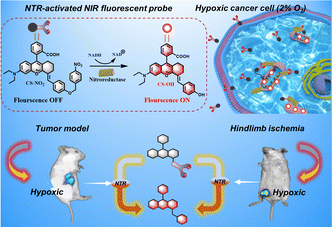
Monitoring mitochondrial nitroreductase activity in tumors and a hind-limb model of ischemia in mice using a novel activatable NIR fluorescent probe†
Xiaosheng
Liu‡
a,
Shuang
Zeng‡
a,
Ming
Zhang
a,
Maojun
Jiang
b,
Yves S.
Kafuti
a,
Pingping
Shangguan
a,
Yichu
Yu
a,
Qixian
Chen
 a,
Jingyun
Wang
a,
Jingyun
Wang
 *ab,
Xiaojun
Peng
*ab,
Xiaojun
Peng
 b,
Juyoung
Yoon
b,
Juyoung
Yoon
 *c and
Haidong
Li
*c and
Haidong
Li
 *ab
*ab
aSchool of Bioengineering, Dalian University of Technology, 2 Linggong Road, Hi-tech Zone, Dalian 116024, China. E-mail: wangjingyun67@dlut.edu.cn; lihd@dlut.edu.cn
bState Key Laboratory of Fine Chemicals, Frontiers Science Center for Smart Materials Oriented Chemical Engineering, Dalian University of Technology, 2 Linggong Road, Hi-tech Zone, Dalian 116024, China
cDepartment of Chemistry and Nanoscience, Ewha Womans University, 52 Ewhayeodae-gil, Seodaemun-gu, Seoul 03760, Korea. E-mail: jyoon@ewha.ac.kr
First published on 9th September 2022
Abstract
We report a mitochondria-targeted nitroreductase (NTR)-activated near-infrared fluorescent probe: CS-NO2. Overexpressed NTR in mitochondria was measured with high sensitivity. More importantly, the probe CS-NO2 successfully monitored NTR activity in solid tumors and a hind-limb model of ischemia in mice. This novel finding indicates the promising function of our probe for the diagnosis of solid tumors and hypoxia-associated diseases.
Solid tumors are distinguished by their hypoxic microenvironment, which results from an unbalanced relationship between oxygen delivery and oxygen uptake. Indeed, 50% to 60% of solid tumors that show locally progression have an uneven distribution of hypoxia.1,2 A hypoxic microenvironment can stimulate epithelial–mesenchymal transition, promote angiogenesis, and increase the risk of tumor metastasis.3,4 Tumors in a hypoxic microenvironment become more aggressive under metabolic stress, inhibit anti-tumor immunity, and reduce chemosensitivity, thereby hindering treatment.5–10 Moreover, hypoxia is associated intimately with inflammation and organ damage.11–13 Thus, detection of a hypoxic microenvironment is crucial for the diagnosis, monitoring, and cure of cancers and several other disorders.14,15
Traditional medical-imaging methods, such as magnetic resonance imaging, computed tomography, and positron emission tomography, are challenging to use when diagnosing cancer at the molecular level. Methods based on fluorescence imaging are utilized extensively for disease diagnosis and detection of physiological processes at cellular, tissue, and molecular levels and in live organisms because of their non-invasiveness, strong biocompatibility, and high spatial and temporal resolution.16–18 Near-infrared (NIR) fluorescence allows deep penetration of tissue and little interference, and could be applied for clinical use.19–24
Nitroreductases (NTRs) are flavin-containing enzymes. They can reduce nitro compounds if nicotinamide adenine dinucleotide (NADH) is used as an electron donor. NTRs can be divided into types I and II based on whether or not the catalytic reaction is sensitive to oxygen. An increased NTR level is a biomarker of tumor cells.25–27 Type-I NTRs have been detected in cancer cells incubated under normoxia.28 However, most probes used for cancer diagnoses based on the NTR response are activated in hypoxic cancer cells, which suggests that NTRs exist mainly in the oxygen-sensitive form (type II) in hypoxic cancer cells.29 The reduction mechanism of type-II NTRs involves continuous transfer of single electrons, and mitochondria are the main sites for redox reactions in eukaryotes.30–32 Therefore, we suspect that type-II NTR-catalyzed reductions take place in hypoxic mitochondria. However, most fluorescent probes dependent on NTR activation lack the capacity to target organelles. Also, monitoring NTR activity in a subcellular structure under a hypoxic microenvironment is difficult (especially in mitochondria).33–37 In addition, oxygen in cells is consumed mainly in the mitochondrial respiratory chain, and the structure and function of mitochondria in a hypoxic microenvironment will be affected.38,39 Studies have shown that hypoxia can affect the morphological changes of mitochondria, causing changes in the composition of the electron transport chain (ETC) complex. Such changes reshape the ETC and reduce the activity of the tricarboxylic acid cycle to adapt to hypoxia, and mitochondrial metabolic pathways and hypoxia-inducible factor (HIF) pathways carry out crosstalk under hypoxia.40 Therefore, tracking and labeling mitochondria in cancer cells is important for the study of cellular metabolic pathways under hypoxia.
