Open Access Article
Open Access ArticleInfluence of peptide chirality on their protein-triggered supramolecular hydrogelation†
Shahaji H.
More
abc,
Tetiana
Dorosh
d,
Jean-Yves
Runser
abc,
Alexis
Bigo-Simon
 a,
Rachel
Schurhammer
a,
Rachel
Schurhammer
 d,
Vincent
Ball
bc,
Leandro
Jacomine
d,
Vincent
Ball
bc,
Leandro
Jacomine
 a,
Marc
Schmutz
a,
Marc
Schmutz
 a,
Alain
Chaumont
a,
Alain
Chaumont
 d,
Pierre
Schaaf
*abc and
Loïc
Jierry
d,
Pierre
Schaaf
*abc and
Loïc
Jierry
 *a
*a
aUniversité de Strasbourg, CNRS, Institut Charles Sadron (UPR22), 23 Rue du Loess, 67034 Strasbourg Cedex 2, BP 84047, France
bInstitut National de la Santé et de la Recherche Médicale, INSERM Unité 1121, CRBS, 1 Rue Eugène Boeckel, 67000 Strasbourg Cedex, CS 60026, France
cUniversité de Strasbourg, Faculté de Chirurgie Dentaire, 8 Rue Sainte Elisabeth, 67000 Strasbourg, France
dUniversité de Strasbourg, Faculté de Chimie, UMR7140, 1 Rue Blaise Pascal, 67008 Strasbourg Cedex, France. E-mail: schaaf@unistra.fr; ljierry@unistra.fr
First published on 15th February 2025
Abstract
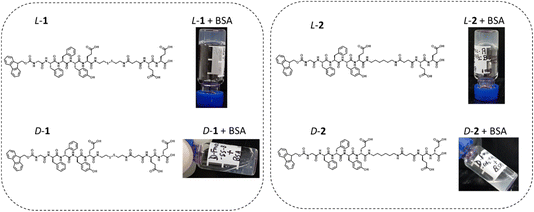
Many articles describe the use of enzymes to induce the formation of a supramolecular hydrogel. These enzymes catalyze the transformation of water-soluble precursors, often short peptides, into hydrogelators. The use of non-enzymatic proteins to induce or stabilize peptide self-assembly is a rarely reported phenomenon, which raises fundamental questions: how can a protein induce peptide self-assembly? How is the peptide recognized and how does it, or the peptide assembly, interact with the protein? The heptapeptide Fmoc-GFFYE–NH–(CH2)2–s-s–(CH2)2–NH–CO–(CH2)2–CO–EE–OH, called L-1 (L = natural chiral amino acids), is a water-soluble compound leading to an increasingly viscous solution over time due to the formation of nanofibers, but does not result in hydrogel (at least not within 3 months). When bovine serum albumin (BSA) is added to a freshly prepared solution of L-1, a hydrogel is obtained in less than 10 min. The variation in the L-1/BSA ratio has an impact on the gelation rate and the mechanical properties of the resulting hydrogel. Thus, the protein appears to act as (i) a catalyst and (ii) a cross-linking point. Strikingly, if the enantiomer D-1 (D = unnatural chiral amino acids) is used instead of L-1, the mixture with BSA remains liquid and non-viscous. Similar behavior is also observed for other proteins. Spectroscopic analyses (CD, fluorescence) and electronic microscopy images confirm that the L-1 peptide self-assembles in nanofibers of 10 nm diameter through β-sheet organization, which is not the case for the peptide D-1. A molecular dynamics study shows that BSA is capable of interacting with both enantiomer peptides L-1 and D-1. However, interaction with L-1 tends to unfold the peptide backbone, making the interaction with the protein more stable and promoting the assembly of L-1 peptides. Conversely, the interaction between BSA and D-1 is more dynamic and appears to be less spatially localized on the BSA. Furthermore, in this interaction, the D-1 peptide keeps its globular conformation. These results highlight the impact of a short peptide’s chirality on protein-triggered supramolecular hydrogelation.
Introduction
Chirality and self-assembly are two features intrinsically related, which characterize all living systems. Firstly, almost all natural amino-acids besides glycine are left-handed (L) resulting in chiral proteins. Secondly, all biological organisms are delimited by self-assembled walls, and cellular metabolism relies on self-assembled structures such as actin or myosin fibers, for instance. The interactions of such self-assembled structures with proteins relies on recognition processes that are possible thanks to the chiral nature of their constituents.1 Thus, chirality plays a crucial role in molecular recognition.2 Understanding how chirality drives self-assembly is thus both of fundamental and practical importance for development in a wide variety of areas.3Over the last two decades, amino acids and short peptide self-assembly have attracted considerable attention from the supramolecular scientific community.4 The resulting assemblies can be induced in various ways, for example by annealing, solvent exchange, light adsorption, pH change or the addition of metal ions.5 In 2004, Xu's group illustrated the possibility of initiating the self-assembly process by another route, which involves an enzyme to transform an amphiphilic precursor compound into a hydrogelator.6 In a very simplified view, the enzyme cuts a chemical bond of the precursor, by hydrolysis or redox process, generating a less soluble peptide capable of self-assembling and leading to the production of organic nanostructures that support the formation of supramolecular hydrogels.7 Examples of enzymes capable of inducing the formation of peptide hydrogels by peptide bond formation have also been reported.5,8 In separate work, it was shown that the role of the enzyme is not limited solely to its catalytic role towards the precursor, but that it is also involved in the nucleation process of peptide assemblies and their resulting nanostructures.9,10 In a chemical system where peptides are able to self-assemble without the enzymatic trigger, Sefcik and Ulijn showed in 2013 that the presence of protein clusters, even at low concentration, can result in a reversal of the chiral peptide organization.11 In addition, this cooperative interaction between peptide self-assembly and proteins can also impact the morphological and mechanical properties of the resulting nanostructured hydrogels. In 2020, Roy and collaborators showed for the first time the hydrogelation of nongelator peptides thanks to protein addition.12 Proteins can interact through non-covalent bonds with aggregate-like peptide assemblies and thus induce fibrillar nanostructure formation. Proteins also interact with self-assembled structures, and it has been shown that the chirality of the self-assembled structure can play an important role in this interaction. For example, L/D glutamic-acid-based amphiphiles which possess molecular chirality and which can be self-assembled in left/right handed nanofibers through solvent effects, present an adsorption efficiency with bovine serum albumin (BSA) that depends mainly on the self-assembled chirality.13 Molecular dynamics simulations show a 2.6–2.8-fold higher interaction energy of BSA with left, compared to right-handed, helical nanofibers. The influence of the chirality of the self-assembled structure also shows up by a different interaction between chiral self-assembled structures with cells and bacteria. It is therefore quite clear that proteins can induce self-assembly even in the absence of enzymatic action and peptide self-assemblies interact with proteins in a chiral dependent manner. However, as far as we know, the influence of the chirality of the peptides on their self-assembly induced by a protein, has never been investigated.
