Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorinated nanobodies for targeted molecular imaging of biological samples using nanoscale secondary ion mass spectrometry†
Selda
Kabatas
 *ab,
Paola
Agüi-Gonzalez
*ab,
Paola
Agüi-Gonzalez
 ab,
Rena
Hinrichs
ab,
Sebastian
Jähne
ab,
Rena
Hinrichs
ab,
Sebastian
Jähne
 ab,
Felipe
Opazo
ab,
Felipe
Opazo
 ab,
Ulf
Diederichsen
ab,
Ulf
Diederichsen
 c,
Silvio O.
Rizzoli
c,
Silvio O.
Rizzoli
 *ab and
Nhu T. N.
Phan
*ab and
Nhu T. N.
Phan
 *ab
*ab
aCenter for Biostructural Imaging of Neurodegeneration, University Medical Center Göttingen, von-Siebold-Straße 3a, 37075 Göttingen, Germany. E-mail: skabata@gwdg.de; srizzol@gwdg.de; thi.phan@med.uni-goettingen.de
bDepartment of Neuro- and Sensory Physiology, University Medical Center Göttingen, Humboldtallee 23, 37073 Göttingen, Germany
cInstitute for Organic and Biomolecular Chemistry, University of Göttingen, Tammannstrasse 2, 37077 Göttingen, Germany
First published on 24th April 2019
Abstract
Molecular imaging of targeted large biomolecules has been restricted in SIMS due to the limited number of probes containing SIMS-detectable isotopes. We introduce here new 19F-containing molecules that can be conjugated in a site-specific manner to nanobodies able to recognize fluorescent proteins (FPs) or mouse immunoglobulins (Igs). In this work, we demonstrate that it is possible to use the 19F-nanobodies to reveal the location of several cellular proteins previously tagged with FPs or Igs. This enables specific bio-imaging in SIMS for a vast repertoire of biomolecules, offering new opportunities to study specific structural and functional molecular interactions in biological specimens.
Secondary ion mass spectrometry (SIMS) has been widely known as a powerful imaging technique due to its high molecular specificity, high sensitivity, and spatial resolution. SIMS functions by probing the samples using a primary ion beam, such as Cs+, which causes the sputtering of secondary ions, which are then identified by using mass spectrometry detectors. The implemented ion source has a huge influence on the detected species, varying from elemental ions to molecular fragments with sizes of up to 2000 Da.1 The lateral resolution is also influenced by the primary beam. For example, nanoscale SIMS (nanoSIMS) utilizes highly reactive monatomic primary ion sources allowing the detection of elemental and small ions with a spatial resolution of around 50 nm.2 As a consequence of this high resolution, nanoSIMS has found increasing applications in biological studies. It allows the study of the biological composition of a sample at a spatial resolution comparable to that of super-resolution fluorescence microscopy and electron microscopy.
One drawback of nanoSIMS, which is common to other SIMS tools in general, is that it is difficult to identify specific cellular structures and organelles, with the exception of a few organelles having specific morphologies (like the nucleus or some large vacuoles).2–4 There has been a need in biological studies to identify different cellular structures and to localize specific proteins, in order to interpret the data accurately, and to understand the underlying molecular mechanisms of cellular processes.