We developed and synthesized a mitochondrial-targeted NIR fluorescent probe, CS-NO2, for localization of mitochondria in hypoxic cells. We applied this probe in a mouse model of solid tumors and hind-limb ischemia (Scheme 1). Rhodamine is a water-soluble cationic dye that can be inserted into mitochondria through the mitochondrial membrane potential.41–44 By expanding the conjugated structure of rhodamine B, we obtained the NIR-fluorescence group CS-OH. We introduced p-nitrobenzyl as the recognition site of NTR to produce CS-NO2. We quenched fluorescence emission by inhibiting intramolecular charge transfer. The nitro unit in CS-NO2 was reduced by NTR to an amino motif in the presence of NADH. This action was followed by 1,6-self-elimination, which released a free CS-OH with intense fluorescence (Fig. S10, ESI†) for hypoxia imaging in vitro and in vivo.
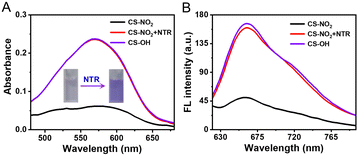
The response of CS-NO2 (10 μM) to NTR (10 μg mL−1) in a HEPES buffer system (pH 7.4; 10% DMSO) was studied. Absorbance at 580 nm increased significantly (Fig. 1A), and the solution changed from colourless to purple after the probe CS-NO2 had been incubated with NTR. Accordingly, the fluorescence intensity at 670 nm of CS-NO2 increased significantly after incubation with NTR (Fig. 1B) in the presence of NADH, and the fluorescence quantum yield (Φf) increased from 0.52% to 10.38%. We verified the activation process of CS-NO2 stated above by high-resolution mass spectrometry (Fig. S11, ESI†).
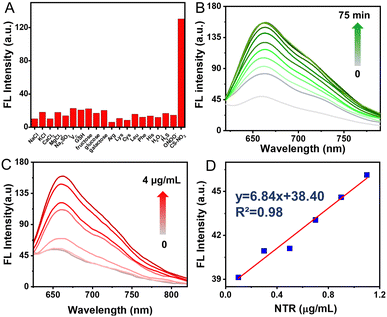
Selectivity assessment of CS-NO2 to NTR was carried out to further verify that CS-NO2 produces a specific response during biomonitoring. The fluorescence changes of CS-NO2 with various potential-interfering species found commonly in organisms were investigated under identical conditions. A significant fluorescence increase was not detected with CS-NO2 upon exposure to potential interfering species (Fig. 2A). When CS-NO2 was incubated only with NTR (10 μg mL−1) and NADH (0.5 mM), the fluorescence intensity increased sharply, indicating that CS-NO2 had excellent selectivity for NTR: this feature is extremely beneficial for monitoring of hypoxic regions in vivo.