Here, we have studied the ability of short peptides made of L or D amino acids, to self-assemble in nanostructures resulting in supramolecular hydrogels. Since cystamine-based peptides, and their disulfide reduced forms, are known to interact with proteins to produce stable hydrogels, we have used the following peptide sequence Fmoc-GFFYE–NH–(CH2)2–s-s–(CH2)2–NH–CO–(CH2)2–CO–EE–OH as the model peptide (G, glycine; F, phenylalanine; Y, tyrosine; E, glutamic acid).14L-1 refers to the peptide sequence constituting amino acids having natural L chirality, and D-1 the peptide sequence constituting amino acids having non-natural D chirality (Fig. 1). First, we studied the impact of the presence of BSA, and others proteins, on aqueous solutions of L-1 or D-1 using spectroscopic methods, dynamic rheology and electronic microscopy. Then, we monitored the L-1 or D-1 self-assembly over time, in the absence or presence of BSA, using transmission electronic microscopy (TEM). The impact of BSA on already formed self-assembled L-1 or D-1 nanofibers is also investigated. Thus, based on these investigations and also considering the influence of the ratio between BSA and L-1 on hydrogel formation and on mechanistic insights arising from molecular dynamics simulations, a route for protein-induced self-assembled peptide nanofibers is discussed.
Experimental methods
Materials
The materials, synthesis and characterization of peptides L-1, D-1, L-2 and D-2 are given in ESI.†PBS buffer preparations: one tablet of commercially available PBS (P4417 from Sigma-Aldrich) was dissolved in 200 mL of ultrapure water (Milli-Q Plus system, Millipore, Billerica, MA) leading to 0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride. The buffer was adjusted to pH 7.4 by addition of a few drops of HCl (0.01 M) or NaOH (0.01 M) solution. The pH value was monitored using a calibrated pH meter LAQUAtwin® from HORIBA.
Hydrogel formation triggered by BSA: appropriate quantity of the peptide was dissolved in PBS buffer (pH = 7.4) to obtain the relevant concentration (usually 1, 5, 7.5 or 10 mg mL−1). Samples were sonicated in an ultrasound bath for 1 minute and agitated until a clear solution was obtained. The resulting solution was used for the rest of the analyses. BSA solution was added. The inverted vial test was performed starting from 10 min.
HPLC analyses
Analytical high-performance liquid chromatography (HPLC) was carried out with a 1100 Series from Agilent Technologies. Chromatograms were recorded and analyzed by the OpenLab ChemStation Agilent 1100 software. Samples with and without BSA were prepared in PBS (pH = 7.4) and all sample solutions were filtered before injection. The HPLC column Supelcosil ABZ+Plus (cat#: 59194C30) product of Sigma-Aldrich with dimensions 15 cm × 4.6 mm, 3 μm particle size was used. Solvent A: acetonitrile (100%) and solvent B: Milli-Q water with 0.1% v/v trifluoroacetic acid. Gradient: from 30% (0 min) solvent A to 70% (20 min) solvent A. Flow rate: 0.5 mL min−1. Injection volume: 20 μL. Concentration: 1 mg mL−1.Rheology
Rheological properties were measured with a Kinexus ultra+ Malvern rheometer using a cone-plate geometry of 40 mm. Generally, 1080 μL of peptide were mixed with 120 μL of BSA in 1.5 mL Eppendorf and mixed 3–5 times with the pipette. The gelation process was followed at a fixed frequency of 1 Hz and 2.5% strain for 120 minutes. Subsequently, strain measurements were carried out from 0.01% to 100% at 1 Hz and frequency sweeps were performed from 0.01 Hz to 20 Hz at a fixed strain of 0.5% to determine the linear viscoelastic domain of the hydrogels.Transmission electron microscopy
The TEM images were obtained with samples prepared in liquid (diluted solutions or diluted hydrogels). 5 μL of the solutions (hydrogels) were deposited onto a fresh glow discharge carbon-covered grid (400 mesh). The drop is left for 1 minute and the grid is negatively stained with 5 μL uranyl acetate (2% in water) for another minute and finally dried using filter paper. The grids were observed at 200 kV with a Tecnai G2 (FEI) microscope. Images were acquired with a ssCCD camera Eagle 2k (FEI).Circular dichroism
Circular dichroism (CD) spectra were recorded using a Jasco J-1700 spectropolarimeter. A 400 μL volume cuvette of 1 mm pathlength was used. Before measuring the peptide + BSA solutions, spectra of solutions containing only BSA were measured as a baseline. Therefore, the contribution of BSA is subtracted from the CD spectra. CD spectra were recorded between 320 and 200 nm using the following parameters: data pitch 0.5 nm, D.I.T. 1 s, bandwidth 1 nm, scanning speed 50 nm min−1 and with 3 accumulation. Spectra were measured at 25 °C using a Peltier apparatus with an accuracy of ±0.2 °C.Fluorescence spectroscopy
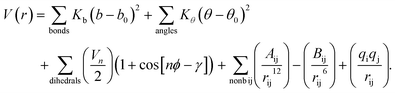 | (1) |
Force field parameters for BSA and the amino acids of peptides 1 were taken from the ff14SB force field,16 while those for all other groups were taken from the GAFF force field.17 Water is represented by the TIP3P of Jorgensen et al.18 and FF parameters of Na+ and Cl− were taken from the work of Joung and Cheatham19
The initial structure for BSA was taken from the crystal structure 3V03 reported by Majorek et al.20 The BSA was initially inserted in a box of pure water of about 150 Å length. Water molecules within 2 Å of the BSA were then removed from the box. Additionally, 1 or 4 Fmoc-ASSB peptides were then placed in the vicinity of the BSA, again removing water molecules within 2 Å of the peptide. Finally, Na+ and Cl− ions were added to the system to neutralize it.