Up to now, the ability to isotopically label specific proteins for SIMS imaging has been the major limiting factor in the mass spectrometry imaging field. The reason for this is the strict criteria of a suitable isotopic probe, which should be highly specific to the target and must have good ionization ability, as well as rare natural existence in the sample under study. Ideally the method to incorporate the probe into a defined biomolecule should be straightforward. Previous studies have already employed colloidal gold immunoprobes in which the gold was conjugated to an antibody able to recognize cellular actin and synaptophysin proteins.5 Another study utilized colloidal gold nanoparticles covered with a fluorinated monolayer, attached to a secondary antibody, for immunolabeling and imaging the influenza hemagglutinin virus in cells.6 Unfortunately, it has also been noticed that 197Au changes the ionization of the surrounding ions in the sample.7197Au is a heavy metal ion and it seems to hinder the simultaneous detection of other smaller biological ions such as 12C2, 12C14N, and 12C15N (in the case of nanoSIMS with 5 parallel detectors). In addition, the immunolabeling using primary and secondary antibodies coupled to colloidal gold could undermine the spatial resolution of the nanoSIMS imaging due to the possible clustering of antibodies and the large distance between the placed colloidal gold particles and the positions of the protein of interest.8,9 Alternatively, lanthanides coupled to antibodies have also been utilized in cellular specimens.10–12 However, lanthanide probes have the tendency to precipitate depending on the polymer backbone coupled to the metal.13 In addition, the large size of the polymer backbone coupled to the antibody results in an even larger distance between the detectable part of the probe (lanthanide) and the position of the proteins of interest. Elements with a small mass and size are more suitable for labeling probes in the complex cellular environment.8
Recently, different strategies using boron-containing probes have been proposed for the detection of positive secondary ions using nanoSIMS,14 including strategies of coupling boron to small immunoaffinity probes, which place the rare isotopes in the immediate vicinity of the epitopes of interest. This has not yet been possible for negative ions, however. These ions, which include the relevant species for C, N, O, P or S, are the most relevant for biological samples, and therefore optimal labeling probes are essential. We have developed in the past a fluorinated (19F) azide-containing probe named SK155 to label alkyne-modified proteins through copper(I)-catalyzed click chemistry,15 which enables 19F-labeling of the proteins of interest, for negative ions. As a proof of concept, SK155 was successfully applied to image the synaptic proteins syntaxin 1, syntaxin 13, and SNAP 25 in mammalian cells.15
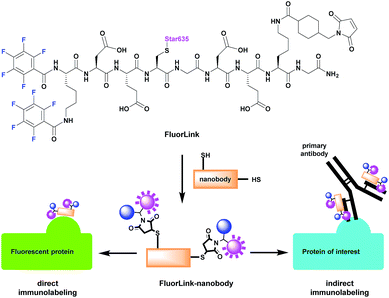
However, the azide-based technique is not very flexible, since the protein of interest needs to be genetically modified to an azide-reactive unnatural amino acid (UAA), the cells have to be fed with such amino acids, and other constructs need to be co-expressed, to help perform the UAA incorporation.14–16 This procedure is relatively difficult, and it cannot be performed for samples that are already fixed or not genetically modified, such as human pathology samples. To solve this problem, we have adapted our fluorine probe to make it suitable not only for click chemistry, but also for commonly used immunostaining techniques that use antibodies and nanobodies.14 The latter are especially relevant, since they are very small in size (∼3 nm). This helps to increase the labeling density and improves the spatial precision, as the 19F isotopes are placed closer to the intended proteins, in comparison to experiments in which conventional antibodies (12–15 nm in size) are used. Therefore, we conjugated our fluorine probe named FluorLink directly to nanobodies (Fig. 1).
To expand the applicability of this approach we used a nanobody that recognizes fluorescent proteins (widely used in biomedical research) and a nanobody that binds specifically to mouse immunoglobulins (commonly used for specific detection of a protein of interest). Finally, we have applied the fluorinated nanobodies to various target proteins in cells, to demonstrate that a broad spectrum of specific proteins can now be revealed using nanoSIMS.
Results and discussion
Our FluorLink molecule consists of a soluble peptide with a fluorinated N-terminus, a fluorophore on the cysteinyl side chain and a maleimide group on the lysine side chain (Fig. 1). The fluorophore as an additional feature allows the use of fluorescence microscopy before the nanoSIMS measurements, and the correlation of the images obtained by these two modalities. The synthesis was similar to previously published procedures14,15 starting with amino acid coupling on a solid support (ESI Section 3†). The peptide was synthesized from the C- to the N-terminus on an acid-sensitive resin applying standard fluorenylmethyloxycarbonyl-solid phase peptide synthesis (Fmoc-SPPS). After coupling of the last amino acid, Fmoc–Lys(Fmoc)–OH, the Fmoc groups were deprotected by basic treatment. Then, 1,2,3,4,5-pentafluorobenzoic acid was attached to the two amino groups of the N-terminal lysine. The completed peptide was cleaved from the solid support under acidic conditions. In the next step, the fluorophore Star635 maleimide was coupled to the cysteine side chain by a sulfhydryl–maleimide reaction. Finally, a maleimide-linker was attached to the lysine side chain to generate FluorLink with 13x 19F atoms per molecule.Afterwards, FluorLink was conjugated to two different nanobodies to be used in immunocytochemical experiments (ESI Section 4†). The chosen nanobodies have two ectopic cysteines to react with FluorLink molecules in a sulfhydryl–maleimide reaction, which result in doubling the 19F amount per detected target of interest.