Next, we tested the time-dependent response of the probe CS-NO2 (10 μM) to NTR (10 μg mL−1). The absorbance and fluorescence intensity of the reaction system was measured at different incubation times using a UV-vis spectrophotometer and fluorescence spectrometer. With an increase in incubation time, the fluorescence and absorbance changed (Fig. 2B and Fig. S12A, ESI†) and reached a plateau in ∼1 h (Fig S13, ESI†), These data suggested the sensitivity of CS-NO2 to NTR.
Moreover, the sensing behavior of probe CS-NO2 (10 μM) to various equivalents of NTR (0–4 μg mL−1) was tested. The maximum fluorescence intensity and absorbance of CS-NO2 increased as the NTR dose increased (Fig. 2C and Fig. S12B, ESI†). In an NTR concentration range of 0–1.2 μg mL−1 (Fig. 2D), the connection between the emission intensity of CS-NO2 and NTR dose was linear, with a linear correlation coefficient of 0.9758. These data indicated that CS-NO2 could be used to detect NTR in vitro.
We wished to further understand the possible interaction between our probe and enzymes and the catalytic mechanism of NTR. We used Auto-Dock tools to calculate docking. The crystal structure of NTR from the PDB database (PDB code: 4DN2) was selected as the research model. CS-NO2 could fit into the hydrophobic cavity of an enzyme due to hydrophobic interactions with the NTR protein and aromatic π–π interactions (Fig. 3A). In addition, the ribbon-band model established by Pymol showed that the oxygen atom of the nitro group of CS-NO2 could form three hydrogen bonds with the amino-acid residues (Phe-132, Ser-12 and Arg-10) of NTR. These hydrogen bonds anchored CS-NO2 firmly to NTR (Fig. 3B).
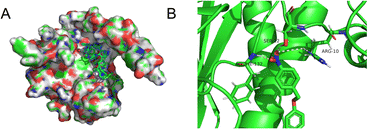 | ||
| Fig. 3 (A) Calculated binding model of CS-NO2 with NTR. (B) The hydrogen bonds between CS-NO2 and amino-acid residues (Phe-132, Ser-12, and Arg-10) in NTR. | ||
Before cell imaging, the biological compatibility of CS-NO2 was detected using the MTT assay (Fig. S14, ESI†). This showed that CS-NO2 had weak toxicity to 4T1 cells and MCF-7 cells, and was an appropriate probe for NTR detection in live cells.
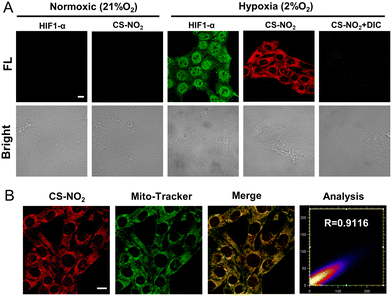
The sensing ability of CS-NO2 to NTR was tested in 4T1 cells and MCF-7 cells incubated in a hypoxic environment (Fig. 4A and Fig. S15, ESI†). After 8 h of incubation under hypoxia (2% O2), 4T1 cells showed obvious red fluorescence, whereas cells incubated in a normal environment did not show fluorescence. After incubation in a hypoxic environment and treatment with the NTR inhibitor dicumarol, the red fluorescence disappeared. These data further validated the selectivity of CS-NO2 to NTR in cells. HIF-α is not expressed in normal cells, and it can be expressed stably only under hypoxic conditions. Expression of this protein is positively correlated with the degree of cell hypoxia. To further evaluate the hypoxia of cells under two culture conditions, we measured expression of HIF-1α protein by immunofluorescence staining and compared it with the confocal fluorescence microscopy of probe CS-NO2. The opening and closing of the immunofluorescence channel and NIR fluorescence channel of the probe were synchronous. These data verified the specificity of probe opening under hypoxia, and indicated that activation of probe CS-NO2 was caused by type-II NTR (oxygen-sensitive) catalysis. To investigate the location of this probe further, we used confocal fluorescence microscopy to study the distribution of intensity in cells (Fig. 4B and Fig. S16, ESI†). First, we used the commercial organelle-targeting dyes Mito-Tracker™ Green, Lyso-Tracker™ Green, ER-Tracker™ Green, and Hoechst 33342 to pre-label mitochondria, endoplasmic reticula, lysosomes, and nuclei in cells, respectively, and then undertook co-incubation with CS-NO2. Through co-localization analyses of probe CS-NO2 with different dyes, the Pearson's colocalization coefficient used to describe the intensity distribution between the two channels was calculated to be 0.9116, 0.6185, 0.4724, and 0.0274, for Mito-Tracker Green, Lyso-Tracker Green, ER-Tracker Green, and Hoechst 33342, respectively. These results showed that probe CS-NO2 could locate the mitochondria of hypoxic cells specifically. Most of the NTR-based fluorescent probes reported so far can be used only to monitor cellular hypoxia: few fluorescent probes can also label mitochondria selectively.