We first performed 5000 minimisation steps (the first 2500 steps were done using the steepest descent method and the final 2500 steps using the conjugate gradient method), which was then followed by a 1 ns MD simulation during which the system was slowly heated to the desired temperature of 298.15 K. During this simulation both the position of the protein and the peptide were constrained. A further 2 ns of MD in the NVT ensemble at 298.15 K was undertaken where only water molecules and ions were allowed to move, in order to relax the solvent around both the peptide and the protein. This was then further followed, by an MD simulation of 3 ns in the NVT ensemble, during which only the protein backbone was constrained and the protein side chains as well as the peptide were allowed to move. Finally, a 2 ns simulation in the NVT ensemble was performed in order to allow the system to fully relax.
Additionally, a 2 ns trajectory in the NPT ensemble was performed to reach the correct density of the system. Once the system fully equilibrated, production runs of 0.25–0.75 μs were then performed. Each system was run 3 times with slightly modified starting conditions to check reproducibility. During the simulation the temperature was controlled via the weak coupling algorithm with a coupling constant of 1 ps.21 During all simulations we applied 3D periodic boundary conditions and a 12 Å cut-off for both van der Waals and electrostatic interactions. Long-range electrostatic calculations were then calculated using the PME summation. Analyses were done using either our in-house MDS software or the VMD software. Typical snapshots shown were also produced using the VMD software.22
Results and discussion
Triggering natural chirality peptide self-assembly and supramolecular hydrogel formation by proteins
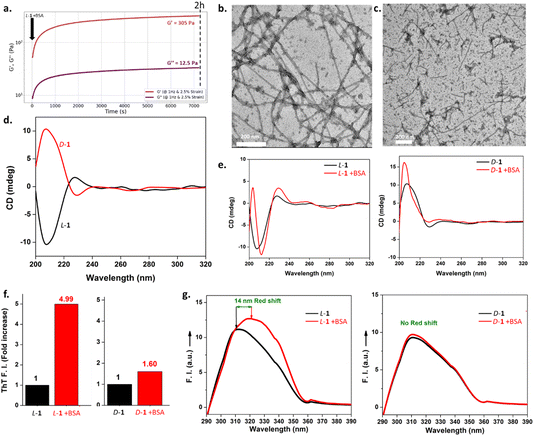
Each L-1 and D-1 enantiomer's peptide (5 mg mL−1) is totally soluble in phosphate-buffered saline solution (PBS, 10 mM) at pH 7.4 and 25 °C resulting in a homogeneous and transparent solution. When BSA (1 mg mL−1, Mw = 66 kDa) is added to each of the freshly prepared L-1 and D-1 solutions, the L-1 sample immediately shows an increase in viscosity and after less than 10 min, the entire viscous liquid has become a hydrogel, as checked by a simple inverted vial test (Fig. 1). On the other hand, the D-1 solution supplemented with BSA, remains liquid and we do not observe any increase in its viscosity. HPLC analysis of both samples shows no cleavage of the disulfide bridge, meaning that the protein must be a trigger of the hydrogelation process only in the case of the peptide with natural chirality L-1 (Fig. S1a†). We also synthesized a peptide 2, identical to peptide 1 except by the presence of an ethylene group instead of the disulfide bridge (Fig. 1). When the enantiomers L-2 and D-2 solutions (5 mg mL−1) in PBS (10 mM, pH 7.4, 25 °C) are mixed with BSA, hydrogelation is observed only in the case of the L-2 peptide.The rheological studies confirm that the D-1 solution remains liquid despite the addition of BSA, whereas the L-1 solution switches to hydrogel within the time needed to perform the rheological measurement (Fig. 2a). The values of storage G′ and loss G′′ moduli measured after 2 h are very low, 305 and 12.5 Pa respectively, highlighting the low mechanical robustness of such a material. However, these values still increase with time, meaning that complete hydrogelation is not yet achieved. TEM images of the gel sample L-1 + BSA show unambiguously the presence of nanofibers several hundreds of nanometers long, entangled together, having a diameter of roughly 10 nm in the case of the peptide of natural chirality L-1 (Fig. 2b). This nanofibrous network thus underpins the hydrogel formation. In the case of D-1 + BSA, amorphous organic matter and a few short-length nanofibers are observed (Fig. 2c). It must be noted that these TEM images given in Fig. 2b and c were recorded one week after the mixing of peptides L-1 or D-1 with BSA. Circular dichroism (CD) spectroscopy was used to monitor the supramolecular organization triggered by the presence of BSA. To this end, it was necessary to use very low concentrations of peptides (0.2 mg mL−1) in order to obtain a suitable high-tension value lower than 500 V for almost the entire spectral window studied (Fig. S2†). In these conditions, the CD spectra intensities of L-1 and D-1 peptide are low but obviously they are mirror images (Fig. 2d). After the addition of BSA (0.04 mg mL−1) in each enantiomer solution, the CD spectra is immediately recorded. Peptide L-1 alone showed a less intense positive band at 228 nm and one high intense negative band with a minimum centered at 209 nm, suggesting a disordered conformation. After BSA addition, the CD spectra changes since a new strong positive band at 204 nm appeared with crossover at 206 nm. Moreover, the original negative CD band is red shifted by 5 nm with a new increased negative minimum appearing at 213 nm (Fig. 2e). This profile of CD bands is typically assigned to the β-sheet conformation and the increase of the band intensities correlated with the increased peptide assemblies. To estimate the conformational change from disordered to ordered β-sheet conformation triggered by the BSA addition, CD spectra were subjected to BESTSEL online software available for secondary structure and fold recognition estimation. In the spectra of L-1 + BSA the estimated antiparallel β-sheet conformation was 72%. Whereas the peptide alone showed only 30% of antiparallel β-sheet content and the major contribution comes from disordered structure (Fig. S3†). In the case of D-1, only slight variations of the CD spectra are observed after the addition of BSA which does not correspond to a β-sheet but still shows interactions with the protein (Fig. 2e). To confirm the β-sheet peptide organization in the presence of BSA, thioflavine (ThT) is added to L-1 and D-1 solutions before being brought into contact with BSA. ThT is a dye well- known to stain and specifically reveal β-sheets in solution, in particular for short peptide hydrogels.23 Compared to L-1 alone, the mixture L-1 + BSA displayed a 5-fold increase in ThT fluorescence emission intensity (Fig. 2f). In the case of D-1 + BSA, the fluorescence emission intensity in presence of ThT is almost equivalent to the one measured without BSA. Finally, Fmoc bearing peptides are known to be stacked through π–π interactions in peptide self-assemblies.24 We measure a characteristic Fmoc excimer fluorescence emission for L-1, which displayed a maximum fluorescence intensity at 309 nm whereas the L-1 + BSA mixture exhibits a fluorescence intensity at 323 nm (Fig. 2g). This 14 nm red shift with enhancement of the intensity indicates the stacking of Fmoc groups when BSA is added. On the contrary, the fluorescence emission spectra of D-1 almost overlapped with the one recorded after the addition of BSA. All these investigations confirm that the presence of BSA induces the self-assembly of peptide L-1 through β-sheets and enables the structuring into nanofibers, leading to the formation of a hydrogel. In contrast, BSA is not able to induce the peptide D-1 self-assembly, explaining the absence of hydrogelation in this case.