Unfortunately, only a few nanobodies recognizing endogenous proteins present in cells are currently commercially available. However, nanobodies against fluorescent proteins (FPs) are readily available and they are known to have strong affinities.17 This is beneficial to our study, since they can be easily combined with cells producing the POI fused to a FP. We selected the nanobodies against a green fluorescent protein (GFP)18 and red fluorescent protein (mCherry)19 to validate the direct approach explained in Fig. 1. In order to increase the specific fluorine signal in nanoSIMS, we used two specific nanobodies able to bind simultaneously to their respective FPs (#1 and #2). Therefore, we were able to decorate each FP with up to 52x 19F atoms. For the indirect labeling approach, a nanobody against the kappa (κ) light chain of mouse immunoglobulins (κIg) was selected. Here, the advantage is the freedom to select any primary antibody raised in mice and label it afterwards with the FluorLink–nanobody anti-mouse κlg. This approach allows the detection of endogenous target proteins, avoiding the genetic manipulation to obtain POI fused to FPs or bearing clickable amino acids.14
The performance of FluorLink–nanobodies for both approaches was evaluated prior to nanoSIMS measurements in regard to any disturbing effects caused by FluorLink to the affinity of the nanobodies (ESI Section 6†). As FluorLink contains a fluorophore, it is possible to image the labeled cells using conventional epifluorescence microscopy. In all cases, it seems that the nanobody function was not impaired by conjugating them to two FluorLink moieties. The control images suggest that the nanobodies are functional and highly specific to their intended targets (ESI Section 6†).
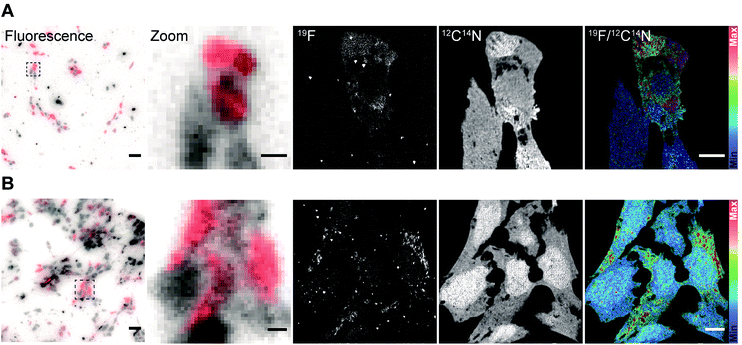
After validating the functionality and specificity of the FluorLink–nanobdies with the help of fluorescence microscopy, we tested them in nanoSIMS imaging (Fig. 2). To confirm the consistency of the nanobodies, we have targeted the same protein. First, human embryonic kidney (HEK293) cells were transfected with a plasmid encoding a mitochondrial membrane protein, TOM70 fused to a GFP. Afterwards, TOM70-GFP was detected using the combination of FluorLink–nanobody anti-GFP #1 and FluorLink–nanobody anti-GFP #2.
A similar procedure was carried out for labeling the same protein with the next set of nanobodies. In this case, the cells were transiently transfected with a vector coding for TOM70 fused to mCherry followed by immunolabeling with FluorLink–nanobodies anti-mCherry #1 and #2. Transient transfections of cells result in a mixed population of cells producing and not producing the chimera POI. As expected, the nanoSIMS images showed that the signal of 19F was significantly higher in the cells that displayed a significant fluorescence signal, compared to non-fluorescent signal in non-transfected cells, and it was much higher than the cell-free background signal. Additionally, a homogeneous signal of 12C14N was observed all over the cells proving that no topographical effect exists on the sample. The ratio image of 19F/12C14N showed that the 19F signal was localized in the cytoplasm, in round or tubule-like organelles, as expected for the mitochondria localization of TOM70-GFP (Fig. 2A) and TOM70-mCherry (Fig. 2B).
In addition, there was a good agreement between the fluorescence and nanoSIMS images in that the 19F signal was mainly detected in the strongly transfected cells, demonstrating that FluorLink–nanobody, as a direct labeling probe for specific proteins, exhibits high specificity, selectivity, and sufficient sensitivity for the detection with NanoSIMS.
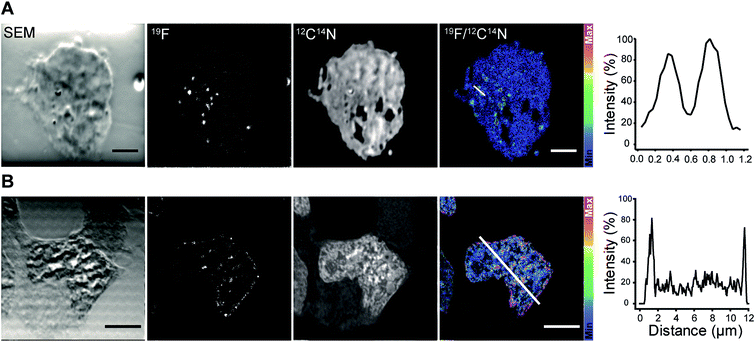
We continued with the indirect immunolabeling strategy. The procedure comprises labeling a target protein with a primary antibody, and subsequent binding to a secondary nanobody conjugated to FluorLink. For example, HEK293 cells were immunostained with an antibody detecting the protein PMP70 located on peroxisomes (Fig. 3). Peroxisome is an organelle present in most mammalian cells responsible for biosynthesis and degradation of different membrane lipids.20,21 This was followed by staining with FluorLink–nanobody anti-mouse κlg. From the nanoSIMS images of peroxisomes in the HEK293 cells labeled with FluorLink (Fig. 3A), 19F signals appear as very bright round shaped dots in the cells, as expected for the typical spherical shape of peroxisomes.
No topographical artifact was observed on the sample as demonstrated by the good and homogeneous signal obtained for the images of 12C14N and secondary electron microscopy (SEM). The line profile across the two adjacent peroxisomes in a cell shows a clearly distinguished 19F signal localized on the peroxisomes compared to the background signal.
Using a similar staining procedure, the FluorLink–nanobody anti-mouse κlg was also examined by detecting a primary antibody against lamin-B2, one of the major architectural proteins of the membrane of the cell nucleus22 (Fig. 3B). The 19F signal was observed at the nucleus edge, where most lamin-B2 molecules are expected to be localized, with a low 19F signal observed inside the nucleus (which is typical of lamin-B immunostaining).
For future applications, especially for low-abundance proteins, one may need to increase the 19F signals by various means. Classically this has been performed in immunohistochemistry by combined antibody packages. One stains the samples first with, for example, rabbit antibodies for the protein of interest. This is followed by incubations with goat anti-rabbit antibodies, which are then revealed by mouse anti-goat antibodies, thereby increasing manifold the number of antibodies per epitope. This approach, however, places the 19F signals far from the structures of interest, and therefore a more elegant application, such as the use of nanobodies engineered to contain multiple 19F probes, will probably be more efficient.
Conclusions
We have successfully synthesized a next-generation fluorine probe FluorLink, which can be conjugated to any surface or protein possessing a thiol group (cysteine). Here we have used this FluorLink to conjugate nanobodies in a site-directed fashion on two engineered cysteines. In order to make our approach as broad as possible, we have chosen two nanobodies that are commercially available and that detect molecules that are extensively used for imaging in the biomedical field (GFP-like proteins and mouse antibodies). These isotopic probes are well suited for imaging of various target proteins with subcellular resolution due to their high sensitivity of detection and high spatial precision of labeling. In addition to the beneficial labeling method with nanobodies, there is a possibility to use these FluorLink–nanobodies in both fluorescence and non-fluorescence imaging. These fluorinated nanobodies enable the imaging of specific protein distributions in correlation with the molecular turnover of the cells using non-optical imaging techniques like nanoSIMS. Importantly, they can also be used in other SIMS techniques such as ToF-SIMS, allowing simultaneous imaging of proteins of interest and other small species such as metabolites, phosphor peptides, sugars or lipids present in the same sample. Therefore, these new tools can be very useful for the understanding of cellular and molecular processes where typically multiple types of molecules are involved.Conflicts of interest
S. O. R and F. O are shareholders of NanoTag Biotechnologies GmbH. All other authors declare no conflict of interest.Acknowledgements
NanoSIMS measurements for Fig. 2 were obtained at the Chemical Imaging Infrastructure (CII), Chalmers University of Technology and University of Gothenburg, Sweden, which is supported by the Knut and Alice Wallenberg Foundation. We thank the nanoSIMS facility at the CII for the service of using their infrastructure. The work was supported by grants to S. O. R. from the European Research Council (ERC-2013-CoG NeuroMolAnatomy) and from the Deutsche Forschungsgemeinschaft (DFG) 1967/7-1 and by grants from DFG (SFB1286/B1) and VR (Swedish Research Council) to N. T. N. P.References
- H. Tian, L. J. Sparvero, A. A. Amoscato, A. Bloom, H. Bayır, V. E. Kagan and N. Winograd, Anal. Chem., 2017, 89, 4611–4619 CrossRef CAS PubMed.
- S. K. Saka, A. Vogts, K. Kröhnert, F. Hillion, S. O. Rizzoli and J. T. Wessels, Nat. Commun., 2014, 5, 3664 CrossRef PubMed.
- A. A. Legin, A. Schintlmeister, M. A. Jakupec, M. Galanski, I. Lichtscheidl, M. Wagner and B. K. Keper, Chem. Sci., 2014, 5, 3135–3143 RSC.
- J. Lovrić, J. Dunevall, A. Larsson, L. Ren, S. Andersson, A. Meibom, P. Malmberg, M. E. Kurczy and A. G. Ewing, ACS Nano, 2017, 11, 3446–3455 CrossRef PubMed.
- G. Thiery-Lavenant, C. Guillermier, M. Wang and C. Lechene, Surf. Interface Anal., 2014, 46, 147–149 CrossRef CAS PubMed.
- R. L. Wilson, J. F. Frisz, W. P. Hanafin, K. J. Carpenter, I. D. Hutcheon, P. K. Weber and M. L. Kraft, Bioconjugate Chem., 2012, 23, 450–460 CrossRef CAS PubMed.
- J. Pett-Ridge and P. K. Weber, Methods Mol. Biol., 2012, 881, 375–408 CrossRef CAS PubMed.
- M. Maidorn and S. O. Rizzoli, Biochem. J., 2016, 473, 3385–3399 CrossRef CAS PubMed.
- M. Mikhaylova, B. M. C. Cloin, K. Finan, R. van den Berg, J. Teeuw, M. M. Kijanka, M. Sokolowski, E. A. Katrukha, M. Maidorn, F. Opazo, S. Moutel, M. Vantard, F. Perez, P. M. P. van Bergen enn Henegouwen, C. C. Hoogenraad, H. Ewers and L. C. Kapitein, Nat. Commun., 2015, 6, 793 Search PubMed.
- X. Lou, G. Zhang, I. Herrera, R. Kinach, O. Ornatsky, V. Baranov, M. Nitz and M. A. Winnik, Angew, Chem. Int. Ed., 2007, 46, 6111–6114 ( Angew. Chem. , 2007 , 119 , 6223–6226 ) CrossRef CAS PubMed.
- M. Angelo, S. C. Bendall, R. Finck, M. B. Hale, C. Hitzman, A. D. Borowsky, R. M. Levenson, J. B. Lowe, S. D. Liu, S. Zhao, Y. Natkunam and G. P. Nolan, Nat. Med., 2014, 20, 436–442 CrossRef CAS PubMed.
- S. R. Adams, M. R. Mackey, R. Ramachandra, S. F. Palida Lemieux, P. Steinbach, E. A. Bushong, M. T. Butko, B. N. G. Giepmans, M. H. Ellisman and R. Y. Tsien, Cell Chem. Biol., 2016, 23, 1417–1427 CrossRef CAS PubMed.
- Nolan Lab Cytobank, http://www.cytobank.org/nolanlab, accessed March 2019.
- S. Kabatas, P. Agüi-Gonzalez, K.-A. Saal, S. Jähne, F. Opazo, S. O. Rizzoli and N. T. N. Phan, Angew. Chem., Int. Ed., 2019, 58, 3438–3443 ( Angew. Chem. , 2019 , 131 , 3476–3481 ) CrossRef CAS PubMed.
- I. C. Vreja, S. Kabatas, S. K. Saka, K. Kröhnert, C. Höschen, F. Opazo, U. Diederichsen and S. O. Rizzoli, Angew. Chem., Int. Ed., 2015, 54, 5784–5788 ( Angew. Chem. , 2015 , 127 , 5876–5880 ) CrossRef CAS PubMed.
- S. Kabatas, I. C. Vreja, S. K. Saka, C. Höschen, K. Kröhnert, F. Opazo, S. O. Rizzoli and U. Diederichsen, Chem. Commun., 2015, 51, 13221 RSC.
- J. Ries, C. Kaplan, E. Platonova, H. Eghlidi and H. Ewers, Nat. Methods, 2012, 9, 582–584 CrossRef CAS PubMed.
- R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509–544 CrossRef CAS PubMed.
- N. C. Shaner, R. E. Campbell, P. A. Steinbach, B. N. G. Giepmans, A. E. Palmer and R. Y. Tsien, Nat. Biotechnol., 2004, 22, 1567–1572 CrossRef CAS PubMed.
- G. M. Cooper, The Cell: A Molecular Approach, Peroxisomes, Sinauer Associates, Sunderland (MA), 2nd edn, 2000 Search PubMed.
- R. J. A. Wanders, S. Ferdinandusse, P. Brites and S. Kemp, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2010, 3, 272–280 CrossRef PubMed.
- T. A. Dittmer and T. Misteli, Genome Biol., 2011, 12, 222 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ja00117d |
| This journal is © The Royal Society of Chemistry 2019 |