was recorded in μmol mL−1. From the activity data, it was observed that compound 4f and 4o, containing a Br-substituted benzene ring with MIC values of 0.0170 μmol mL−1 (P. aeruginosa and E. coli) and 0.0177 μmol mL−1 (B. subtilis and E. coli), respectively, showed significant results as compared to 7-azaindole linked alkyne 2 with an MIC value of 0.0800 μmol mL−1. The bioactivity results revealed that the antibacterial activity of 1,2,3-triazoles is nearly five times better than that of the precursor alkyne. Further modifications can improve the activity of triazoles in the moiety compared to the reference drug ciprofloxacin. The SAR (structure–activity relationship) study showed that all 7-azaindole linked 1,4 disubstituted 1,2,3-triazole moieties reflected better potency than the alkyne, which confirmed that the incorporation of the 1,2,3-triazole unit into 7-azaindole linked alkyne 2 augmented the antibacterial activity to a better extent.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a
a
| S. no | Compounds | M. Wt. | A | B | C | D | E | F |
|---|---|---|---|---|---|---|---|---|
| A: P. aeruginosa; B: B. subtilis; C: E. coli; D: S. aureus; E: C. albicans; and F: A. niger. | ||||||||
| 1. | 2 | 156.18 | 0.0800 | 0.0800 | 0.0800 | 0.0800 | 0.0800 | 0.0400 |
| 2. | 4a | 289.33 | 0.0432 | 0.0432 | 0.0432 | 0.0216 | 0.0216 | 0.0432 |
| 3. | 4b | 307.32 | 0.0407 | 0.0407 | 0.0407 | 0.0407 | 0.0814 | 0.0204 |
| 4. | 4c | 323.78 | 0.0386 | 0.0772 | 0.0193 | 0.0386 | 0.0772 | 0.0386 |
| 5. | 4d | 323.78 | 0.0386 | 0.0193 | 0.0386 | 0.0386 | 0.0193 | 0.0386 |
| 6. | 4e | 323.78 | 0.0386 | 0.0772 | 0.0193 | 0.0386 | 0.0772 | 0.0193 |
| 7. | 4f | 368.23 | 0.0170 | 0.0340 | 0.0170 | 0.0679 | 0.0679 | 0.0170 |
| 8. | 4g | 334.33 | 0.0374 | 0.0374 | 0.0187 | 0.0374 | 0.0748 | 0.0187 |
| 9. | 4h | 303.36 | 0.0412 | 0.0412 | 0.0206 | 0.0412 | 0.0412 | 0.0824 |
| 10. | 4i | 319.36 | 0.0391 | 0.0391 | 0.0196 | 0.0391 | 0.0783 | 0.0783 |
| 11. | 4j | 275.31 | 0.0454 | 0.0454 | 0.0227 | 0.0908 | 0.0908 | 0.0454 |
| 12. | 4k | 293.30 | 0.0426 | 0.0426 | 0.0426 | 0.0852 | 0.0426 | 0.0426 |
| 13. | 4l | 309.75 | 0.0202 | 0.0404 | 0.0404 | 0.0404 | 0.0807 | 0.0404 |
| 14. | 4m | 309.75 | 0.0807 | 0.0202 | 0.0404 | 0.0807 | 0.0807 | 0.0404 |
| 15. | 4n | 309.75 | 0.0202 | 0.0404 | 0.0404 | 0.0404 | 0.0404 | 0.0202 |
| 16. | 4o | 354.20 | 0.0353 | 0.0177 | 0.0177 | 0.0706 | 0.0706 | 0.0353 |
| 17. | 4p | 320.31 | 0.0390 | 0.0781 | 0.0195 | 0.0781 | 0.0781 | 0.0390 |
| 18. | 4q | 305.33 | 0.0409 | 0.0409 | 0.0205 | 0.0409 | 0.0819 | 0.0409 |
| 19. | Ciprofloxacin | 0.0094 | 0.0094 | 0.0094 | 0.0094 | — | — | |
| 20. | Fluconazole | — | — | — | — | 0.0408 | 0.0408 | |
Evaluation of the antifungal activity
All the newly synthesized 7-azaindole linked 1,4 disubstituted 1,2,3-triazole moieties were subjected to in vitro antifungal activities using C. albicans and A. niger as fungal strains with the help of the standard dilution method.56 The antifungal data were recorded in terms of μmol mL−1. As per the tested results, compounds 4d and 4n having a Cl-moiety as an EWG on the benzene ring with minimum inhibitory concentration (MIC) values of 0.0193 and 0.0404 μmol mL−1, respectively, reflected better activity for the C. albicans fungal strain as compared to the reference drug fluconazole with an MIC value of 0.0408 μmol mL−1. The observations from the results indicated that compound 4d (MIC: 0.0193 μmol mL−1) reflected more than two-fold superior