More strikingly, when one of the following proteins such as alkaline phosphatase (AP) or carbonic anhydrase (CA) is mixed with L-1 in the same conditions as those described here above, a self-supporting hydrogel is observed within 48 h, whereas a liquid state is systematically observed with the D-1 peptide enantiomer (Fig. 3a and b). When urease (U) and β-galactosidase (β-GAL) are used as potential protein triggers, only a viscous liquid is obtained in 48 h with the natural (L) enantiomer (Fig. 3c and d). But whatever the protein considered – AP, CA, U or β-GAL – TEM analyses show the presence of nanofibrous structures when the L-1 peptide is involved, as shown in Fig. 3.
To obtain insight into the role of proteins in the hydrogelation mechanism, we monitored the nanofibrous network formation over time using TEM, when L-1 or D-1 are brought into contact with BSA.
TEM monitoring of BSA-triggered self-assembly overtime
By observing a solution of L-1 and D-1 left at room temperature (21–25 °C) for more than three months, we noticed that these two solutions were viscous. These two samples are called “aged samples”. Despite the time spent, the two peptides remained intact according to HPLC analyses, meaning that no peptide bond hydrolysis occurred (Fig. S1b†). TEM analysis of these solutions showed a very high degree of entanglement of the long fibers with a diameter of 10 nm (Fig. 4a), a value equivalent to the one observed when L-1 is mixed with BSA. Thus, it shows that each enantiomer L and D, of peptide 1, can self-assemble spontaneously, albeit very slowly, leading to the formation of a nanofibrous network, but unable to support hydrogel formation, at least not within 3 months. Monitoring of the L-1 peptide (5 mg mL−1, PBS 10 mM pH 7.4) solution evolution was realized by TEM at t = 1 h, 24 h and 48 h. Typical TEM images are given in Fig. 4b and show the presence of![[left double angle bracket]](https://www.rsc.org/images/entities/char_27ea.gif) dots
dots![[right double angle bracket]](https://www.rsc.org/images/entities/char_27eb.gif) of 1–2 nm in diameter at t = 1 h, which must correspond to the formation of micelles or aggregates inherent to the amphiphilic nature of peptide L-1. However, we already observe a few nanofibers of 10 nm in diameter but they are short in length, i.e. less than one hundred nanometers. After 24 hours in solution, the number of peptide L-1 aggregates still appears to be very high, but there are also many so-called “nanofiber nuclei”, some of which are still growing. Some nanofibers are more than one hundred nanometers long. After 48 hours, we still observe a few peptide aggregates but mainly more growing fibers. The TEM image series of the D-1 peptide (5 mg mL−1) in PBS solution (10 mM, pH 7.4) is quasi-identical to the one of the L-1 peptide. To assess the impact of the presence of BSA on the L-1 peptide nanofiber assembly process, we carried out the same TEM monitoring over time by mixing peptide L-1 and BSA at t = 0. After one hour (t = 1 h), nanofibers of several hundred nanometers were visible (Fig. 4c), in contrast to what is observed without BSA. We notice that there are two nanofiber diameters: nanofibers with a diameter of 10 nanometers, which are present in the majority of cases; and more rarely, thinner nanofibers with a diameter of 6–7 nanometers. Peptide micelles, easily visible in the case without BSA, seem to have given way to larger globular objects that appear to be made of associations of several micelles and probably protein clusters as well. Magnification of the TEM images reveals the characteristic presence of aggregated proteins, which is not observed in the absence of BSA. More interestingly, many of these globular objects are present at the ends of the growing nanofibers. After 24 hours, the nanofibers are clearly longer than in the absence of BSA. Again, a few 7 nm thick nanofibers remain present. We also note that located very near to the end of the nanofibers, there are either very small pieces of growing fibers and/or globular objects. Two days after mixing BSA and L-1 together, a mat of entangled nanofibers several micrometers long is observed, the ends of which seem impossible to find. Despite careful checking of the TEM grids, we were unable to find any thin nanofibers, with all nanofibers having the same diameter of around 10 nm. When BSA is mixed with D-1 peptide in PBS (10 mM, pH 7.4), we do not observe over time t = 1, 24 and 48 h, the same strong promoting effect towards nanofibers formation than with the naturally chiral L-1 peptide (Fig. 4c). More strikingly, when BSA is added to the aged samples of L-1 or D-1 peptide solution, we observe a completely different behavior. In the case of the L-1, a self-supporting hydrogel is formed in less than 2 min and a very dense nanofibrous network is observed by TEM (Fig. 4d). In the case of D-1, 48 h later, it seems that this aged sample solution has become less viscous, and broken nanofibers are observed by TEM. These simple experiments suggest that BSA is not only a catalyst for L-1 peptides self-assembly and a trigger of hydrogelation, but it is also a disassembling agent for the assembly of the non-natural D-1 peptides.
