Open Access Article
Open Access ArticleControlling the thermally-driven crystallization of DNA-coated nanoparticles with formamide†
Theodore
Hueckel
,
Seungyeon
Woo
 and
Robert J.
Macfarlane
and
Robert J.
Macfarlane
 *
*
Department of Materials Science and Engineering, Massachusetts Institute of Technology (MIT), 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA. E-mail: rmacfarl@mit.edu
First published on 7th August 2024
Abstract
DNA-coated nanoparticles, also known as programmable atom equivalents (PAEs), facilitate the construction of materials with nanoscopic precision. Thermal annealing plays a pivotal role by controlling DNA hybridization kinetics and thermodynamics, which ensures the formation of intended structures. While various design handles such as particle size, DNA design, and salt concentration influence the stability of the DNA duplexes linking PAEs in a lattice, their influence on the system's melting temperature (Tm) often follows complicated trends that make rational tuning of self-assembly challenging. In this work, the denaturant formamide is used to precisely tune the thermal response of PAEs. Our results reveal a clear and predictable trend in the PAEs’ response to formamide, enabling rational control over the Tm of a diverse set of PAE systems. Unlike adjustments made through alterations to PAE design or solution parameters such as ionic strength, formamide achieves its temperature shift without impacting the kinetics of assembly. As a result, PAEs can be rapidly crystallized at ambient temperatures, producing superlattices with similar quality to PAE crystals assembled through standard protocols that use higher temperatures. This study therefore positions formamide as a useful tool for enhancing the synthesis of complex nanostructures under mild conditions.
Introduction
Colloidal self-assembly is a powerful method for creating materials with intricate nanoscale features, which can exhibit unique properties for applications in structural materials, electronics, and photonics.1–11 The primary advantage of self-assembly is its ability to produce these features through a rational, programmable approach using building block design and processing conditions to control the formation of different structural motifs.12–18 In particular, DNA hybridization provides a robust method for customizing interparticle interactions for a wide range of particle sizes, shapes, and compositions.19–24 The highly ordered superlattices generated by DNA-directed nanoparticle crystallization have resulted in the moniker of “programmable atom equivalents” (PAEs) to describe these DNA-grafted colloids.25–27The thermal response of PAEs is crucial for controlling particle interactions, thereby enabling the assembly of diverse crystalline structures through thermal annealing.28–30 While a variety of design handles (e.g. solution ionic strength, DNA design, particle size, particle shape) have been studied as a means to tailor PAEs’ thermal response,31–33 modulation of these parameters often leads to complex assembly phenomena that can be difficult to rationally and finely control in laboratory experiments. For example, ionic strength affects the system's melting temperature (Tm),34 but also changes the interaction pair potential,32 as well as the crystallization kinetics and final crystal quality.35 Particle size/curvature and DNA grafting density also affect Tm, but these cannot be easily manipulated after PAEs are synthesized.36 Thus, it would be beneficial to have a singular design handle that could be applied post-synthetically to modulate DNA interactions and thermal response in a straightforward manner.
In this work, we examine how formamide, a known denaturant for DNA, can be used to predictably control the thermodynamics and kinetics of PAE assembly. Formamide disrupts DNA's hydration shell, interfering with DNA duplexation, thereby lowering PAE dissociation temperature.37 Importantly, increasing formamide concentration linearly lowers the temperature at which DNA strands dissociate, regardless of DNA nucleobase sequence. Thus, formamide could be an excellent addition to the toolbox of PAE self-assembly as a means of more finely controlling assembly behavior. However, dense brushes of DNA grafted to a nanoparticle surface can alter the local ion distribution around DNA duplexes and also the local concentration of DNA duplexes between particles.34 These structural differences make it unclear if formamide will have the same reliability in altering PAEs’ Tm. By investigating how varying concentrations of formamide affect the melting transitions of PAEs with different particle and DNA designs, we demonstrate that it provides significantly enhanced level of control over the thermal response of PAEs compared to existing methods. This enhanced control then enables the formation of high-quality crystal structures at significantly lower temperatures than are typically required for PAE crystallization.