antifungal activity compared to the reference drug fluconazole (MIC: 0.0408 μmol mL−1). However, for the A. niger fungal strain, almost all 1,2,3-triazole compounds (4b–4g and 4l–4q) except for 4a and 4h–4k compounds showed better antifungal activity as compared to the standard reference drug fluconazole (MIC: 0.0408 μmol mL−1). However, compound 4f with an MIC value of 0.0170 μmol mL−1 was found to be more influential among all 7-AI linked 1,2,3-triazoles against P. aeruginosa and E. coli bacterial strains along with the A. niger fungal strain. In addition, compounds 4d and 4n containing a Cl-moiety on the benzene ring also exhibited better activity with MIC values of 0.0193, 0.0193, and 0.0386 and 0.0202, 0.0404, and 0.0202 μmol mL−1, for B. subtilis, C. albicans, and A. niger and P. aeruginosa, C. albicans, and A. niger, respectively, as compared to the reference antibacterial drug ciprofloxacin (MIC: 0.0094 μmol mL−1) and antifungal drug fluconazole (MIC: 0.0408 μmol mL−1). After reviewing the antifungal data and their structure–activity relationship, an augmentation in the biological activity was observed due to the incorporation or linkage of the 7-azaindole moiety with different 1,2,3-triazole units. Substantially, the test results revealed that compounds 4d and 4n are most potent against both fungal strains and can be a better substitute for the pre-existing antifungal drug fluconazole.Fluorescence activity
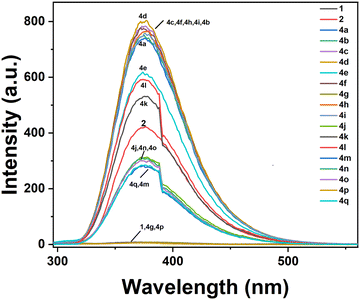
The UV-Vis spectrum of 7-AI (1), 7-AI-linked alkyne (2), and 7-azaindole-linked aryl 1,2,3-triazoles (4a–4i), as shown in Fig. SI-2 (ESI†), showed an absorbance peak from 265 to 285 nm which could be due to the presence of the 7-azaindole moiety in MeOH (concentration 2.7–8.4 mmol). The fluorescence change in compounds 7-AI (1), 7-AI-linked alkyne (2), and 7-azaindole-linked aryl 1,2,3-triazoles (4a–4q) was checked under UV light as shown in Fig. 3, which revealed that most of the synthesized 1,2,3-triazoles appeared fluorescent under UV light through naked eyes except for the 4-NO2 (a well-known fluorescence quencher)57 benzyl substituted 1,2,3-triazole 4g. However, 7-AI-linked alkyne 2 seemed to be the most robust fluorescent compound through naked eyes under UV light. Also, a relative fluorescence intensity change in compounds 7-AI (1), 7-AI-linked alkyne (2), and 7-azaindole-linked aryl 1,2,3-triazoles (4a–4q) in MeOH (concentration 2.7–8.4 mmol) with λex = 285 nm was monitored, as shown in Fig. 4. The relative decreasing order in the fluorescence intensity change in compounds 7-AI (1), 7-AI-linked alkyne (2), and 7-azaindole-linked aryl 1,2,3-triazoles (4a–4q) as shown below revealed that compound 4d having a 3-Cl benzyl-substituted 1,2,3-triazole with 799.1815 a.u. intensity is highly fluorescent with the least intensity shared by compound 4p having a 4-NO2 (a well-known fluorescence quencher) phenyl-substituted 1,2,3-triazole moiety. | ||
| Fig. 3 A naked eye-detectable fluorescence change in 7-AI (1), 7-AI-linked alkyne (2), and 7-azaindole-linked aryl 1,2,3-triazoles (4a–4q) under UV light. | ||
Decreasing order of the fluorescence intensity of compounds (1, 2, and 4a–4q): 4d (799.1815) > 4c (781.8753) > 4f (774.9273) > 4h (763.4579) > 4i (754.3394) > 4b (750.0651) > 4a (737.9546) > 4e (619.8882) > 4l (592.2746) > 4k (528.8380) > 2 (424.1054) > 4j (311.8661) > 4n (306.0840) > 4o (298.9996) > 4q (284.8308) > 4m (279.5175) > 1 (08.3395) > 4g (07.3845) > 4p (05.8384).