of 1–2 nm in diameter at t = 1 h, which must correspond to the formation of micelles or aggregates inherent to the amphiphilic nature of peptide L-1. However, we already observe a few nanofibers of 10 nm in diameter but they are short in length, i.e. less than one hundred nanometers. After 24 hours in solution, the number of peptide L-1 aggregates still appears to be very high, but there are also many so-called “nanofiber nuclei”, some of which are still growing. Some nanofibers are more than one hundred nanometers long. After 48 hours, we still observe a few peptide aggregates but mainly more growing fibers. The TEM image series of the D-1 peptide (5 mg mL−1) in PBS solution (10 mM, pH 7.4) is quasi-identical to the one of the L-1 peptide. To assess the impact of the presence of BSA on the L-1 peptide nanofiber assembly process, we carried out the same TEM monitoring over time by mixing peptide L-1 and BSA at t = 0. After one hour (t = 1 h), nanofibers of several hundred nanometers were visible (Fig. 4c), in contrast to what is observed without BSA. We notice that there are two nanofiber diameters: nanofibers with a diameter of 10 nanometers, which are present in the majority of cases; and more rarely, thinner nanofibers with a diameter of 6–7 nanometers. Peptide micelles, easily visible in the case without BSA, seem to have given way to larger globular objects that appear to be made of associations of several micelles and probably protein clusters as well. Magnification of the TEM images reveals the characteristic presence of aggregated proteins, which is not observed in the absence of BSA. More interestingly, many of these globular objects are present at the ends of the growing nanofibers. After 24 hours, the nanofibers are clearly longer than in the absence of BSA. Again, a few 7 nm thick nanofibers remain present. We also note that located very near to the end of the nanofibers, there are either very small pieces of growing fibers and/or globular objects. Two days after mixing BSA and L-1 together, a mat of entangled nanofibers several micrometers long is observed, the ends of which seem impossible to find. Despite careful checking of the TEM grids, we were unable to find any thin nanofibers, with all nanofibers having the same diameter of around 10 nm. When BSA is mixed with D-1 peptide in PBS (10 mM, pH 7.4), we do not observe over time t = 1, 24 and 48 h, the same strong promoting effect towards nanofibers formation than with the naturally chiral L-1 peptide (Fig. 4c). More strikingly, when BSA is added to the aged samples of L-1 or D-1 peptide solution, we observe a completely different behavior. In the case of the L-1, a self-supporting hydrogel is formed in less than 2 min and a very dense nanofibrous network is observed by TEM (Fig. 4d). In the case of D-1, 48 h later, it seems that this aged sample solution has become less viscous, and broken nanofibers are observed by TEM. These simple experiments suggest that BSA is not only a catalyst for L-1 peptides self-assembly and a trigger of hydrogelation, but it is also a disassembling agent for the assembly of the non-natural D-1 peptides.
Influence of the molar ratio L-1 peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) BSA, on hydrogelation
BSA, on hydrogelation
The conditions used so far are based on 5 mg mL−1 of L-1 peptide and 1 mg mL−1 of BSA, which correspond to a molar ratio of L-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA ≈ 240
BSA ≈ 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Keeping a constant L-1 peptide concentration of 5 mg mL−1, we varied the proportion of BSA and thus evaluated the effect of the L-1
1. Keeping a constant L-1 peptide concentration of 5 mg mL−1, we varied the proportion of BSA and thus evaluated the effect of the L-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA molar ratio, first on the formation, or not, of self-supported hydrogels by a simple inverted vial test. When the BSA concentration is 0.0001, 0.001, 0.01, 0.1, 0.25, 0.5, 1, 2.5, 5, 10 or 20 mg mL−1, the corresponding BSA
BSA molar ratio, first on the formation, or not, of self-supported hydrogels by a simple inverted vial test. When the BSA concentration is 0.0001, 0.001, 0.01, 0.1, 0.25, 0.5, 1, 2.5, 5, 10 or 20 mg mL−1, the corresponding BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L-1 molar ratio is 1
L-1 molar ratio is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240
240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960, 1
960, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480, 1
480, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240, 1
240, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96, 1
96, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48, 1
48, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24 and 1
24 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12, respectively. 24 hours after mixing the L-1 peptide with each of the different proportions of BSA, we observe the formation of a self-supporting hydrogel in all cases, except in the extreme ones when the BSA concentration is less than 0.1 mg mL−1 or higher than 20 mg mL−1 (Fig. 5a). Regular monitoring over time of self-supported hydrogel formation when the BSA concentration is 0.0001, 0.5, 1, 2.5, 10 and 20 mg mL−1 indicates that a self-supported hydrogel is observed in less than 10 min for BSA concentrations of 1 and 2.5 mg mL−1 (Fig. S4†). Apart from these two concentrations, obtaining a self-supported hydrogel is slowed down when the L-1/BSA ratio is increased or decreased: when the BSA concentration is 0.5 or 10 mg mL−1, it takes more than 1 h or 3 h, respectively, to obtain a self-supported hydrogel. A concentration of 0.0001 or 20 mg mL−1 of BSA leads to the formation of a hydrogel after three months only in the case where the BSA concentration is the highest, i.e. at 20 mg mL−1: in this case, the self-assembled L-1 peptide nanofibers are almost completely covered with organic matter, as observed by TEM (Fig. S5†), that can be assumed to be BSA. The use of a rheometer equipped with a cone-plane geometry allows us to determine the gel point and the evolution of mechanical features such as G′ and G′′. Five BSA concentrations were studied: 0.0001, 0.5, 1, 2 and 20 mg mL−1. With the exception of the lowest BSA concentration, the other four concentrations (0.5, 1, 2 and 20 mg mL−1) show that the value of G′ is greater than that of G′′ in less than 2 min (Fig. S6†). As these two values continued to evolve over time beyond several hours as already mentioned, we plotted the value of G′ measured after 2 h for each BSA concentration (Fig. 5b): a bell-shaped curve can thus be observed, showing that a more mechanically robust hydrogel was obtained for BSA concentration values of the order >0.5 to <2 mg mL−1, when 5 mg mL−1 of L-1 is used.
12, respectively. 24 hours after mixing the L-1 peptide with each of the different proportions of BSA, we observe the formation of a self-supporting hydrogel in all cases, except in the extreme ones when the BSA concentration is less than 0.1 mg mL−1 or higher than 20 mg mL−1 (Fig. 5a). Regular monitoring over time of self-supported hydrogel formation when the BSA concentration is 0.0001, 0.5, 1, 2.5, 10 and 20 mg mL−1 indicates that a self-supported hydrogel is observed in less than 10 min for BSA concentrations of 1 and 2.5 mg mL−1 (Fig. S4†). Apart from these two concentrations, obtaining a self-supported hydrogel is slowed down when the L-1/BSA ratio is increased or decreased: when the BSA concentration is 0.5 or 10 mg mL−1, it takes more than 1 h or 3 h, respectively, to obtain a self-supported hydrogel. A concentration of 0.0001 or 20 mg mL−1 of BSA leads to the formation of a hydrogel after three months only in the case where the BSA concentration is the highest, i.e. at 20 mg mL−1: in this case, the self-assembled L-1 peptide nanofibers are almost completely covered with organic matter, as observed by TEM (Fig. S5†), that can be assumed to be BSA. The use of a rheometer equipped with a cone-plane geometry allows us to determine the gel point and the evolution of mechanical features such as G′ and G′′. Five BSA concentrations were studied: 0.0001, 0.5, 1, 2 and 20 mg mL−1. With the exception of the lowest BSA concentration, the other four concentrations (0.5, 1, 2 and 20 mg mL−1) show that the value of G′ is greater than that of G′′ in less than 2 min (Fig. S6†). As these two values continued to evolve over time beyond several hours as already mentioned, we plotted the value of G′ measured after 2 h for each BSA concentration (Fig. 5b): a bell-shaped curve can thus be observed, showing that a more mechanically robust hydrogel was obtained for BSA concentration values of the order >0.5 to <2 mg mL−1, when 5 mg mL−1 of L-1 is used.
In addition, when a BSA concentration of 1 mg mL−1 is maintained and the L-1 peptide concentration is varied by 0.625, 1.25, 2.5, 5 and 10 mg mL−1, corresponding to an L-1/BSA molar ratio of 30/1, 60/1, 120/1, 240/1, and 480/1, respectively, a self-supporting hydrogel is obtained in less than 10 min only in the case of the highest L-1 peptide concentrations, i.e., 5 and 10 mg mL−1 (Fig. 5c). In the other cases, even after 24 hours, the mixtures of L-1 peptide and BSA remain liquid.
Molecular dynamics simulations
To further gain insight into the interactions between BSA and L-1 or D-1 peptide at the molecular scale, we have performed molecular dynamics simulations (MD). Initially, a single peptide 1 was placed at about 15 Å from three different sites around the BSA, corresponding to well-known adsorption sites of the latter commonly called Sudlow's site I, II and III (Fig. S7†).25 Each condition was replicated three times, called Run 1, 2 and 3. After 0.25 μs of MD, we observe that in all simulations realized, both L-1 or D-1 are found to adsorb on the protein surface. While in most cases after 0.25 μs the peptide is found on the surface region of the protein close to where the peptide was initially positioned, in some cases the peptide moved to other regions of the protein, pointing to the fact that both the L-1 and D-1 strongly interact with the BSA protein but that there is no preferential or specific area of interaction between the surface of the BSA and the two enantiomeric peptides. On average, the L-1 peptide forms about 1.8 ± 0.8 hydrogen bonds with the BSA while the D-1 is found to only form 1.4 ± 0.6, thus highlighting that the peptide of natural chirality interacts somewhat stronger with the protein than the unnatural one. For both L-1 and D-1 peptides, hydrogen bonds are mainly found between the glutamic acids (there are three in the peptide 1 sequence) and the arginine or lysine amino acids on the surface of the protein. To a lesser extent, we also observe the formation of hydrogen bonds between the tyrosine residue of the peptide 1 and either glutamic acid and asparagine amino acids of the BSA protein. Visual inspection of the various trajectories leads to the assumption, that even if the behavior of peptide 1 is rather dynamic at the BSA surface, the L-1 peptide on average tends to adopt a more elongated conformation at the surface of the BSA protein while on the other hand, the D-1 peptide keeps its globular shape (Fig. 6). This is somewhat confirmed by the average end-to-end distance of the peptide, calculated between the first carbon atom linked to the N-terminal position and the alpha carbon of the last glutamic acid at the C-terminal position, taken over all trajectories which is seen to be longer in the case of the L-1 peptide (21.1 Å ± 3.9) then for the D-1 peptide (17.7 Å ± 4.6). While the elongated conformation of L-1 should favor the formation of aggregates, the globular shape of D-1 rather hinders this formation. To test this hypothesis, we have undertaken further MD simulations starting from the final structure of the peptide L-1, or D-1, adsorbed at site II of BSA, through three independent runs. Then, we have added three additional peptides L-1, or D-1, in the vicinity of the first peptide L-1, or D-1, already interacting with BSA (Fig. S8†). This let to a remarkable difference in behavior between the L-1 and D-1: where in the former case we observe the formation of entangled tetramer assembly of four L-1 peptides in all three independent MD trajectories (Fig. 6); while on the other hand, in the case of the D-1 peptide, we observe in two cases that the four peptides are found to interact at different positions on the surface of the BSA protein, while in the third case we observe the formation of a juxtaposed tetramer.Model suggestion based on experimental and simulated data
In the absence of BSA, whatever the peptide enantiomer under consideration L-1 or D-1, due to its amphiphilic nature, peptide 1 spontaneously forms globular micelles of the order of approximately 6–7 nm in diameter in water (Fig. 4b). This phenomenon is well known in the field of low molecular weight peptides involved in supramolecular hydrogel formation. Peptide 1 micelles appear to assemble to form nanofibers 10 nm in diameter that grow very slowly over time, with longer nanofibers appearing in the middle of a large number of very small nanofibers constituting only a few peptide 1 micelles. This observation suggests that the nanofibers are formed and deformed at their ends: in fact, after several days, despite the presence of long nanofibers of several hundred nanometers, we still observe the presence of a very large number of peptide 1 micelles. If the growth mechanism of the nanofibers was not dynamic, we should have observed a drastic reduction in the density of micelles after a few days. TEM monitoring therefore seems to suggest end-to-end growth kinetics.In the presence of BSA, the observation of L-1 peptide-based micelles and nanofibers can already be seen after a short time (t = 1 h) (Fig. 4c). We also observed the presence of BSA clusters, of the order 8–9 nm, in agreement with the value found in solution (10 mM PBS, pH 7.4) by dynamic light scattering (Fig. S9†) and in agreement with the literature. It appears that at the ends of some nanofibers, there is a bulky aggregate that could be a BSA cluster. On the other hand, BSA clusters are observed at some nanofiber intersections and some nanofibers grow from BSA clusters. In the presence of BSA, two types of nanofiber were also observed: nanofibers 10 nm in diameter and some thinner nanofibers of the order 6 to 7 nm. Curiously, in the absence of BSA, only large diameter nanofibers are observed. It does not appear that large nanofibers are formed by association with thinner ones, otherwise we would observe bifurcations between nanofibers, which is not the case. We can therefore assume that these are two different structures (polymorphs), which come and go, the thicker ones being the most stable. It thus appears that BSA acts as a nucleating agent for the thick fibers. For peptide D-1 in the presence of BSA, only rare nanofibers are observed and no peptide cluster is present anymore (Fig. 4c).
Solutions of L-1 and D-1 peptides buffered at pH 7.4 (10 mM PBS), stored at room temperature for three months (in the absence of BSA), show numerous identical entangled nanofibers 10 nm in diameter. However, we should be reminded that these samples are not gels but viscous liquids. When BSA was added to each of the enantiomeric peptide 1 solutions, it was observed for the L-1 peptide sample that some nanofibers were decorated with BSA clusters and other BSA clusters were found at the nodes of several nanofibers (Fig. 4d). However, when BSA was added to the solution containing D-1 peptides, the nanofibers were still decorated with BSA but were shorter. This suggests that by interacting with the D-1 peptide fibers, the BSA destabilizes the nanofibers. These observations are consistent with the formation of a hydrogel when BSA is added to the viscous fluid containing the L-1 peptides and with the observed decrease in the viscosity of the solution containing the D-1 peptide.
These observations suggest that BSA acts as a nucleating agent for L-1 peptides but inhibits self-assembly nucleation for the D-1 peptide. For the L-1 peptide, in agreement with molecular dynamics simulations, the interaction between the peptide and BSA could stabilize the peptide in an ‘unfolded’ conformation, which could then promote interaction with other L-1 peptides in a conformation close to those found in the self-assembled fibrillar nanostructure. However, for the D-1 peptide, interaction with BSA induces a more globular peptide conformation that does not result in self-assembly. In addition, small peptide clusters present in solution can interact with BSA. In the case of peptide L-1, this would lead to a change in conformation that rearranges the peptide clusters into a conformation that allows the initiation of self-assembly. In the case of D-1 peptides, the interaction between BSA and the peptide clusters destabilizes these clusters and the peptides return to a free state in solution. As BSA can interact with peptides at several sites, BSA or BSA clusters can be starting points for several self-assembled peptide nanofibers, as observed experimentally. Finally, when BSA was added to peptide solutions after 90 days, the BSA interacts with the nanofibers for both L and D peptides. For L peptides, this interaction did not destabilize the fibers and led to cross-linking nodes between fibers. In the case of D peptides, this interaction leads to a change in the conformation of the peptide in the self-assembly, which destabilizes it. The nanofibers are cut and by interacting with the nanofiber ends, the BSA leads to the release of the fiber-end peptides, which become free again in solution.
Conclusion
We have investigated the effect of proteins, mainly BSA, on the self-assembling behavior of the amphiphilic L-1 peptide and its enantiomer, D-1 peptide. The presence of proteins in a solution of L-1 peptides clearly catalyzes self-assembly in nanofibers and induces supramolecular hydrogel formation, while in a solution of its D-1 enantiomer peptide, BSA destabilizes peptide self-assemblies and assemblies. Molecular dynamics simulations show that both L-1 and D-1 peptides interact non-specifically with BSA but in different ways: whereas L-1 peptides unfold over the protein surface, D-1 peptides adopt a more globular conformation when in contact with BSA. The unfolding of the L-1 peptides favors interaction with other peptides also in an unfolded conformation, thus promoting L-1 peptide self-assembly; whereas the globular conformation of the D-1 dipeptide inhibits long self-assembled nanostructure formation and destabilizes assembled D-1 peptide nanofibers. Further studies are necessary to fully validate this mechanism and to better understand the nature of the peptide/protein interactions. One could for example use fluorescence resonance energy transfer experiments to validate the unfolding of the peptide sequence in the presence of BSA. More extensive molecular dynamics simulations should give a better insight into the amino-acid sequences that are interacting with the peptides, and this information could then allow the design of other peptides that self-assemble in the presence of proteins, maybe in specific ways. The control of the unfolding process of peptides could also be of interest in the design of cryptic site biomacromolecules which play a major role in biology.26Data availability
All the data is presented in the manuscript and the ESI.†Author contributions
SHR: formal analysis, methodology, validation, writing. TD: software, methodology, formal analysis. J-YR: formal analysis, methodology, supervision, validation, writing. AB-S: methodology, formal analysis, visualization, writing. RS: software, methodology, formal analysis, supervision. VB: conceptualization, methodology, formal analysis, validation, writing. LeJ: formal analysis, methodology, supervision, validation. MS: methodology, validation, formal analysis, visualization. AC: software, methodology, formal analysis, supervision, resources, funding, writing. PS: conceptualization, methodology, validation, formal analysis, investigation, resources, funding, writing, visualization, supervision. LJ: conceptualization, methodology, validation, formal analysis, investigation, resources, funding, writing, visualization, supervision.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
S. H. M. and T. D. acknowledge the financial support from “Agence Nationale de la Recherche” (CASH, ANR-21-CE06-0033) for postdoctoral fellowship and PhD funding, respectively. J.-Y. R. acknowledges his postdoc fellowship from the “Fondation Jean-Marie Lehn” (Labex Chimie des Systèmes Complexes, Project ITI-CSC-PSC-22). A. B.-S. acknowledges the doctoral school ED222 for his PhD funding. We also gratefully acknowledge the financial support from “Agence Nationale de la Recherche” (CASH, ANR-21-CE06-0033), the “Fondation Jean-Marie Lehn”, “ITI Chimie des Systèmes Complexes” (Project ITI-CSC-PSC-24), and the SATT Conectus Alsace (Project SUPRAFOAM). The ICS characterization, microscopy and rheology platforms CARMAC, PLAMICS and INCA respectively, are acknowledged. François Courtier (ICS UPR22-CNRS), Soraya Saad-Guermeche (ICS UPR22-CNRS) and Karim Benmlih (INSERM U1121) are acknowledged for their technical contribution. We also thank Dr Jean-Marc Strub (LSMBO/IPHC UMR7178) for the mass spectrometry analysis.Notes and references
- (a) K. Lv, L. Zhang, W. Lu and M. Liu, ACS Appl. Mater. Interfaces, 2014, 6, 18878–18884 CrossRef CAS PubMed; (b) M. Conejero-Muriel, J. A. Gavira, E. Pineda-Molina, A. Belsom, M. Bradley, M. Moral, J. de Dios García-López Durán, A. Luque González, J. J. Díaz-Mochón, R. Contreras-Montoya, Á. Martínez-Peragón, J. M. Cuerva and L. Álvarez de Cienfuegos, Chem. Commun., 2015, 51, 3862–3865 RSC.
- J.-M. Lehn, Supramolecular Chemistry – Concepts and Perspectives, VCH, Weinheim, Germany, 1995 Search PubMed.
- R. J. Williams, T. E. Hall, V. Glattauer, J. White, P. J. Pasic, A. B. Sorensen, L. Waddington, K. M. McLean, P. D. Currie and P. G. Hartley, Biomaterials, 2011, 32, 5304–5310 CrossRef CAS PubMed; M. C. Manas-Torres, S. Illescas-Lopez, J. A. Gavira, L. Álvarez de Cienfuegos and S. Marchesan, Isr. J. Chem., 2022, e202200018 Search PubMed.
- (a) R. J. Williams, R. J. Mart and R. V. Ulijn, Pept. Sci., 2010, 94, 107–117 CrossRef CAS PubMed; (b) B. Yang, D. J. Adams, M. Marlow and M. Zelzer, Langmuir, 2018, 34, 15109–15125 CrossRef CAS PubMed; (c) H. He, W. Tan, J. Guo, M. Yi, A. N. Shy and B. Xu, Chem. Rev., 2020, 120, 9994–10078 CrossRef CAS PubMed.
- X. Du, J. Zhou, J. Shi and B. Xu, Chem. Rev., 2015, 115, 13165–13307 Search PubMed.
- Z. Yang, H. Gu, D. Fu, P. Gao, J. K. Lam and B. Xu, Adv. Mater., 2004, 16, 1440–1444 CrossRef CAS.
- A. R. Hirst, S. Roy, M. Arora, A. K. Das, N. Hodson, P. Murray, S. Marshall, N. Javid, J. Sefcik, J. Boekhoven, J. H. van Esch, S. Santabarbara, N. T. Hunt and R. V. Ulijn, Nat. Chem., 2010, 2, 1089–1094 CrossRef CAS PubMed; R. J. Williams, J. Gardiner, A. B. Sorensen, S. Marchesan, R. J. Mulder, K. M. McLean and P. G. Hartley, Aust. J. Chem., 2013, 66, 572–578 CrossRef; C. Muller, A. Ontani, A. Bigo-Simon, P. Schaaf and L. Jierry, Adv. Colloid Interface Sci., 2022, 304, 102660 CrossRef PubMed.
- S. Toledano, R. J. Williams, V. Jayawarna and R. V. Ulijn, J. Am. Chem. Soc., 2006, 128, 1070–1071 CrossRef CAS PubMed; X. Qin, W. C. Xie, S. Tian, J. L. Cai, H. Yuan, Z. Yu, G. L. Butterfoss, A. C. Khuong and R. A. Gross, Chem. Commun., 2013, 49, 4839–4841 Search PubMed; C. Vigier-Carriere, D. Wagner, A. Chaumont, B. Durr, P. Lupattelli, C. Lambour, M. Schmutz, J. Hemmerlé, B. Senger, P. Schaaf, F. Boulmedais and L. Jierry, Langmuir, 2017, 33, 8267–8276 CrossRef PubMed.
- Y. Xie, R. Huang, W. Qi, Y. Wang, R. Su and Z. He, J. Mater.Chem. B, 2016, 4, 844–851 RSC.
- Z. Wang, C. Liang, Y. Shang, S. He, L. Wang and Z. Yang, Chem. Commun., 2018, 54, 2751–2754 RSC.
- N. Javid, S. Roy, M. Zelzer, Z. Yang, J. Sefcik and R. V. Ulijn, Biomacromolecules, 2013, 14, 4368–4376 CrossRef CAS PubMed.
- R. Jain, V. K. Pal and S. Roy, Biomacromolecules, 2020, 21, 4180–4193 Search PubMed.
- X. Liu, Z. Zhao, C. Gao, X. Sun, S. Li and J. Han, Int. J. Biol. Macromol., 2024, 283, 137613 Search PubMed.
- (a) D. Li, H. Wang, D. Kong and Z. Yang, Nanoscale, 2012, 4, 3047–3049 RSC; (b) C. Yang, D. Li, Q. FengZhao, L. Wang, L. Wang and Z. Yang, Org. Biomol. Chem., 2013, 11, 6946–6951 Search PubMed; (c) J. Rodon Fores, M. Criado-Gonzalez, M. Schmutz, C. Blanck, P. Schaaf, F. Boulmedais and L. Jierry, Chem. Sci., 2019, 10, 4761–4766 RSC.
- D. A. Case, H. M. Aktulga, K. Belfon, I. Y. Ben-Shalom, J. T. Berryman, S. R. Brozell, D. S. Cerutti, T. E. Cheatham III, G. A. Cisneros, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, G. Giambasu, M. K. Gilson, H. Gohlke, A. W. Goetz, R. Harris, S. Izadi, S. A. Izmailov, K. Kasavajhala, M. C. Kaymak, E. King, A. Kovalenko, T. Kurtzman, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, M. Machado, V. Man, M. Manathunga, K. M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, K. A. O'Hearn, A. Onufriev, F. Pan, S. Pantano, R. Qi, A. Rahnamoun, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, A. Shajan, J. Shen, C. L. Simmerling, N. R. Skrynnikov, J. Smith, J. Swails, R. C. Walker, J. Wang, J. Wang, H. Wei, R. M. Wolf, X. Wu, Y. Xiong, Y. Xue, D. M. York, S. Zhao and P. A. Kollman, Amber 2018, University of California, San Francisco, 2022 Search PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Haeuser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 Search PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 Search PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- I. S. Joung and T. E. Cheatham, J. Phys. Chem. B, 2008, 112, 9020–9041 CrossRef CAS PubMed.
- K. A. Majorek, P. J. Porebski, M. Chruszcz, S. C. Almo and W. Minor, Mol. Immunol., 2012, 52, 174–182 Search PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- T. N. Tikhonova, N. N. Rovnyagina, Z. A. Arnon, B. P. Yakimov, Y. M. Efremov, D. Cohen-Gerassi, M. Halperin-Sternfeld, N. V. Kosheleva, V. P. Drachev, A. A. Svistunov, P. S. Timashev, L. Adler-Abramovich and E. A. Shirshin, Angew. Chem., Int. Ed., 2021, 60, 25339–25345 CrossRef CAS PubMed.
- A. M. Smith, R. J. Williams, C. Tang, P. Coppo, R. F. Collins, M. L. Turner, A. Saiani and R. V. Ulijn, Adv. Mater., 2008, 20, 37–41 CrossRef CAS.
- A. J. F. C. Fernandes, L. de Carvalho Bertozo, A. P. R. Povinelli, G. Zazeri, A. R. de Souza, N. H. Morgon and V. F. Ximenes, J. Photochem. Photobiol., A, 2022, 433, 114197 CrossRef CAS.
- S. Sharma, S. Subramani and I. Popa, FEBS J., 2021, 288, 1742–1758 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: List of chemicals used, synthesis and characterization of peptides L-1, D-1, L-2 and D-2. Additional Fig. S1 to S9. See DOI: https://doi.org/10.1039/d5fd00007f |
| This journal is © The Royal Society of Chemistry 2025 |