Due to the high sensitivity and specificity of CS-NO2 to NTR, a tumor-bearing model and hind-limb ischemic model in mice was used to test the capability of probe CS-NO2 to monitor NTR in vivo. Both models showed enhanced fluorescence signals over time (Fig. 5). Compared with the control group (phosphate-buffered saline), probe CS-NO2 distinguished the tumor or hind-limb-operation site from surrounding normal tissue within 45 min upon local injection. These findings implied that CS-NO2 could be used as a NIR fluorescent probe in the monitoring of solid-tumor markers and other hypoxia-related diseases.
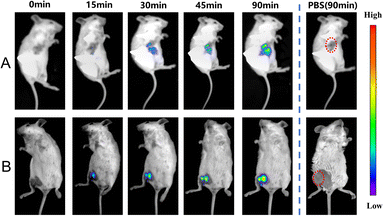 | ||
| Fig. 5 In vivo fluorescence imaging of hypoxia in mice (A) bearing tumors and with (B) hind-limb ischemia. λex = 550 nm, λem= 760 nm. | ||
In conclusion, we designed and synthesized a rhodamine-based fluorescent probe for sensing endogenous NTR activity. The reactive probe CS-NO2, quenched by the p-nitrobenzyl group, had high selectivity and sensitivity. The specific response of NTR to CS-NO2 was verified via spectroscopic characterization and molecular docking analysis. Imaging of cell lines (4T1 and MCF-7) under hypoxia further verified the excellent monitoring ability of the probe CS-NO2 towards endogenous NTR activity. More importantly, CS-NO2 could target mitochondria selectively, thereby providing a potent instrument for researching the metabolism of nitro compounds in cells under hypoxia. CS-NO2 could monitor hypoxic regions in a tumor-bearing mouse model. It could also distinguish hypoxic regions in normal tissues and surgical sites in a hind-limb model of ischemia in mice; this has not been demonstrated before. This efficient NIR fluorescent probe may provide a new strategy for diagnosing and monitoring hypoxia-related diseases.
We acknowledge financial support provided by the National Natural Science Foundation of China (No. 22078050). We also thank the National Research Foundation of Korea for grants funded by the Korean government (MSIT) (No. 2022R1A2C3005420), and the Fundamental Research Funds for the Central Universities (Nos DUT22RC(3)025 and DUT22LAB601).
Conflicts of interest
There are no conflicts to declare.Notes and references
- S. B. Bader, M. W. Dewhirst and E. M. Hammond, Cancers, 2020, 13, 27 CrossRef PubMed.
- Y. Li, Y. Sun, J. Li, Q. Su, W. Yuan, Y. Dai, C. Han, Q. Wang, W. Feng and F. Li, J. Am. Chem. Soc., 2015, 137, 6407–6416 CrossRef CAS PubMed.
- A. L. Harris, Nat. Rev. Cancer, 2002, 2, 38–47 CrossRef CAS PubMed.
- J. P. Joseph, M. K. Harishankar, A. A. Pillai and A. Devi, Oral Oncol., 2018, 80, 23–32 CrossRef CAS PubMed.
- S. K. Daniel, K. M. Sullivan, K. P. Labadie and V. G. Pillarisetty, Clin. Transl. Med., 2019, 8, 10 CAS.
- C. Riera-Domingo, A. Audige, S. Granja, W. C. Cheng, P. C. Ho, F. Baltazar, C. Stockmann and M. Mazzone, Physiol. Rev., 2020, 100, 1–102 CrossRef CAS.