Molecular docking
The studies of antimicrobial activity results revealed that the incorporation of the 1,2,3-triazole moiety improved the antifungal activities of almost all the synthesized triazoles more efficiently for both fungal strains than the antibacterial activities. The docking simulations of the predecessor alkyne 2 and 7-azaindole N-linked triazoles (4a–4q) were studied by performing molecular docking with an active site of the fungal enzyme DNA gyrase lanosterol 14α-demethylase (PDB ID: 1EA1) by using software Auto dock Vina open-source58–61 as shown in Table SI-2 (ESI†). The fungal enzyme 14α-demethylase (PDB ID: 1EA1) was obtained from the RCSB protein data bank (https://www.rcsb.org) in PDB format. CHEMSKETCH was used to draw 3D-structures of the ligands at www.acdlabs.com, and SPDBV software was used to perform their energy minimization. AutoDockTools-1.5.7 software was used to prepare the ligands and receptors in pdbqt format. During the preparation, water molecules were removed, Gasteiger charges were included, and only polar H-atoms were added. The results were visualized using a Discovery studio visualizer.62In docking simulations, different conformations of the hybrid molecules were obtained, and the energetically most favorable docked structure was studied. It was evident that the incorporation of a triazole unit into the alkyne 2 (with a docking score of −6.11 kcal mol−1) showed improvement in the docking score (−8.38 to −9.28 kcal mol−1) as apparent from 7-azaindole N-linked triazoles (4a–4q). For reference, we have considered the hybrid molecule 4d and its alkyne 2 possessing binding energies of −8.81 and −6.11 kcal mol−1, respectively. The alkyne 2 showed only one H-bond between the N-atom of the pyridine ring and THR260 (Fig. 5). The pi–electron density of the 7-AI ring system interacted with ALA400 and PRO320. Additionally, pi–Sigma interactions were observed with THR260. However, in the case of triazole 4d, the N-atom present in the triazole ring is involved in three donor hydrogen bond interactions with CYS394, VAL395, and GLY396, denoted by a green-colored dotted line. The pi–orbitals of the pyridine ring formed pi–alkyl interactions with LEU105, LEU152, and ALA256. The chlorine-linked phenyl ring has interacted with LEU100 and ALA256 through pi–alkyl and Sigma alkyl bonds, respectively. The pi-orbitals of the triazole ring system also formed pi–alkyl interactions with CYS394, VAL395, and ALA256. The chlorine atom, present on the benzene ring, is involved in hydrophobic interactions with LEU100 and MET99. The pyrrole ring system is also involved in pi–alkyl-type interactions with LEU100. However, pi–Sigma interactions are formed between the pi–orbital of the pyrrole ring and LEU105 and ALA256. All triazoles exhibited various interactions; however, the dominant interactions for the binding process are conventional hydrogen bonding with THR260, CYS394, SER261, GLY72, ARG96, ARG326, VAL395, HIS259, HIS392, and GLN72, as presented in Fig. SI-3, ESI† and listed in Table SI-2, ESI.† The smaller value of bond length and the more significant negative value of the docking score are the leading indicators of the stability of hybrid molecules. Hence, theoretical data obtained through molecular docking seemed to agree with the experimentally obtained data, i.e., biological activity testing. For reference, the cartoon images of dipolarophile 2 and the more promising potent 1,2,3-triazole 4d docked in the active site of DNA gyrase lanosterol 14-α demethylase enzyme (PDBID:1EA1) are shown in Fig. 6.