Results and discussion
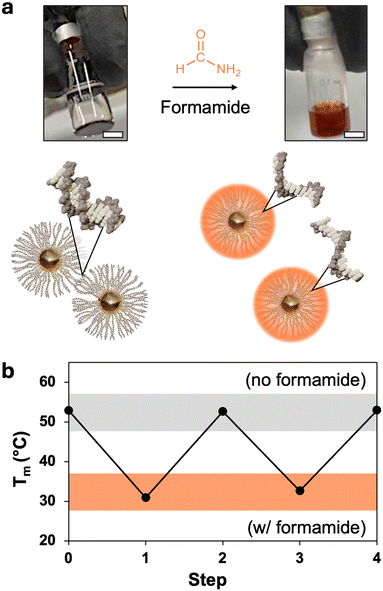
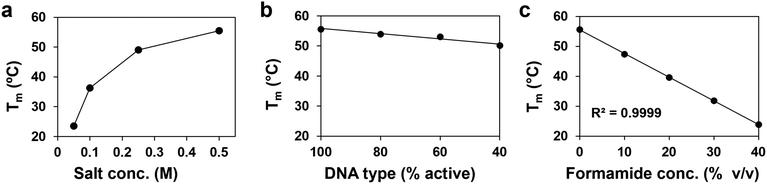
Formamide is a denaturant that is widely used in molecular biology to reduce the melting temperature of DNA (Fig. 1a).38 Its mild yet effective action allows for the controlled separation of genetic material while preserving the integrity of nucleic acids,39 making it a useful tool for applications such as Southern blots, polyacrylamide gel electrophoresis (PAGE), and polymerase chain reactions (PCR).40,41 It is also reliable enough in its effect on DNA hybridization to be used as a substitute for thermal annealing in the complex folding process of DNA-origami nanostructures.42,43 This work demonstrates that formamide similarly serves as a denaturant for PAEs, which is an effective means to increase our control over PAE self-assembly.Adding formamide to PAE assemblies comprising gold nanoparticle cores bound through hybridized DNA results in their rapid dissolution into a homogeneous nanoparticle suspension at temperatures significantly lower than their native Tm (Fig. 1a). Crucially, the nucleobases are kept intact during denaturation, allowing the nanoparticles to return to their original binding capacity upon formamide removal (Fig. 1b). While other processing conditions can similarly be used to reversibly affect DNA binding to disperse or assemble PAEs, formamide potentially offers much finer control over self-assembly than existing methods. In particular, formamide is noteworthy because DNA duplex Tm decreases linearly with increasing concentrations of formamide, and this behavior is consistent across various DNA designs, with trends ranging from 0.6 to 0.8 °C per volume percentage of formamide (% v/v).37 In comparison, the impact of variables such as ionic strength on DNA can have more complicated, non-linear effects. Existing predictive models can provide general information to tune Tm using these design handles,44 but precise modulation remains challenging. These complications are further exacerbated with PAE assembly due to the larger number of design variables and the high local concentration of polyanionic DNA strands around the nanoparticle core.32 Adopting this ability to tune thermal response is essential for PAEs, as there is currently no method to precisely set the Tm of a suspension without extensive empirical testing of conditions. For example, like free DNA duplexes, PAE Tms respond non-linearly to solution ionic strength, and trends can vary significantly between systems (Fig. 2a).34 The grafting density of DNA strands on each PAE can also be used to alter Tm. In practice, PAE assembly is typically induced by hybridizing “linker” strands to the DNA that is covalently bonded to the nanoparticle core (Fig. S1, ESI†). Thus, the number of DNA interactions between any given pair of bonded PAEs can be controlled by altering the ratio of linker strands to PAEs. PAEs can also be co-loaded with both linker strands and non-participatory “dummy” strands, which lack the unpaired sticky ends necessary to form DNA duplexes between PAEs. Reduction of linker density also lowers PAEs’ Tm, but offers only a narrow window of control (Fig. 2b), and the quality of crystals can drop significantly with low loading of active linkers. In contrast to both of these design handles (ionic strength and linker density), formamide allows Tm to be tuned over a wide window (>30 °C), and follows a linear and predictable trend (Fig. 2c). In principle, this linearity allows any new Tm to be selected without meticulously optimizing assembly conditions, if the original Tm is known (0% v/v formamide).
While this initial result is promising for the use of formamide as a means to modulate PAE assembly thermodynamics, it is important to note that PAEs exhibit unique multivalency-driven thermal behaviors that are affected by the number and arrangement of the numerous oligonucleotides grafted to the nanoparticles’ surfaces.34 Such effects could alter the slope or disrupt the linearity of the PAEs’ formamide response. Thus, more quantitative examination of formamide's effects on various systems must also be performed in order for this approach to yield a useful tool for modulating PAE assembly.
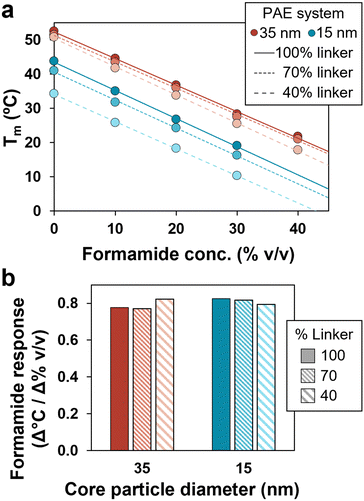
Experiments were conducted to investigate multivalency effects by varying core sizes and grafting density of PAEs. Two particle sizes were prepared (15 and 35 nm) (Fig. S2, ESI†), containing approximately 200 and 900 DNA linkers at maximum grafting density, respectively.45,46 The grafting density of PAEs was then modified independently of particle size by co-loading PAEs with controlled ratios of active linkers with dummy linkers, ultimately providing a wide variety of multivalent architectures to explore. Notably, with formamide, all systems demonstrated a similar reliable linear trend in Tm reduction across a wide temperature range (Fig. 3a). Additionally, the slope of these plots was measured to be very similar across all systems, showing a decrease of approximately 0.8 ± 0.02 °C per volume percent of formamide (Fig. 3b). This formamide response is consistent with prior examinations of formamide's effects on free DNA strands.37 These trends were also observed for different DNA designs, including DNA linkers of different lengths, and both binary and unary PAE assemblies (Fig. S3, ESI†). These results confirm that the formamide response is consistent across multiple PAE designs, emphasizing its utility as a tool for tuning PAE self-assembly.
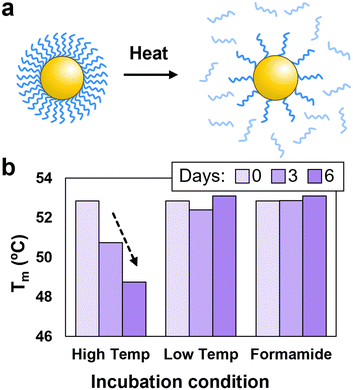
To further demonstrate the advantages of using formamide to modulate PAE assembly, it is important to consider the potential benefits associated with conducting assembly at lower temperatures. Typically, to grow large single crystals, PAEs must undergo slow cooling to prevent excessive nucleation.28,31 This process often requires maintaining the particles at elevated temperatures for extended periods,47 which can result in the detachment of DNA grafts thereby degrading the DNA brush and diminishing assembly quality (Fig. 4a).45,48 An indication of this degradation is a drop in Tm after the PAEs are held at elevated temperatures for significant periods of time, which is attributed to a reduced number of linkers binding the particles together. For example, when PAEs were incubated at 60 °C, a 4 °C decrease in Tm was observed after six days (Fig. 4b). In contrast, PAEs maintained at room temperature display a consistent Tm over the same time period. Similarly, PAEs incubated in 25% v/v formamide (at room temperature) show no decrease in Tm, indicating that formamide does not degrade DNA over extended periods. Therefore, formamide could alleviate DNA desorption issues that would be expected during standard slow cooling experiments to produce single crystals, as the addition of formamide would permit assembly to be conducted at temperatures where DNA grafts remain stable. However, it is important to note that formamide can oxidize into formic acid which can cleave nucleobases,49 thus requiring the use of fresh formamide stored in at low temperatures (additional details in ESI,† Section S1.5).
As noted above, solution ionic strength would be expected to induce many of the same effects observed from the addition of formamide, albeit with a non-linear correlation between solution salt concentration and Tm. Nevertheless, even if the correlation between solution salt concentration and PAE Tm were meticulously characterized, formamide still provides a major advantage because it only alters DNA hybridization thermodynamics. Conversely, lowering PAE Tm by altering solution ionic strength drastically changes both assembly thermodynamics and assembly kinetics, which has adverse effects on the quality of the resulting PAE crystals. The Tm of a given PAE system is the temperature at which the rate of DNA association (C0kon) and dissociation (koff) are equal, where C0 is the initial concentration of unhybridized DNA, and kon and koff are the rate constants of association and dissociation, respectively.35,36 One of the effects of low ionic strength on DNA is to decrease the rate of kon,35,50 which slows down particle reconfiguration around Tm (Fig. 5a). Consequently, even with careful control of Tm by altering solution salt concentration, assembling high-quality crystals at lower temperatures remains challenging due to sluggish particle kinetics. In contrast, evidence suggests that formamide's denaturation effects are due mainly to the destabilization the double helix,37,38 that is, increasing koff without significantly affecting kon.51 Therefore, the kinetics of assembly are expected to remain high across all formamide concentrations, even at lower Tms (Fig. 5b).
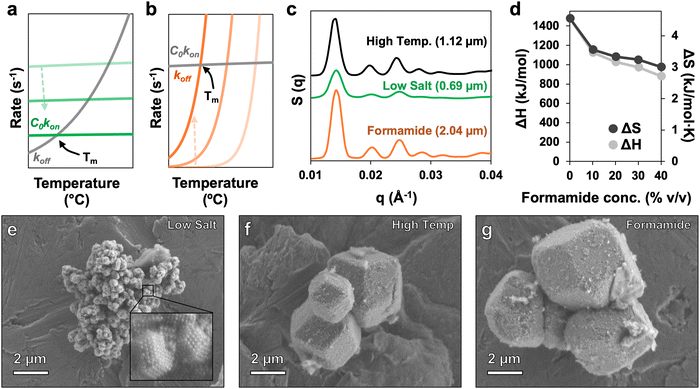 | ||
| Fig. 5 Utility of formamide for PAE assembly. (a) Heuristic image showing the trends of C0kon (green) and koff (grey) at various salt concentrations (light green: high salt, dark green: low salt). Tm is represented by the intersection of C0kon and koff for each trace; decreasing C0kon with lower salt concentrations leads to a drop in Tm. At lower Tms, rates for koff and C0kon drop, leading to poor crystallization kinetics.35,36,50 (b) Heuristic image showing the trends of C0kon (grey) and koff (orange) at various formamide concentrations (light orange: low formamide, dark orange: high formamide). Increasing koff with higher formamide concentrations leads to a drop in Tm. At all Tms, rates for koff and C0kon remain high, allowing for rapid reconfiguration and large crystal growth.51 (c) 1D SAXS patterns for traditional high temperature annealing of PAEs (black; Tm = 48 °C), low temperature annealing using a low salt concentration to lower Tm (green; Tm = 25 °C), and low temperature annealing using formamide to lower Tm (orange; Tm = 25 °C). All samples cooled from 60 °C to 10 °C at a rate of 0.1 °C min−1. Included is the average crystallite size determined via Williamson–Hall analysis. (d) ΔH and ΔS for PAE binding at varied formamide concentration determined via Van’t Hoff analysis. A drop in ΔH indicates broadening of the melt curve at higher formamide concentrations, which can help facilitate the growth of larger crystals. (e)–(g) SEM images of crystals formed via low temperature annealing with low salt, traditional high temperature annealing, and low temperature annealing using formamide, respectively. | ||
To further investigate the effects of kinetics, annealing experiments were conducted to assess the quality of crystals formed under different processing conditions, including traditional high temperature annealing and lower temperature annealing where Tm adjustments were made using either ionic strength or formamide. SAXS data confirm that while the control “high temperature” and “formamide” samples have a high degree of crystallinity, the “low salt” sample displays only a few broad peaks indicating a lower quality of crystallization (Fig. 5c and Fig. S4, S5, ESI†). Williamson–Hall analysis of SAXS patterns also indicate that assembling crystals under low salt conditions produces smaller crystallites, while assembly in the presence of formamide produces crystallites that actually exceed the size of crystals assembled using standard high-temperature methods. One possibility for the relative increase in crystal size under formamide conditions is broadening of the melt curve, which provides particles more time to nucleate and assemble into fewer, larger crystals. Melt broadening, characterized by a decrease in ΔH, has been previously observed in formamide-treated DNA systems,37 while a similar trend emerged for PAEs, as determined by Van’t Hoff analysis of melt curves for different formamide concentrations (Fig. 5d and Fig. S6, ESI†). High resolution SEM images indeed show that crystals formed in low salt conditions are substantially smaller compared to the other annealing methods (Fig. 5e). Additionally, both the standard high temperature method and assembly with formamide-tuned Tm successfully produced crystals with well-defined rhombic dodecahedral habits, confirming their higher quality indicated by SAXS (Fig. 5f–g). While the low salt sample clearly does display some order, and likely could be improved in quality by increasing the duration of cooling, formamide provides a far more robust and simple means to achieve crystallization at ambient conditions, without the need to extensively optimize PAE assembly protocols.
Conclusions
The use of formamide to tune PAE Tm represents a significant advance for the field of self-assembly. While numerous colloidal interactions can be used to modulate DNA-driven assembly thermodynamics, formamide offers a rare design handle that allows precise and rational control. The predictability and consistency of Tm reduction with formamide, as opposed to the non-linear effects observed with other variables such as ionic strength, present a clear advantage in designing and implementing controlled self-assembly processes. This research enhances our understanding of PAE behavior under varying thermal conditions and offers a practical method to achieve rapid crystallization at ambient temperatures. Such conditions not only prolong the lifespan of traditional PAE systems utilizing gold-thiol bonds but could also be crucial for utilizing PAEs with DNA ligands bound through weaker chemistries that are even more sensitive to elevated temperatures.52,53 Additionally, this methodology may be similarly applicable for controlling the interactions of DNA-coated colloidal particles on the micron scale.22 High-temperatures currently pose challenges for detailed analysis using higher-magnification oil immersion optics, which act as heat sinks. In contrast, room temperature assembly could facilitate the use of these lenses for enhanced imaging at higher magnifications. Precise Tm control also enables applications for sensing, wherein functional nanostructures respond programmatically to thermal fluctuations near biologically relevant temperatures.Author contributions
All authors designed experiments and analyzed data. TH and SW synthesized materials and collected data. All authors wrote and edited the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by funding from the Department of the Navy, Office of Naval Research under Contract No. N00014-19-1-2213, the Air Force Office of Scientific Research's (AFOSR) Natural Materials and Systems Program (FA9550-23-1-0210), and the US Army Research Office under Cooperative Agreement No. W911NF-19-2-0026 for the Institute for Collaborative Biotechnologies.References
- F. Li, S. F. Liu, W. Liu, Z. W. Hou, J. Jiang and Z. Fu, et al., 3D printing of inorganic nanomaterials by photochemically bonding colloidal nanocrystals, Science, 2023, 381(6665), 1468–1474 CrossRef CAS PubMed.
- T. Hueckel, X. Luo, O. F. Aly and R. J. Macfarlane, Nanoparticle Brushes: Macromolecular Ligands for Materials Synthesis, Acc. Chem. Res., 2023, 56(14), 1931–1941 CrossRef CAS PubMed.
- T. Hueckel, G. M. Hocky and S. Sacanna, Total synthesis of colloidal matter, Nat. Rev. Mater., 2021, 6(11), 1053–1069 CrossRef CAS.
- Z. Cai, Z. Li, S. Ravaine, M. He, Y. Song and Y. Yin, et al., From colloidal particles to photonic crystals: advances in self-assembly and their emerging applications, Chem. Soc. Rev., 2021, 50(10), 5898–5951 RSC.
- S. Dhulipala, D. W. Yee, Z. Zhou, R. Sun, J. E. Andrade and R. J. Macfarlane, et al., Tunable Mechanical Response of Self-Assembled Nanoparticle Superlattices, Nano Lett., 2023, 23(11), 5155–5163 CrossRef CAS PubMed.
- R. L. Li, C. J. Thrasher, T. Hueckel and R. J. Macfarlane, Hierarchically Structured Nanocomposites via a “Systems Materials Science” Approach, Acc. Mater. Res., 2022, 3(12), 1248–1259 CrossRef CAS.
- L. Z. Zornberg, D. J. Lewis, A. Mertiri, T. Hueckel, D. J. D. Carter and R. J. Macfarlane, Self-Assembling Systems for Optical Out-of-Plane Coupling Devices, ACS Nano, 2023, 17(4), 3394–3400 CrossRef CAS PubMed.
- D. J. Lewis, D. J. D. Carter and R. J. Macfarlane, Using DNA to Control the Mechanical Response of Nanoparticle Superlattices, J. Am. Chem. Soc., 2020, 142(45), 19181–19188 CrossRef CAS PubMed.
- M. A. Boles, M. Engel and D. V. Talapin, Self-Assembly of Colloidal Nanocrystals: From Intricate Structures to Functional Materials, Chem. Rev., 2016, 116(18), 11220–11289 CrossRef CAS PubMed.
- S. C. Glotzer and M. J. Solomon, Anisotropy of building blocks and their assembly into complex structures, Nat. Mater, 2007, 6(8), 557–562 CrossRef PubMed.
- Z. Li, Q. Fan and Y. Yin, Colloidal Self-Assembly Approaches to Smart Nanostructured Materials, Chem. Rev., 2022, 122(5), 4976–5067 CrossRef CAS PubMed.
- T. Hueckel, G. M. Hocky, J. Palacci and S. Sacanna, Ionic solids from common colloids, Nature, 2020, 580(7804), 487–490 CrossRef CAS PubMed.
- J. Opdam, R. Tuinier, T. J. Hueckel, T. Snoeren and S. Sacanna, Selective colloidal bonds via polymer-mediated interactions, Soft Matter, 2020, 16(32), 7438–7446 RSC.
- R. Klajn, K. J. M. Bishop and B. A. Grzybowski, Light-controlled self-assembly of reversible and irreversible nanoparticle suprastructures, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(25), 10305–10309 CrossRef CAS PubMed.
- A. M. Kalsin, M. Fialkowski, M. Paszewski, S. K. Smoukov, K. J. M. Bishop and B. A. Grzybowski, Electrostatic Self-Assembly of Binary Nanoparticle Crystals with a Diamond-Like Lattice, Science, 2006, 312(5772), 420–424 CrossRef CAS PubMed.
- P. F. Damasceno, M. Engel and S. C. Glotzer, Predictive Self-Assembly of Polyhedra into Complex Structures, Science, 2012, 337(6093), 453–457 CrossRef CAS PubMed.
- P. J. M. Swinkels, Z. Gong, S. Sacanna, E. G. Noya and P. Schall, Visualizing defect dynamics by assembling the colloidal graphene lattice, Nat. Commun., 2023, 14(1), 1524 CrossRef CAS PubMed.
- C. J. Thrasher, F. Jia, D. W. Yee, J. M. Kubiak, Y. Wang and M. S. Lee, et al., Rationally Designing the Supramolecular Interfaces of Nanoparticle Superlattices with Multivalent Polymers, J. Am. Chem. Soc., 2024, 146(16), 11532–11541 CAS.
- R. J. Macfarlane, B. Lee, M. R. Jones, N. Harris, G. C. Schatz and C. A. Mirkin, Nanoparticle Superlattice Engineering with DNA, Science, 2011, 334(6053), 204–208 CrossRef CAS PubMed.
- Y. Zhang, F. Lu, K. G. Yager, D. van der Lelie and O. Gang, A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems, Nat. Nanotech., 2013, 8(11), 865–872 CrossRef CAS PubMed.
- W. B. Rogers, W. M. Shih and V. N. Manoharan, Using DNA to program the self-assembly of colloidal nanoparticles and microparticles, Nat. Rev. Mater., 2016, 1(3), 1–14 Search PubMed.
- Y. Wang, Y. Wang, X. Zheng, É. Ducrot, J. S. Yodh and M. Weck, et al., Crystallization of DNA-coated colloids, Nat. Commun., 2015, 6(1), 7253 CrossRef CAS PubMed.