- N. E. Scharping, D. B. Rivadeneira, A. V. Menk, P. D. A. Vignali, B. R. Ford, N. L. Rittenhouse, R. Peralta, Y. Wang, Y. Wang, K. DePeaux, A. C. Poholek and G. M. Delgoffe, Nat. Immunol., 2021, 22, 205–215 CrossRef CAS PubMed.
- J. Tao, G. Yang, W. Zhou, J. Qiu, G. Chen, W. Luo, F. Zhao, L. You, L. Zheng, T. Zhang and Y. Zhao, J. Hematol. Oncol., 2021, 14, 14 CrossRef PubMed.
- B. Wang, Q. Zhao, Y. Zhang, Z. Liu, Z. Zheng, S. Liu, L. Meng, Y. Xin and X. Jiang, J. Exp. Clin. Cancer Res., 2021, 40, 24 CrossRef CAS PubMed.
- Y. Yang, X. Liu, W. Ma, Q. Xu, G. Chen, Y. Wang, H. Xiao, N. Li, X. J. Liang, M. Yu and Z. Yu, Biomaterials, 2021, 265, 120456 CrossRef CAS PubMed.
- A. Emami Nejad, S. Najafgholian, A. Rostami, A. Sistani, S. Shojaeifar, M. Esparvarinha, R. Nedaeinia, S. Haghjooy Javanmard, M. Taherian, M. Ahmadlou, R. Salehi, B. Sadeghi and M. Manian, Cancer Cell Int., 2021, 21, 62 CrossRef CAS PubMed.
- S. Konisti, S. Kiriakidis and E. M. Paleolog, Nat. Rev. Rheumatol., 2012, 8, 153–162 CrossRef CAS PubMed.
- J. W. Lee, J. Ko, C. Ju and H. K. Eltzschig, Exp. Mol. Med., 2019, 51, 1–13 Search PubMed.
- J. Sun, Z. Hu, R. Wang, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 1384–1390 CrossRef CAS PubMed.
- X. Zhao, S. Long, M. Li, J. Cao, Y. Li, L. Guo, W. Sun, J. Du, J. Fan and X. Peng, J. Am. Chem. Soc., 2020, 142, 1510–1517 CrossRef CAS PubMed.
- H. Li, Q. Yao, W. Sun, K. Shao, Y. Lu, J. Chung, D. Kim, J. Fan, S. Long, J. Du, Y. Li, J. Wang, J. Yoon and X. Peng, J. Am. Chem. Soc., 2020, 142, 6381–6389 CrossRef CAS PubMed.
- J. Yin, L. Huang, L. Wu, J. Li, T. D. James and W. Lin, Chem. Soc. Rev., 2021, 50, 12098–12150 RSC.
- Y. Zhang, G. Zhang, Z. Zeng and K. Pu, Chem. Soc. Rev., 2022, 51, 566–593 RSC.
- H. Li, Q. Yao, J. Fan, N. Jiang, J. Wang, J. Xia and X. Peng, Chem. Commun., 2015, 51, 16225–16228 RSC.
- H. Li, H. Kim, F. Xu, J. Han, Q. Yao, J. Wang, K. Pu, X. Peng and J. Yoon, Chem. Soc. Rev., 2022, 51, 1795–1835 RSC.
- H. Li, Y. Li, Q. Yao, J. Fan, W. Sun, S. Long, K. Shao, J. Du, J. Wang and X. Peng, Chem. Sci., 2019, 10, 1619–1625 RSC.
- J. Mu, M. Xiao, Y. Shi, X. Geng, H. Li, Y. Yin and X. Chen, Angew. Chem., Int. Ed., 2022, 61, e202114722 CAS.
- M. Ling, R. Sun, G. Li, M. Z. Syeda, W. Ma, Z. Mai, L. Shao, L. Tang and Z. Yu, Nano Res., 2022, 15, 6288–6296 CrossRef CAS.