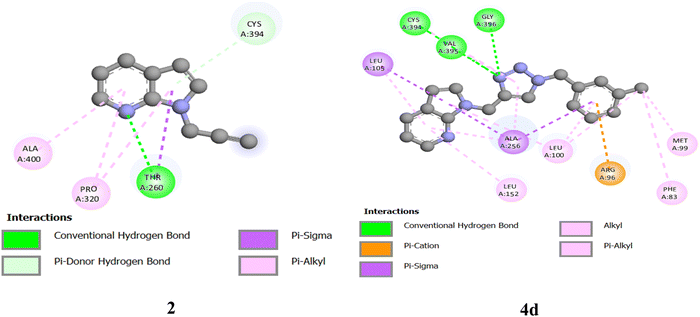 | ||
| Fig. 5 2D images of alkyne 2 and 1,2,3-triazole hybrid 4d showing conventional H-bonding and other interactions with an active site of DNA 14α-demethylase lanosterol fungal enzyme. | ||
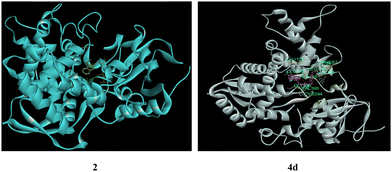 | ||
| Fig. 6 Cartoon images of 2 and 4d docked in the active site of DNA gyrase lanosterol 14-α demethylase enzyme (PDBID:1EA1). | ||
ADME prediction
The in silico ADME (Absorption, Distribution, Metabolism, and Excretion) properties of all the newly synthesized 7-azaindole linked 1,4 disubstituted 1,2,3 triazole hybrid molecules (4a–q) were predicted by using the Molinspiration online software (https://www.molinspiration.com/cgi-bin/properties) as shown in Table SI-3 (ESI†). Here, we have studied various parameters such as %Absorbance (%ABS), lipophilicity (miLogP), Total Polar Surface Area (TPSA), Molecular Weight (MW), number of H-bond acceptors, number of H-bond donors, etc. In addition to them, the number of violations to Lipinski's rule of five (Nviol)63 and the drug-likeness score were also ascertained, which are helpful for the selection of a particular bio-active compound from the library of synthesized compounds.64–66 The drug-likeness property also helps to study various pharmacokinetic properties such as absorption, distribution, metabolism, and excretion of the synthesized drug/compound in human beings. All the synthesized compounds 2 and 4a–q exhibited a TPSA (Total Polar Surface Area) value of <160 Å2 (i.e., 18–95 Å2) and also the absorption value in the favorable range, i.e. %ABS = 76.44 to 102.84, which was calculated using the mathematical expression %ABS = 109 − (0.345 × TPSA).67 It is observed from the data that an orally active drug should follow at least four or more points in Lipinski's rule of five.68,69According to Lipinski's rule of five, an orally active drug should satisfy these points:
(1) Molecular weight should be ≤500
(2) Octane–water partition coefficient ≤5
(3) No. of H-bond acceptors ≤10 and
(4) No. of H-bond donors ≤5.
The synthesized alkyne 2 and triazole compounds (4a–4q) strictly adhere to the Lipinski's rule with zero violation as is observed from Table SI-3 (ESI†), wherein the molecular weight of all the compounds lies in the range 156.17–368.24, i.e., less than 500, which showed their easy absorption, diffusion, and transportation from the human body. The miLogP value of all compounds is also less than 5 (i.e., 1.49–3.48). The number of hydrogen bond acceptor atoms, i.e., O and N atoms, lies in the range 2 to 8, i.e., less than 10. Additionally, the number of rotatable bonds is 1 to 5, i.e., less than 10, indicating their easily binding flexibility. Hence, all these parameters confirmed effective drug-like properties.
In general, if the bioactivity score (G protein-coupled receptor (GPCR) ligand, a kinase inhibitor, ion channel modulator, nuclear receptor ligand, protease inhibitor, and enzyme inhibitor) of the synthesized compounds is >−0.5, then the drug is biologically active, but if the score is <−0.5, then the drug is not active. The bioactivity scores, as provided in Table SI-4 (ESI†), showed that alkyne 2 and triazole compounds 4a–4q are active and confirmed their binding flexibilities.
DFT studies
The Density Functional Theory (DFT) method acts as a potent tool for the interpretation of reactivity parameters of the synthesized drug compounds,70 invaluable information, complementary to the experimental data, about molecular systems and processes. Thus, to confirm the stability and the geometry optimization of the proposed 7-azaindole linked 1,2,3-triazole hybrid molecule, DFT calculations were carried out in the gas phase at the B3LYP 6-311G(d,p) level by using the Gaussian09 program package.71According to the Frontier Molecular Orbital (FMO) theory, the higher energy orbital HOMO (Highest Occupied Molecular Orbital) and the lower energy orbital LUMO (Lowest Unoccupied Molecular Orbital) participate in the bond formation between the precursors (alkyne and azides). The energy gap between these orbitals contributes to the bioactivity of the synthesized hybrid molecules (4a–4q). With the help of this method, various energy parameters such as energy gap, chemical potential, and chemical hardness and chemical softness as represented by symbols ELUMO–HOMO, μ, and η, respectively, were calculated using Koopman's theorem.72 Additionally, the electrophilicity index represented by ω was calculated using Robert G. Parr approximation.73–75
The formulation is represented below to show the relationships among these parameters as-
• μ = (EHOMO + ELUMO)/2
• η = (ELUMO − EHOMO)/2
• ω = μ2/2η
It is well known that the higher the energy band gap, the harder the compound, and correspondingly, the lower the band gap, the softer, towards the chemical reactivity, and less stable the compound.76–78 Hence, from our calculation data table, as shown in Table SI-5 (ESI†), it was observed that the incorporation of the triazole unit into the alkyne considerably decreases the bandgap energy (ΔELUMO–HOMO) of the resultant triazoles 4a–4q (3.35–5.44 eV) except for compound 4f (5.44 eV) as compared to the initial alkyne (5.11 eV). Also, compounds 4g and 4p having –NO2 as an EWG on the benzyl and phenyl rings showed the least energy band gap, i.e., 3.35 eV. This indicates that nitro as a functional group decreased the band gap and favored the easy movement of electrons. Thus, it leads to the softness and more reactivity of the compound. The electrophilicity index (ω) indicates the stabilization energy of the compound. Here, the computed data revealed that drugs 4g and 4p showed high electrophilicity index values of 5.97 eV and 6.27 eV, respectively, thus, supporting better results in correlation with the experimental results.
Conclusions
Two new libraries of fluorescence-active 7-azaindole N-linked benzyl 1,2,3-triazole (4a–4i) and 7-azaindole N-linked phenyl 1,2,3-triazole (4j–4q) hybrids have been designed and synthesized via a Cu(I)-catalyzed alkyne-azide cycloaddition (CuAAC) click reaction and successfully characterized using FT-IR, 1H-NMR, 13C-NMR, and HRMS data. The structure of compound 4f was finally supported by its single-crystal X-ray data. Compounds 4d and 4n showed better antifungal activity against C. albicans and A. niger (MIC: 0.0193, 0.0386, 0.0404, and 0.0202 μmol mL−1, respectively) with reference to the existing drug fluconazole (MIC: 0.0408 μmol mL−1). Compound 4f was found to be a suitable alternative to the existing antifungal drug fluconazole for A. niger with the least MIC: 0.0170 μmol mL−1 along with considerable antibacterial activity against P. aeruginosa and E. coli (MIC: 0.0170 μmol mL−1, each) for the reference drug ciprofloxacin (MIC: 0.0094 μmol mL−1). The most promising compounds 4d, 4f, and 4n showed good binding energies of −8.81, −8.76, and −8.79 kcal mol−1 with DNA 14α-demethylase lanosterol enzyme (PDB ID:1EA1) and showed good agreement with the experimental and theoretical results. The results of the studies on the DFT and ADME properties reasonably supported the experimental results.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We acknowledge the Department of Chemistry, NIT Kurukshetra for the research facilities and financial assistance to KS. We also acknowledge the Central Instrumentation Laboratory GJUS&T, Hisar, Haryana 125001 for NMR analysis and IIT Ropar for ESI-MS facilities.References
- L. Trovato, M. Calvo, G. Migliorisi, M. Astuto, F. Oliveri and S. Oliveri, Respir. Med. Case Rep., 2021, 32, 101367 Search PubMed.
- S. Narayanan, J. V. Chua and J. W. Baddley, Clin. Infect. Dis., 2022, 74, 1279 CrossRef CAS PubMed.
- P. Koehler, O. A. Cornely, B. W. Böttiger, F. Dusse, D. A. Eichenauer and F. Fuchs, Mycoses, 2020, 63, 528 CrossRef CAS PubMed.
- N. T. Raveendran, A. Mohandas, R. R. Menon, A. S. Menon, R. Biswas and R. Jayakumar, ACS Appl. Bio Mater., 2019, 2, 243 CrossRef PubMed.
- N. Nehra, R. K. Tittal, V. D. Ghule, N. Naveen and K. Lal, J. Mol. Struct., 2021, 1245, 131013 CrossRef CAS.
- N. Nehra, R. K. Tittal and V. D. Ghule, ACS Omega, 2021, 6, 27089 CrossRef CAS PubMed.
- A. V. Smirnov, D. S. English, R. L. Rich, J. Lane, L. Teyton, A. W. Schwabacher, S. Luo, R. W. Thornburg and J. W. Petrich, J. Phys. Chem. B, 1997, 101, 2758 CrossRef CAS.
- M. Négrerie, F. Gai, S. M. Bellefeuille and J. W. Petrich, J. Phys. Chem., 1991, 95, 8663 CrossRef.
- R. Gelabert, M. Moreno and J. M. Lluch, J. Phys. Chem. A, 2006, 110, 1145 CrossRef CAS PubMed.
- O. H. Kwon and D. J. Jang, J. Phys. Chem. B, 2005, 109, 8049 CrossRef CAS PubMed.
- K. Harkala, L. Eppakayala, S. Sripelly and T. C. Maringanti, Indian J. Heterocycl. Chem., 2015, 25, 165 CAS.
- M. P. Clark, M. W. Ledeboer, I. Davies, R. A. Byrn, S. M. Jones, E. Perola, A. Tsai, M. Jacobs, K. Nti-Addae, U. K. Bandarage, M. J. Boyd, R. S. Bethiel, J. J. Court, H. Deng, J. P. Duffy, W. A. Dorsch, L. J. Farmer, H. Gao, W. Gu, K. Jackson, D. H. Jacobs, J. M. Kennedy, B. Ledford, J. Liang, F. Maltais, M. Murcko, T. Wang, M. W. Wannamaker, H. B. Bennett, J. R. Leeman, C. McNeil, W. P. Taylor, C. Memmott, M. Jiang, R. Rijnbrand, C. Bral, U. Germann, A. Nezami, Y. Zhang, F. G. Salituro, Y. L. Bennani and P. S. Charifson, J. Med. Chem., 2014, 57, 6668 CrossRef CAS PubMed.
- M. J. Boyd, U. K. Bandarage, H. Bennett, R. R. Byrn, I. Davies, W. Gu, M. Jacobs, M. W. Ledeboer, B. Ledford, J. R. Leeman, E. Perola, T. Wang, Y. Bennani, M. P. Clark and P. S. Charifson, Bioorg. Med. Chem. Lett., 2015, 25, 1990 CrossRef CAS PubMed.
- R. A. Byrn, S. M. Jones, H. B. Bennett, C. Bral, M. P. Clark, M. D. Jacobs, A. D. Kwong, M. W. Ledeboer, J. R. Leeman, C. F. McNeil, M. A. Murcko, A. Nezami, E. Perola, R. Rijnbrand, K. Saxena, A. W. Tsai, Y. Zhou and P. S. Charifson, Antimicrob. Agents Chemother., 2015, 59, 1569 CrossRef PubMed.
- J. Liang, J. E. Cochran, W. A. Dorsch, L. Davies and M. P. Clark, Org. Process Res. Dev., 2016, 20, 965 CrossRef CAS.
- R. I. Benhamou, M. Bibi, K. B. Steinbuch, H. Engel, M. Levin, Y. Roichman, J. Berman and M. F. Orcid, ACS Chem. Biol., 2017, 12, 1769 CrossRef CAS PubMed.
- K. Bozorov, J. Zhao and H. A. Aisa, Bioorg. Med. Chem., 2019, 27, 3511 CrossRef CAS PubMed.
- B. Schulze and U. S. Schubert, Chem. Soc. Rev., 2014, 43, 2522 RSC.
- D. Ashok, P. Chiranjeevi, A. V. Kumar, M. Sarasija, V. S. Krishna, D. Sriram and S. Balasubramanian, RSC Adv., 2018, 8, 16997 RSC.
- W. S. Horne, M. K. Yadav, C. D. Stout and M. R. Ghadiri, J. Am. Chem. Soc., 2004, 126, 15366 CrossRef CAS PubMed.
- P. D. Jarowski, Y. L. Wu, W. B. Schweizer and F. Diederich, Org. Lett., 2008, 10, 3347 CrossRef CAS PubMed.
- D. Dheer, V. Singh and R. Shankar, Bioorg. Chem., 2017, 71, 30 CrossRef CAS PubMed.
- S. Shafia, M. M. Alama, N. Mulakayala, C. Mulakayala, G. Vanaja, A. M. Kalle, R. Pallud and M. S. Alam, Eur. J. Med. Chem., 2012, 49, 324 CrossRef PubMed.
- N. S. Vatmurge, B. G. Hazra, V. S. Pore, F. Shirazi, M. V. Deshpande, S. Kadreppa and S. Chattopadhyay, Org. Biomol. Chem., 2008, 6, 3823 RSC.
- N. Nehra, R. K. Tittal, V. D. Ghule, N. Kumar, A. K. Paul, K. Lal and A. Kumar, ChemistrySelect, 2021, 6, 685 CrossRef CAS.
- N. Naveen, R. K. Tittal, V. D. Ghule, P. Rani, K. Lal and A. Kumar, ChemistrySelect, 2020, 5, 6723 CrossRef.
- K. Lal, N. Poonia, P. Rani, A. Kumar and A. Kumar, J. Mol. Struct., 2020, 1215, 128234 CrossRef CAS.
- P. Yadav, K. Lal, A. Kumar, S. Bhushan, S. K. Guru and S. Jaglan, Eur. J. Med. Chem., 2017, 126, 944 CrossRef CAS PubMed.
- V. D. Da Silva, B. M. de Faria, E. Colombo, L. Ascari, G. P. A. Freitas, L. S. Flores, Y. Cordeiro, L. Romao and C. D. Buarque, Bioorg. Chem., 2019, 83, 87 CrossRef CAS PubMed.
- Y. W. He, C. Z. Dong, J. Y. Zhao, L. L. Ma, Y. H. Li and H. A. Aisa, Eur. J. Med. Chem., 2014, 76, 245 CrossRef CAS PubMed.
- F. Silva, M. C. B. V. Souza, I. I. P. Frugulhetti, H. C. Castro, S. L. Souza, T. M. Souza, D. Q. Rodrigues, A. M. T. Souza, P. A. Abreu, F. Passamani, C. R. Rodrigues and V. F. Ferreira, Eur. J. Med. Chem., 2009, 44, 373 CrossRef PubMed.
- M. Whiting, J. Muldoon, J. C. Lin, S. M. Silverman, W. Lindstorm, A. J. Olson, H. C. Kolb, M. G. Finn, K. B. Sharpless, J. H. Elder and V. V. Fokin, Angew. Chem., Int. Ed., 2006, 45, 1435 CrossRef CAS PubMed.
- F. Jabeen, S. A. Shehzadi, M. Q. Fatmi, S. Shaheen, L. Iqbal, N. Afza, S. S. Pandad and F. L. Ansari, Bioorg. Med. Chem. Lett., 2015, 26, 1029 CrossRef PubMed.
- K. K. Angajala, S. Vianala, R. Macha, M. Raghavender, M. K. Thupurani and P. J. Pathi, SpringerPlus, 2016, 5, 423 CrossRef PubMed.
- R. Raj, P. Singh, P. Singh, J. Gut, R. J. Rosenthal and V. Kumar, Eur. J. Med. Chem., 2013, 62, 590 CrossRef CAS PubMed.
- D. R. Buckle, D. J. Outred, C. J. M. Rockell, H. Smith and B. A. Spicer, J. Med. Chem., 1983, 26, 251 CrossRef CAS PubMed.
- M. A. Bonache, S. M. Fernandez, M. Miguel, B. Sabater-Munoz and R. Gonzalez-Muniz, ACS Comb. Sci., 2018, 20, 694 CrossRef CAS PubMed.
- J. Brahmi, S. Bakari, S. Nasri, H. Nasri, A. Kadri and K. Aouadi, Mol. Biol. Rep., 2019, 46, 679 CrossRef CAS PubMed.
- R. Huisgen, Angew. Chem., Int. Ed. Engl., 1963, 2, 565 CrossRef.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596 CrossRef CAS PubMed.
- T. R. Chan, R. Hilgraf, K. B. Sharpless and V. V. Fokin, Org. Lett., 2004, 6, 2853 CrossRef CAS PubMed.
- P. Wu, A. K. Feldman, A. K. Nugent, C. J. Hawker, A. Scheel, B. Voit, J. Pyun, J. M. Frchet, K. B. Sharpless and V. V. Fokin, Angew. Chem., Int. Ed., 2004, 43, 3928 CrossRef CAS PubMed.
- M. Twhiting, J. C. Tripp, Y. C. Lin, W. Lindstrom, A. J. Olson, J. H. Elder, K. B. Sharpless and V. V. Fokin, J. Med. Chem., 2006, 49, 7697 CrossRef PubMed.
- E. J. Yoo, M. Ahlquist, S. H. Kim, I. Bae, V. V. Fokin, K. B. Sharpless and S. Chang, Angew. Chem., Int. Ed., 2007, 46, 1730 CrossRef CAS PubMed.
- W. Christian, T. C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057 CrossRef PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046 CrossRef CAS PubMed.
- J. C. Jewett, E. M. Sletten and C. R. Bertozzi, J. Am. Chem. Soc., 2010, 132, 3688 CrossRef CAS PubMed.
- J. C. Jewett and C. R. Bertozzi, Chem. Soc. Rev., 2010, 39, 1272 RSC.
- S. Deswal, N. Naveen, R. K. Tittal, V. D. Ghule, K. Lal and A. Kumar, J. Mol. Struct., 2020, 1209, 127982 CrossRef CAS.
- N. Naveen, R. K. Tittal, V. D. Ghule, N. Kumar, L. Kumar, K. Lal and A. Kumar, J. Mol. Struct., 2020, 1209, 127951 CrossRef.
- N. Naveen, R. K. Tittal, V. D. Ghule, P. Yadav, K. Lal and A. Kumar, Steroids, 2020, 161, 108675 CrossRef PubMed.
- P. K. Naikawadi, L. Mucherla, R. Dandela, M. Sambari and K. S. Kumar, Adv. Synth. Catal., 2021, 363, 3796 CrossRef CAS.
- L. Jing, Wu, X. Hao, F. A. Olotu, D. Kang, C. H. Chen, K. H. Lee, M. E. S. Solima, X. Liu, Y. Song and P. Zhan, Eur. J. Med. Chem., 2019, 183, 111696 CrossRef CAS PubMed.
- M. I. Mangione, R. A. Spanevello and M. B. Anzardi, RSC Adv., 2017, 7, 47681 RSC.
- N. Naveen, R. K. Tittal, P. Yadav, K. Lal and A. Kumar, New J. Chem., 2019, 43, 8052 RSC.
- S. Deswal, R. K. Tittal, P. Yadav, K. Lal, V. D. Ghule and N. Kumar, ChemistrySelect, 2019, 4, 759–764 CrossRef CAS.
- C. X. Zhao, T. Liua, M. Xua, H. Lina and C. Zhang, J. Chin Chem Lett., 2021, 32, 1925 CrossRef CAS.
- N. Ö. Can, U. Acar Çevik, B. N. Sağlık, S. Levent, B. Korkut, Y. Özkay, Z. A. Kaplancıklı and A. S. Koparal, J. Chem., 2017, 9387102 Search PubMed.
- L. M. Podust, T. L. Poulos and M. R. Waterman, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(6), 3068 CrossRef CAS PubMed.
- J. Eberhardt, D. S. Martins, A. F. Tillack and S. Forli, J. Chem. Inf. Model., 2021, 61(8), 3891 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comp. Chem., 2010, 31(2), 455 CAS.
- Dassault System BIOVIA, Discovery Studio Visualizer v17.2.0.16349, San Diego, 2016 Search PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 2001, 46(1), 3 CrossRef CAS PubMed.
- B. Shah, P. Modi and S. R. Sagar, Life Sci., 2020, 252, 117652 CrossRef CAS PubMed.
- T. Kampmann, R. Yennamalli, P. Campbell, M. J. Stoermer, D. P. Fairlie, B. Kobe and P. R. Young, Antiviral Res., 2009, 84(3), 234 CrossRef CAS PubMed.
- F. Shaikh, Y. Zhao, L. Alvarez, M. Iliopoulou, C. Lohans, C. J. Schofield, S. Padilla–Parra, S. W. I. Siu, E. E. Fry, J. Ren and D. I. Stuart, J. Med. Chem., 2019, 62(6), 2928 CrossRef CAS PubMed.
- MolinspirationChemoinformaticsBrastislava, Slovak Republic, available from: http://www.Molinspiration.com/cgibin/properties, 2014.
- A. Kumar, K. Lal, L. Kumar, A. Kumar, N. Naveen and R. K. Tittal, Res. Chem. Intermed., 2022, 48, 5089 CrossRef CAS.
- V. Kumar, K. Lal, A. Kumar, R. K. Tittal, M. B. Singh and P. Singh, Res. Chem. Intermed., 2023, 49, 917–937 CrossRef CAS.
- (a) L. H. Mendoza-Huizar and C. H. Rios-Reyes, J. Mex. Chem. Soc., 2011, 55, 142 CAS; (b) I. Piyanzina, B. Minisini, D. Tayurskii and J. F. Bardeau, J. Mol. Model., 2015, 21, 34 CrossRef PubMed; (c) M. Elango, R. Parthasarathi, V. Subramanian, U. Sarkar and P. K. Chattaraj, J. Mol. Struct., 2005, 723, 43 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision B.01, Gaussian Inc., Wallingford, 2010 Search PubMed.
- T. Koopmans, Physica, 1934, 1(1), 104 CrossRef.
- R. G. Parr, L. V. Szentpály and S. Liu, J. Am. Chem. Soc., 1999, 121(9), 1922 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37(2), 785 CrossRef CAS PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98(7), 5648 CrossRef CAS.
- Richa, N. Kushwaha, S. Negi, A. Kumar, E. Zangrando, R. Kataria and V. Sain, Dalton Trans., 2021, 50, 13699 RSC.
- Richa, S. Kumar, J. Sindhu, P. Choudhary, S. Jaglan, E. Zangrando, R. Kumar, S. C. Sahoo, V. Kumar, S. K. Mehta and R. Kataria, J. Mol. Str., 2021, 1228, 129460 CrossRef CAS.
- S. Kumar, V. Saini, I. K. Maurya, J. Sindhu, M. Kumari, R. Kataria and V. Kumar, PLoS ONE, 2018, 13(4), 0196016 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2192737 (4f). For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3nj00223c |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2023 |