- A. Hensley, W. M. Jacobs and W. B. Rogers, Self-assembly of photonic crystals by controlling the nucleation and growth of DNA-coated colloids, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(1), e2114050118 CrossRef CAS PubMed.
- L. Ding, X. Chen, W. Ma, J. Li, X. Liu and C. Fan, et al., DNA-mediated regioselective encoding of colloids for programmable self-assembly, Chem. Soc. Rev., 2023, 52(16), 5684–5705 RSC.
- D. Samanta, W. Zhou, S. B. Ebrahimi, S. H. Petrosko and C. A. Mirkin, Programmable Matter: The Nanoparticle Atom and DNA Bond, Adv. Mater., 2022, 34(12), 2107875 CrossRef CAS PubMed.
- P. A. Gabrys, L. Z. Zornberg and R. J. Macfarlane, Programmable Atom Equivalents: Atomic Crystallization as a Framework for Synthesizing Nanoparticle Superlattices, Small, 2019, 15(26), 1805424 CrossRef PubMed.
- C. Zhang, R. J. Macfarlane, K. L. Young, C. H. J. Choi, L. Hao and E. Auyeung, et al., A general approach to DNA-programmable atom equivalents, Nat. Mater, 2013, 12(8), 741–746 CrossRef CAS PubMed.
- E. Auyeung, L. Ting, A. J. Senesi, A. L. Schmucker, B. C. Pals and M. O. de la Cruz, et al., DNA-mediated nanoparticle crystallization into Wulff polyhedra, Nature, 2014, 505(7481), 73–77 CrossRef PubMed.
- R. J. Macfarlane, B. Lee, H. D. Hill, A. J. Senesi, S. Seifert and C. A. Mirkin, Assembly and organization processes in DNA-directed colloidal crystallization, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(26), 10493–10498 CrossRef CAS PubMed.
- R. J. Macfarlane, M. R. Jones, B. Lee, E. Auyeung and C. A. Mirkin, Topotactic Interconversion of Nanoparticle Superlattices, Science, 2013, 341(6151), 1222–1225 CrossRef CAS PubMed.
- D. J. Lewis, L. Z. Zornberg, D. J. D. Carter and R. J. Macfarlane, Single-crystal Winterbottom constructions of nanoparticle superlattices, Nat. Mater., 2020, 19(7), 719–724 CrossRef CAS PubMed.
- S. E. Seo, M. Girard, M. O. de la Cruz and C. A. Mirkin, The Importance of Salt-Enhanced Electrostatic Repulsion in Colloidal Crystal Engineering with DNA, ACS Cent. Sci., 2019, 5(1), 186–191 CrossRef CAS PubMed.
- M. R. Jones, R. J. Macfarlane, B. Lee, J. Zhang, K. L. Young and A. J. Senesi, et al., DNA-nanoparticle superlattices formed from anisotropic building blocks, Nat. Mater., 2010, 9(11), 913–917 CrossRef CAS PubMed.
- R. Jin, G. Wu, Z. Li, C. A. Mirkin and G. C. Schatz, What Controls the Melting Properties of DNA-Linked Gold Nanoparticle Assemblies?, J. Am. Chem. Soc., 2003, 125(6), 1643–1654 CrossRef CAS PubMed.
- R. J. Macfarlane, R. V. Thaner, K. A. Brown, J. Zhang, B. Lee and S. T. Nguyen, et al., Importance of the DNA “bond” in programmable nanoparticle crystallization, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(42), 14995–15000 CrossRef CAS PubMed.
- R. J. Macfarlane, M. R. Jones, A. J. Senesi, K. L. Young, B. Lee and J. Wu, et al., Establishing the Design Rules for DNA-Mediated Programmable Colloidal Crystallization, Angew. Chem., 2010, 122(27), 4693–4696 CrossRef.
- R. D. Blake and S. G. Delcourt, Thermodynamic Effects of Formamide on DNA Stability, Nucleic Acids Res., 1996, 24(11), 2095–2103 CrossRef CAS PubMed.
- L. Levine, J. A. Gordon and W. P. Jencks, The relationship of structure to the effectiveness of denaturing agents for deoxyribonucleic acid, Biochemistry, 1963, 2, 168–175 CrossRef CAS PubMed.
- B. L. McConaughy, C. D. Laird and B. J. McCarthy, Nucleic acid reassociation in formamide, Biochemistry, 1969, 8(8), 3289–3295 CrossRef CAS PubMed.
- Y. L. Sun, Y. Z. Xu and P. Chambon, A simple and efficient method for the separation and detection of small DNA fragments by electrophoresis in formamide containing agarose gels and Southern blotting to DBM-paper, Nucleic Acids Res., 1982, 10(19), 5753–5763 CrossRef CAS PubMed.
- G. Sarkar, S. Kapelner and S. S. Sommer, Formamide can dramatically improve the specificity of PCR, Nucleic Acids Res., 1990, 18(24), 7465 CrossRef CAS PubMed.
- R. Jungmann, T. Liedl, T. L. Sobey, W. Shih and F. C. Simmel, Isothermal Assembly of DNA Origami Structures Using Denaturing Agents, J. Am. Chem. Soc., 2008, 130(31), 10062–10063 CrossRef CAS PubMed.
- Z. Zhang, J. Song, F. Besenbacher, M. Dong and K. V. Gothelf, Self-assembly of DNA origami and single-stranded tile structures at room temperature, Angew. Chem., Int. Ed., 2013, 52(35), 9219–92123 CrossRef CAS PubMed.
- R. Owczarzy, Y. You, B. G. Moreira, J. A. Manthey, L. Huang and M. A. Behlke,
et al., Effects of Sodium Ions on DNA Duplex Oligomers:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Improved Predictions of Melting Temperatures, Biochemistry, 2004, 43(12), 3537–3554 CrossRef CAS PubMed.
Improved Predictions of Melting Temperatures, Biochemistry, 2004, 43(12), 3537–3554 CrossRef CAS PubMed. - J. Lee and S. Lee, Non-Invasive, Reliable, and Fast Quantification of DNA Loading on Gold Nanoparticles by a One-Step Optical Measurement, Anal. Chem., 2023, 95(3), 1856–1866 CrossRef CAS PubMed.
- S. J. Hurst, A. K. R. Lytton-Jean and C. A. Mirkin, Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes, Anal. Chem., 2006, 78(24), 8313–8318 CrossRef CAS PubMed.
- S. Lee, H. A. Calcaterra, S. Lee, W. Hadibrata, B. Lee and E. Oh, et al., Shape memory in self-adapting colloidal crystals, Nature, 2022, 610(7933), 674–6749 CrossRef CAS PubMed.
- N. Bhatt, P. J. J. Huang, N. Dave and J. Liu, Dissociation and Degradation of Thiol-Modified DNA on Gold Nanoparticles in Aqueous and Organic Solvents, Langmuir, 2011, 27(10), 6132–6317 CrossRef CAS PubMed.
- M. S. Lowenthal, E. Quittman and K. W. Phinney, Absolute quantification of RNA or DNA using acid hydrolysis and mass spectrometry, Anal. Chem., 2019, 91(22), 14569–14576 CrossRef CAS PubMed.
- J. G. Wetmur and N. Davidson, Kinetics of renaturation of DNA, J. Mol. Biol., 1968, 31(3), 349–370 CrossRef CAS PubMed.
- J. R. Hutton, Renaturation Kinetics and thermal stability of DNA in aqueous solutions of formamide and urea, Nucleic Acids Res., 1977, 4(10), 3537–3555 CrossRef CAS PubMed.
- K. L. Young, M. B. Ross, M. G. Blaber, M. Rycenga, M. R. Jones and C. Zhang, et al., Using DNA to Design Plasmonic Metamaterials with Tunable Optical Properties, Adv. Mater., 2014, 26(4), 653–659 CrossRef CAS PubMed.
- C. Chen, X. Luo, A. E. K. Kaplan, M. G. Bawendi, R. J. Macfarlane and M. Bathe, Ultrafast dense DNA functionalization of quantum dots and rods for scalable 2D array fabrication with nanoscale precision, Sci. Adv., 2023, 9(32), eadh8508 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. Synthesis, fabrication, and data collection protocols; PAE system designs; (PDF). See DOI: https://doi.org/10.1039/d4sm00854e |
| This journal is © The Royal Society of Chemistry 2024 |