- Z. Wang, L. Yu, Y. Wang, C. Wang, Q. Mu, X. Liu, M. Yu, K. N. Wang, G. Yao and Z. Yu, Adv. Sci., 2022, 9, e2104793 CrossRef PubMed.
- S. Chen, J. Liu, Y. Li, X. Wu, Q. Yuan, R. Yang and J. Zheng, TrAC, Trends Anal. Chem., 2020, 131, 116010 CrossRef CAS.
- X. Meng, F. Yang, H. Dong, L. Dou and X. Zhang, Nano Today, 2021, 38, 101156 CrossRef CAS.
- H. Zhou, M. Guo, J. Li, F. Qin, Y. Wang, T. Liu, J. Liu, Z. F. Sabet, Y. Wang, Y. Liu, Q. Huo and C. Chen, J. Am. Chem. Soc., 2021, 143, 1846–1853 CrossRef CAS PubMed.
- A. Chevalier, Y. Zhang, O. M. Khdour, J. B. Kaye and S. M. Hecht, J. Am. Chem. Soc., 2016, 138, 12009–12012 CrossRef CAS PubMed.
- H. Li, D. Kim, Q. Yao, H. Ge, J. Chung, J. Fan, J. Wang, X. Peng and J. Yoon, Angew. Chem., Int. Ed., 2021, 60, 17268–17289 CrossRef CAS PubMed.
- M. Khacho, R. Harris and R. S. Slack, Nat. Rev. Neurosci., 2019, 20, 34–48 CrossRef CAS PubMed.
- E. M. Williams, R. F. Little, A. M. Mowday, M. H. Rich, J. V. Chan-Hyams, J. N. Copp, J. B. Smaill, A. V. Patterson and D. F. Ackerley, Biochem. J., 2015, 471, 131–153 CrossRef CAS PubMed.
- M. Zhu, R. R. Liu, H. L. Zhai, Y. J. Meng, L. Han and C. L. Ren, Int. J. Biol. Macromol., 2020, 150, 509–518 CrossRef CAS PubMed.
- M. Li, Y. Zhang, X. Ren, W. Niu, Q. Yuan, K. Cao, J. Zhang, X. Gao and D. Su, Chem. Commun., 2022, 58, 819–822 RSC.
- W. Qin, C. Xu, Y. Zhao, C. Yu, S. Shen, L. Li and W. Huang, Chin. Chem. Lett., 2018, 29, 1451–1455 CrossRef CAS.
- X. Wu, W. Shi, X. Li and H. Ma, Acc. Chem. Res., 2019, 52, 1892–1904 CrossRef CAS PubMed.
- F. Xu, M. Fan, S. Kang and X. Duan, Anal. Chim. Acta, 2019, 1088, 131–136 CrossRef CAS.
- S. Zhang, H. Chen, L. Wang, X. Qin, B. P. Jiang, S. C. Ji, X. C. Shen and H. Liang, Angew. Chem., Int. Ed., 2022, 61, e202107076 CAS.
- R. A. Smith, R. C. Hartley, H. M. Cocheme and M. P. Murphy, Trends Pharmacol. Sci., 2012, 33, 341–352 CrossRef CAS PubMed.
- Z. Yang, J. Wang, S. Liu, X. Li, L. Miao, B. Yang, C. Zhang, J. He, S. Ai and W. Guan, Biomaterials, 2020, 229, 119580 CrossRef CAS PubMed.
- D. C. Fuhrmann and B. Brune, Redox. Biol., 2017, 12, 208–215 CrossRef CAS PubMed.
- X. Jing, F. Yang, C. Shao, K. Wei, M. Xie, H. Shen and Y. Shu, Mol. Cancer, 2019, 18, 157 CrossRef PubMed.
- Z. Mi, L. Liu, Y. Zhao and J. Guan, Int. J. Biol. Macromol., 2020, 164, 932–938 CrossRef CAS PubMed.
- L. Yuan, W. Lin, K. Zheng, L. He and W. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- X. Zhang, X. Li, W. Shi and H. Ma, Chem. Commun., 2021, 57, 8174–8177 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc04112j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |