Open Access Article
Open Access ArticleFiber-optic array using molecularly imprinted microspheres for antibiotic analysis†
Sergio
Carrasco
a,
Elena
Benito-Peña
*ab,
David R.
Walt
*b and
María C.
Moreno-Bondi
*a
aDepartment of Analytical Chemistry, Faculty of Chemistry, Complutense University, Ciudad Universitaria s/n, Madrid 28040, Spain. E-mail: elenabp@ucm.es; mcmbondi@ucm.es; Fax: +34-91394-4329; Tel: +34-91394-5147
bDepartment of Chemistry, Tufts University, 62 Talbot Avenue, Medford, MA 02155, USA. E-mail: david.walt@tufts.edu; Fax: +1-617-627-3443; Tel: +1-617-627-3470
First published on 25th March 2015
Abstract
In this article we describe a new class of high-density optical microarrays based on molecularly imprinted microsphere sensors that directly incorporate specific recognition capabilities to detect enrofloxacin (ENRO), an antibiotic widely used for both human and veterinary applications. This approach involves the preparation of highly cross-linked polymer microspheres by thermal precipitation–polymerization in the presence and absence of the target analyte ENRO to generate either molecularly imprinted (MIP) or non-imprinted polymer (NIP) microspheres, respectively. Each polymer type of tailor-made microsphere is fluorescently encoded with either coumarin-30 or tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) dichloride [Ru(dip)3]Cl2 to enable the microspheres to be distinguished. The new MIP-based sensing platform utilizes an optical fiber bundle containing approximately 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individual 3.1 μm diameter fibers that are chemically etched to create microwells in which MIP and NIP microspheres can be deposited and imaged using an epi-fluorescence microscope. The method enables multiplexed detection by independently addressing both types of beads through their separate light channels. The unique response to the presence of ENRO is manifested on the basis of a competitive immunoassay. A red-fluorescent dye-tagged ENRO, labeled with BODIPY® TR Cadaverine, competes with ENRO for specific binding sites. The developed immuno-like assay displayed a limit of detection (LOD) of 0.04 μM (10% binding inhibition) and a dynamic range of 0.29–21.54 μM (20–80% binding inhibition). The selectivity of the assay was evaluated by measuring the cross-reactivity of other fluoroquinolones (ciprofloxacin, norfloxacin, danofloxacin, and flumequine) and non-related antibiotics (penicillin G and doxycycline). This work demonstrates, for the first time, the applicability of MIPs, as an alternative to biomolecule receptors, for the development of multiplexed detection fiber-optic microarrays paving the way for a new generation of biomimetic sensors.
000 individual 3.1 μm diameter fibers that are chemically etched to create microwells in which MIP and NIP microspheres can be deposited and imaged using an epi-fluorescence microscope. The method enables multiplexed detection by independently addressing both types of beads through their separate light channels. The unique response to the presence of ENRO is manifested on the basis of a competitive immunoassay. A red-fluorescent dye-tagged ENRO, labeled with BODIPY® TR Cadaverine, competes with ENRO for specific binding sites. The developed immuno-like assay displayed a limit of detection (LOD) of 0.04 μM (10% binding inhibition) and a dynamic range of 0.29–21.54 μM (20–80% binding inhibition). The selectivity of the assay was evaluated by measuring the cross-reactivity of other fluoroquinolones (ciprofloxacin, norfloxacin, danofloxacin, and flumequine) and non-related antibiotics (penicillin G and doxycycline). This work demonstrates, for the first time, the applicability of MIPs, as an alternative to biomolecule receptors, for the development of multiplexed detection fiber-optic microarrays paving the way for a new generation of biomimetic sensors.
Introduction
Biologically-inspired materials have attracted considerable interest over the last decade and have opened the door to new and important applications such as smart materials that can perform the same tasks as cells or organs.1 Molecular imprinting allows the creation of synthetic materials that function with the selectivity and affinity of biological recognition elements such as antibodies or enzymes.2 Molecularly Imprinted Polymers (MIPs) are artificial materials containing molecularly engineered receptor sites for a large variety of target analytes.3,4 Such specific receptor sites are introduced when a target molecule (template) is mixed with a stoichiometric number of functional monomers and cross-linkers to form complementary cavities to the target molecule's structure, which becomes fixed into the resulted polymer network during copolymerization. MIPs have been designed for the recognition of small molecules, such as toxins, for which it is difficult to obtain a corresponding biological receptor. In contrast to their natural counterparts, MIPs can be used efficiently under non-physiological or harsh conditions, such as high temperature, low pH, or organic solvents.MIPs have been used in a diverse number of applications, such as solid-phase extraction, chromatographic sorbents, polymeric catalysts and sensory materials.3,5 The utility of MIPs is boosted by their versatility and ease of preparation in a plurality of formats such as gels, membranes, filaments or nano/microspheres.6,7 For example, MIPs have been proven to be excellent materials for selective solid-phase extraction of fluoroquinolone antimicrobials (FQs).8 These drugs are widely prescribed to both humans and food-producing animals and their “uncontrolled” use, or disposal, may give rise to the presence of FQ residues in foods and also may lead to resistance that creates challenges for health care systems.9,10 Conventional enzyme-linked immunosorbent assays and microbial inhibition tests are the methods of choice for bulk screening of these antimicrobials;11,12 however, they have inherent limitations as they employ expensive bioreagents, are time-consuming, and require sophisticated laboratory infrastructure and technical expertise.
The direct optical detection of FQs requires UV excitation (λexc 280, 320 nm) and their native fluorescence (ca. λexc 400–420 nm) is influenced by interferences caused by both autofluorescence of the samples and light scattering off the polymer particles. In order to overcome these limitations, we previously reported a proof of concept where two immuno-like detection systems for FQs combining MIPs and either europium-amplified luminescent13 or Förster resonance energy transfer (FRET)-based photochemical sensing.14 Although both approaches worked, the reported LODs and the design scheme required improvements in order to be easily adaptable to cheap and portable systems.15
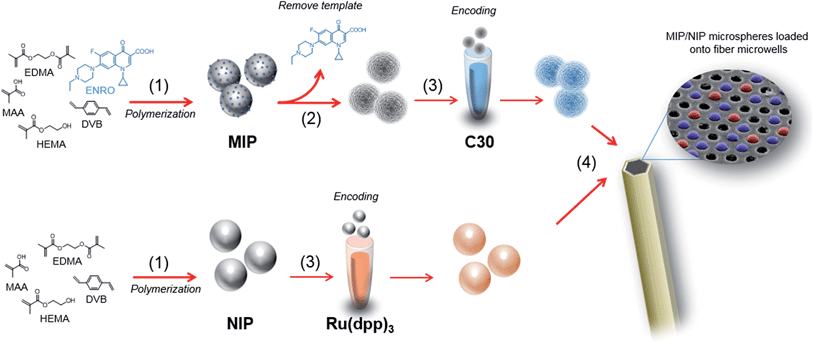
This paper describes a new MIP-based sensing platform that combines MIP microspheres with bead-based microarrays. This combination benefits from the advantages of tailor-made biomimetic materials in addition to a well-established array technology. Our sensing microspheres were prepared by thermal precipitation–polymerization, which is the mainstay method for obtaining polymeric microspheres with extremely uniform sizes and homogeneous binding site distribution.16 Moreover, microspheres feature a high surface-to-volume ratio, which results in better binding kinetics, ease of manipulation, and the ability to multiplex by encoding the microspheres.17 Additionally, to illustrate the multiplexing abilities of the assay, we used both MIP beads and NIP beads (negative control) so we could account for non-specific binding and background.
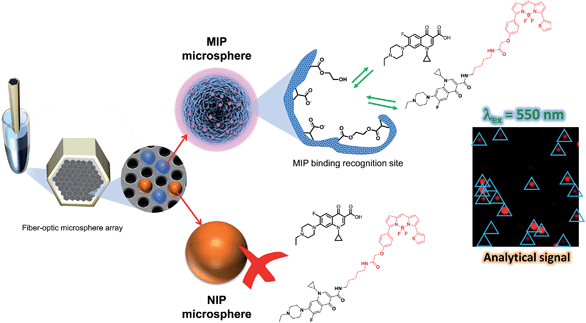
The platform described in this article has its roots in the use of optical fiber bundles and encoded microsphere-based arrays that Walt and colleagues have pioneered as powerful tools for many practical fluorescent biosensing applications.18–21 The optical fiber bundles are chemically etched with an acid solution22 to create a planar array of microwells where fluorescently encoded MIP and NIP bead types are pooled to form a randomly ordered but addressable high density array.23 The bead-based competition assay is then performed by incubating the array in a sample containing both the antibiotic ENRO and a fluorescent analogue of the antibiotic (BODIFLOXACIN), which competes for active sites within the imprinted microspheres. The encoding, assay protocols and data handling were optimized to allow multiplexing while preserving both sensitivity and specificity. Fiber-optic MIP microsphere-based arrays have been successfully applied to the analysis of ENRO in the serum of a healthy sheep after intravenous administration of the drug and the results have been fully validated by HPLC-FLD.
Experimental
Materials and instrumentation
Optical fiber bundles containing approximately 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individual optical fibers (with diameters of 3.1 μm) were purchased from SCHOTT North America, Inc. N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, 98+%) was purchased from Acros Organics (Madrid, Spain). 4-Morpholineethanesulfonic acid (MES, BupH buffered saline packs), phosphate buffered saline (PBS) and tween-20 (Surfact-Amps 20) were purchased from Thermo Scientific (Rockford, IL). 5-(((4-(4,4-Difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-yl)phenoxy)acetyl)amino)pentylamine hydrochloride (BODIPY® TR Cadaverine, >99.5%) was purchased from Molecular Probes Invitrogen (Grand Island, NY). Danofloxacin (DANO, 99%) was obtained from Riedel-de-Häen (Seelze, Germany). Sodium dodecyl sulfate (SDS, 98.5%), enrofloxacin (ENRO, 99.7%), 4-(dimethylamino)pyridine (DMAP, >99%), 2-[4-(2-hydroxyethyl)-1-piperazinyl]-ethanesulfonic acid (HEPES, >99.5%), coumarin-30 (C30, >99%), norfloxacin (NORF, 98%), ciprofloxacin hydrochloride (CIPRO, 99.8%), flumequine (FLUME, 98%), doxycycline (DOXY), penicillin G procaine salt (PGP), methacrylic acid (MAA, 99%), 2-hydroxyethyl methacrylate (HEMA, 97%), ethylene glycol dimethacrylate (EDMA, 98%) and divinylbenzene (DVB, 55% mixture of isomers) were purchased from Sigma-Aldrich (Madrid, Spain). MAA, HEMA and EDMA were passed through a packed column with an ion exchange resin (inhibitor removers, Aldrich) to remove monomethyl ether hydroquinone employed as an inhibitor. The initiator 2,2′-azobis(2,4-dimethylvaleronitrile) (ABDV) from Wako, (Neuss, Germany) was recrystallized from cold absolute ethanol. Potassium chloride (KCl, 99%) was purchased from Probus (Barcelona, Spain). Sodium chloride (NaCl, 99%) was obtained from Scharlau (Badalona, Spain). Oxazine-170 (Ox-170) was purchased from Radiant Dyes (Wermelskirchen, Germany) and tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) dichloride [Ru(dip)3]Cl2 was previously synthesized and characterized by our Research Group at Complutense University.24 Dimethylformamide (DMF), dimethylsulfoxide (DMSO) and tetrahydrofuran (THF, HPLC grade) were obtained from Carlo Erba (Sabadell, Spain). Acetonitrile (MeCN, HPLC grade) and absolute ethanol (EtOH, HPLC grade) were purchased from BDH-Prolabo (Llinars del Vallès, Spain). Methanol (MeOH, HPLC grade) was obtained from SDS (Pepyn, France). Distilled deionized water was obtained with a Milli-Q water purification system Millipore (Bedford, MA). Trifluoroacetic acid (TFA, peptide synthesis) was obtained from Fluorochem (Hadfield, Derbyshire, UK).
000 individual optical fibers (with diameters of 3.1 μm) were purchased from SCHOTT North America, Inc. N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, 98+%) was purchased from Acros Organics (Madrid, Spain). 4-Morpholineethanesulfonic acid (MES, BupH buffered saline packs), phosphate buffered saline (PBS) and tween-20 (Surfact-Amps 20) were purchased from Thermo Scientific (Rockford, IL). 5-(((4-(4,4-Difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-yl)phenoxy)acetyl)amino)pentylamine hydrochloride (BODIPY® TR Cadaverine, >99.5%) was purchased from Molecular Probes Invitrogen (Grand Island, NY). Danofloxacin (DANO, 99%) was obtained from Riedel-de-Häen (Seelze, Germany). Sodium dodecyl sulfate (SDS, 98.5%), enrofloxacin (ENRO, 99.7%), 4-(dimethylamino)pyridine (DMAP, >99%), 2-[4-(2-hydroxyethyl)-1-piperazinyl]-ethanesulfonic acid (HEPES, >99.5%), coumarin-30 (C30, >99%), norfloxacin (NORF, 98%), ciprofloxacin hydrochloride (CIPRO, 99.8%), flumequine (FLUME, 98%), doxycycline (DOXY), penicillin G procaine salt (PGP), methacrylic acid (MAA, 99%), 2-hydroxyethyl methacrylate (HEMA, 97%), ethylene glycol dimethacrylate (EDMA, 98%) and divinylbenzene (DVB, 55% mixture of isomers) were purchased from Sigma-Aldrich (Madrid, Spain). MAA, HEMA and EDMA were passed through a packed column with an ion exchange resin (inhibitor removers, Aldrich) to remove monomethyl ether hydroquinone employed as an inhibitor. The initiator 2,2′-azobis(2,4-dimethylvaleronitrile) (ABDV) from Wako, (Neuss, Germany) was recrystallized from cold absolute ethanol. Potassium chloride (KCl, 99%) was purchased from Probus (Barcelona, Spain). Sodium chloride (NaCl, 99%) was obtained from Scharlau (Badalona, Spain). Oxazine-170 (Ox-170) was purchased from Radiant Dyes (Wermelskirchen, Germany) and tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) dichloride [Ru(dip)3]Cl2 was previously synthesized and characterized by our Research Group at Complutense University.24 Dimethylformamide (DMF), dimethylsulfoxide (DMSO) and tetrahydrofuran (THF, HPLC grade) were obtained from Carlo Erba (Sabadell, Spain). Acetonitrile (MeCN, HPLC grade) and absolute ethanol (EtOH, HPLC grade) were purchased from BDH-Prolabo (Llinars del Vallès, Spain). Methanol (MeOH, HPLC grade) was obtained from SDS (Pepyn, France). Distilled deionized water was obtained with a Milli-Q water purification system Millipore (Bedford, MA). Trifluoroacetic acid (TFA, peptide synthesis) was obtained from Fluorochem (Hadfield, Derbyshire, UK).
The pH of all buffered solutions was adjusted with a GLP 22 pH meter from Crison (Barcelona, Spain). The semi-preparative HPLC system consisted of an HP1200 LC from Agilent Technologies (Palo Alto, CA) equipped with a solvent delivery quaternary pump, an autosampler, diode array detector (DAD), and an analytical scale fraction collector. Chromatographic purification of the new FQ was carried out on a ZORBAX Eclipse XDB-C18 semipreparative column (250 mm × 9.4 mm, 5 μm) from Agilent Technologies (Palo Alto, CA). Chromatographic analysis of the antibiotics was performed with a HP1100 LC from Agilent Technologies (Palo Alto, CA) equipped with a quaternary pump, online degasser, autosampler, automatic injector, and a column thermostat. A fluorescence detector (FLD) system was coupled online with the DAD system. Chromatographic separation was performed on an Aqua C18 analytical column (polar endcapped; 250 mm × 4.6 mm, 5 μm) from Phenomenex (Torrance, CA). Scanning electron microscopy (SEM) measurements were performed with a JEOL JSM-6330F field-emission scanning electron microscope at an acceleration voltage of 5 kV. The samples were coated with a thin gold film before analysis. Optical fiber bundles were imaged with an Olympus BX51 epi-fluorescence microscope (Olympus America Inc. Center Valley, PA). The microscope is equipped with a color charge-coupled device (CCD) camera (Infinity 3, Lumenera Corporation), a 100 W halogen lamp (operating at 6 V to acquire bright field images and 12 V for fluorescence images), and three fluorescence filter sets for C30 (excitation 405/10, dichroic Q414LP, emission 425LP), BODIPY® TR Cadaverine (excitation 550/10, dichroic T600LPXR, emission 590LP) and [Ru(dip)3]Cl2 (excitation 470/10, dichroic 490DCXR, emission 500LP).
Synthesis and encoding of microspheres
Molecularly imprinted microspheres selective to ENRO were prepared following a precipitation–polymerization approach,16 as previously reported.8 Briefly, a mixture of template molecule, ENRO (362 mg, 1 mmol), functional monomer, MAA (350 μL, 4 mmol), and diluent monomer, HEMA (496 μL, 4 mmol) were dissolved in 30 mL of MeCN. The radical initiator, ABDV (25 mg, 0.1 mmol) was dissolved in EDMA (960 μL, 5 mmol) and DVB (1300 μL, 5 mmol). This mixture was then added to the prior solution to yield the pre-polymerization mixture. Prior to polymerization, the mixture was homogenized in an ultrasonic bath for 5 min, transferred to a borosilicate glass reactor, and gently purged with argon for 10 min. A stir blade, coupled to a rotor head, maintained slow stirring (34 rpm) during the reaction, conducted at 60 °C for 24 h. For template removal, polymer microspheres were extensively washed with MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TFA (95
TFA (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v), MeOH, followed by acetone until no trace of the template was detected by HPLC-FLD. Finally, the particles were washed with a mixture of MeOH
5, v/v), MeOH, followed by acetone until no trace of the template was detected by HPLC-FLD. Finally, the particles were washed with a mixture of MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetone (50
acetone (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) and allowed to dry at room temperature for 48 h. The same procedure was followed to obtain NIP beads, but in the absence of the template molecule.
50, v/v) and allowed to dry at room temperature for 48 h. The same procedure was followed to obtain NIP beads, but in the absence of the template molecule.
MIP and NIP microspheres were then encoded with C30 and [Ru(dip)3]Cl2, respectively as follows: 60 μL of each microsphere stock solution (0.1 g mL−1 of microspheres in H2O containing 0.1% SDS as surfactant) were washed by centrifugation/re-suspension with 600 μL of 1 × PBS three times. The MIP microspheres were washed and re-suspended in 600 μL of THF and NIP microspheres in 600 μL of THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN (70
MeCN (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v). 13.26 mg of C30 was dissolved in 3.2 mL THF (11.93 mM) and 37.25 mg of [Ru(dip)3]Cl2 was dissolved in 3.2 mL of THF
30, v/v). 13.26 mg of C30 was dissolved in 3.2 mL THF (11.93 mM) and 37.25 mg of [Ru(dip)3]Cl2 was dissolved in 3.2 mL of THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN (70
MeCN (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v) (9.97 mM). NIP microspheres were re-suspended in 600 μL of [Ru(dip)3]Cl2 solution as well as MIP microspheres using C30 solution. The microspheres were then incubated at room temperature for 24 hours at 2500 rpm, protected from light. After incubation, microspheres were washed with 600 μL of cold methanol six times until no fluorescence signal from dyes could be detected in the supernatant, as monitored by HPLC-FLD. Finally, methanol was evaporated at low pressure to obtain dry microspheres and the microspheres were then stored protected from light.
30, v/v) (9.97 mM). NIP microspheres were re-suspended in 600 μL of [Ru(dip)3]Cl2 solution as well as MIP microspheres using C30 solution. The microspheres were then incubated at room temperature for 24 hours at 2500 rpm, protected from light. After incubation, microspheres were washed with 600 μL of cold methanol six times until no fluorescence signal from dyes could be detected in the supernatant, as monitored by HPLC-FLD. Finally, methanol was evaporated at low pressure to obtain dry microspheres and the microspheres were then stored protected from light.
Microarray fabrication
Both ends of a 3 cm optical fiber bundle were polished with a fiber polisher (Allied High Tech, Ranch Dominguez) using 30, 15, 9, 6, 3, 1, 0.5 and 0.05 μm diamond lapping films (Allied High Tech). The chemical etching was performed as follows:25 one end of the fiber was sonicated for 20 s in DMSO followed by 20 s in EtOH to clean and remove residual materials. Excess of EtOH was removed with a gentle stream of Ar. This distal end was submerged in an HCl (0.025 M) solution with magnetic stirring for 80 s to obtain microwells of approximately 3.1 μm in depth. The fiber was then rinsed and sonicated in distilled water for 10 s, subsequently washed with EtOH and dried with Ar. A suspension of 0.25 mg mL−1 of NIP and MIP microspheres were prepared in MeCN, and a drop of 1 μL of the suspension was deposited onto the etched distal end of the fiber. The fiber was kept in the dark for 5 min to allow complete evaporation of the solvent and appropriate incorporation of the microspheres into the microwells. The process was repeated 10 times to increase microspheres loading efficiency and achieve approximately 1000 microspheres over the entire measuring area of one fiber-optic microarray. Excess microspheres, those not loaded into the microwells, were removed using a cotton swab. The workflow of the assay protocol is depicted in Fig. 1.Assay protocol
ENRO quantification was based on a competitive assay where the target analyte competes with the labeled ENRO (BODIFLOXACIN) to bind to specific binding sites on the MIP microspheres. The distal end of the fiber that holds the microspheres was incubated in 1 mL of (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) MeCN
50, v/v) MeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEPES 25 mM pH 7.5, containing a constant amount of BODIFLOXACIN (250 nM) and increasing amounts of ENRO (0–1000 μM) while shaking for 1 h at 70 rpm. After incubation, the fiber was washed with 1 mL of wash buffer ((50
HEPES 25 mM pH 7.5, containing a constant amount of BODIFLOXACIN (250 nM) and increasing amounts of ENRO (0–1000 μM) while shaking for 1 h at 70 rpm. After incubation, the fiber was washed with 1 mL of wash buffer ((50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) MeCN
50, v/v) MeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEPES (25 mM, NaCl 138 mM, KCl 2.7 mM pH 7.5)) while shaking for 5 min at 70 rpm. The washing step was repeated three times. In the last step, the fiber was dipped in 1 mL of MeCN, shaken for 5 min at 70 rpm, and then dried and mounted onto a specially-designed microscope platform for measuring fiber-optic microarrays. The workflow of the assay protocol is depicted in Fig. 2.
HEPES (25 mM, NaCl 138 mM, KCl 2.7 mM pH 7.5)) while shaking for 5 min at 70 rpm. The washing step was repeated three times. In the last step, the fiber was dipped in 1 mL of MeCN, shaken for 5 min at 70 rpm, and then dried and mounted onto a specially-designed microscope platform for measuring fiber-optic microarrays. The workflow of the assay protocol is depicted in Fig. 2.
Image capturing and data analysis
The fiber-optic microarray was placed vertically with respect to the microscope objective in a home-built fiber holder. Images were acquired with an epi-fluorescence microscope (Olympus BX51) as described before. The fibers were imaged with 20× magnification for different exposure times (Table S2†) as well as sequentially, in the C30, [Ru(dip)3]Cl2, and BODIFLOXACIN channels. All images were analyzed using ImageJ (v.1.44p, Wayne Rasband, National Institutes of Health, USA). Both fluorescence and bright field micrographs (1392 × 1040 pixels) were acquired using a 16 bit output format.For data analysis, fluorescence micrographs were set as RGB images and deconvoluted in their respective channels. Green and blue channels were discarded and the analysis was performed in the red channel. A threshold for intensities (9–255 a.u.) was applied and the microspheres were analyzed by displaying the measurements inside their size range of 30–130 pixel2 with a circularity of 0.7–1.0. The tri-mean criterion (see ESI†) was employed instead of conventional mean statistics to avoid outliers, which gives more importance to the center of the population than to external points and thus provides a more robust analysis.
Microarray fabrication
The experimental data obtained for calibration plots were fitted to eqn (1), which represents a 4-parameter logistic (4 PL) regression model.26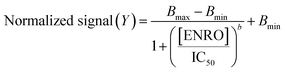 | (1) |
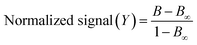 | (2) |
Sheep serum sample collection and preparation
ENRO 10% (p/v) (Baytril® 10%, Bayer, Germany) was administered to a healthy female lactating sheep (ca. 70 kg) in a dose of 5 mg kg−1via injection in the right jugular. Blood samples (3 mL) were collected in the left jugular at 0 and 5 min. after drug administration.27 Blood samples were centrifuged at 3400 rpm for 15 min to obtain serum, which was stored at −20 °C. For ENRO analysis, samples were treated as follows: 100 μL of serum was mixed with 280 μL of MeCN. The mixture was then centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. 285 μL of the supernatant was mixed with 15 μL of 7.5 μM BODIFLOXACIN in MeCN and 150 μL of HEPES (37.5 mM, pH 7.5). For calibrations in serum samples, 100 μL of the serum extracted 0 min after drug administration was mixed with 280 μL of various concentrations of ENRO in MeCN ranging from 0 to 500 μM. After centrifugation (13
000 rpm for 15 min. 285 μL of the supernatant was mixed with 15 μL of 7.5 μM BODIFLOXACIN in MeCN and 150 μL of HEPES (37.5 mM, pH 7.5). For calibrations in serum samples, 100 μL of the serum extracted 0 min after drug administration was mixed with 280 μL of various concentrations of ENRO in MeCN ranging from 0 to 500 μM. After centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 15 min), 285 μL aliquots were mixed with 15 μL of 7.5 μM BODIFLOXACIN in MeCN and 150 μL of HEPES (37.5 mM, pH 7.5).
000 rpm, 15 min), 285 μL aliquots were mixed with 15 μL of 7.5 μM BODIFLOXACIN in MeCN and 150 μL of HEPES (37.5 mM, pH 7.5).
Results and discussion
Bead based optical fiber microarrays utilize fluorescently encoded microparticles, further conjugated with specific biorecognition elements (antibodies, antigens, enzyme substrates, oligonucleotides, etc.) for the simultaneous detection of multiple analytes in a sample. The application of this strategy in combination with imprinted polymers has not been explored so far, although these biomimetics offer an alternative to surmount the problems associated with the use of biological recognition elements such as the cost of in vivo production of some biomolecules and the social concern about experiments with animals. In contrast to biologically-derived recognition elements, MIPs are thermally and chemically stable and have long-term storage stability at room temperature.4,6The preparation of MIP-based microarrays requires imprinted spherical beads with the right size to be deposited in the etched fiber optic microwells. In addition, the polymers should be encoded with fluorescent dyes to enable the identification of each bead type, without compromising the selective recognition properties of the resulting material. Finally, the optical properties of the MIP should change in response to the presence of the target analytes. As a proof of concept, this paper describes the development of microarrays prepared with ENRO-selective MIPs and their corresponding NIPs for preparing multiplexed MIP-based fiber optic arrays.
MIP/NIP beads synthesis, encoding and image capturing
The polymers were prepared by precipitation polymerization using ENRO as template, MAA as functional monomer, HEMA as diluent monomer and a mixture of EDMA and DVB (p-DVB and m-DVB isomer mixture) as cross-linkers (molar ratio, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). The synthesis was optimized to obtain spherical beads with a diameter of 3.22 ± 0.03 μm, as deduced from DLS measurements. As reported in the literature, the core of beads prepared with a mixture of MAA and DVB (p-DVB and m-DVB isomers) monomers is richer in p-DVB. Therefore, in principle, such particles could be encoded with hydrophobic dyes without affecting the recognition properties of the most external layers, where methacrylic monomers and m-DVB units tend to polymerize randomly.28
5). The synthesis was optimized to obtain spherical beads with a diameter of 3.22 ± 0.03 μm, as deduced from DLS measurements. As reported in the literature, the core of beads prepared with a mixture of MAA and DVB (p-DVB and m-DVB isomers) monomers is richer in p-DVB. Therefore, in principle, such particles could be encoded with hydrophobic dyes without affecting the recognition properties of the most external layers, where methacrylic monomers and m-DVB units tend to polymerize randomly.28
The MIP and NIP beads were encoded with C30 or [Ru(dip)3]Cl2, respectively by swelling the beads in a THF solution containing the dyes.29 After incubation, cold methanol was used to deswell the microspheres and physically trap the encoding fluorophores in the polymer matrix. Following this protocol, NIP and MIP each contained a different label and could be easily identified demonstrating, for the first time, the potential of this strategy for the development of encoded MIP-based multiplexed assays.
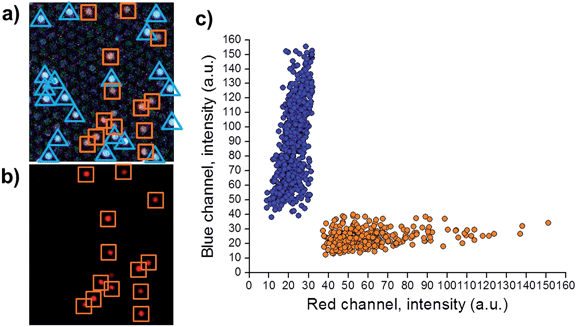
Using the conditions depicted in Table S2,† both encoding dyes can be monitored by exciting microspheres at 405 nm. Fig. 3a shows the emission of red light (due to NIP microspheres, encoded with [Ru(dip)3]Cl2) and blue light (due to MIP microspheres, encoded with coumarin-30). Only the ruthenium complex emits when the sample is excited at 470 nm, (Fig. 3b), verifying the location of NIP microspheres in the previous image. Applying the algorithm described by Nie et al.30 (details in ESI†) two different populations were distinguished (Fig. 3c), demonstrating the capability of multiplexing.
The reproducibility of the encoding protocol and the recognition capabilities of both the MIPs and NIPs were evaluated by detecting 250 nM BODIFLOXICIN in the presence and absence of ENRO using different microspheres batches. The results show no statistical variance between different batches prepared by different researchers during the last five years and highlight the excellent long-term stability of these materials.
The encoded MIP and NIP beads were subjected to the assay conditions (buffers, hydro-organic solvent mixtures, incubation times, temperatures), in the absence of the target molecule, to determine if there was any fluorophore leakage. No detectable amounts of either C30 or Ru(dip)3 were detected in the supernatants by HPLC-FLD, at the detection limit of the technique. These experiments were carried out for different bead batches and over a period of, at least, one year.
Synthesis and characterization of BODIFLOXACIN
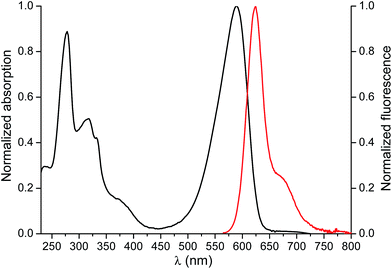
Several approaches have been described for the functionalization of the piperazine ring at position C-7 of the FQs.14 However, as reported previously8 selective interactions between the binding groups in the MIP and ENRO involves this moiety. Therefore, although the reactivity is remarkably lower,31 the antibiotic was labeled with BODIPY® TR Cadaverine by reaction with the carboxyl group at position 3 of ENRO, as described in the ESI.†32–34The absorption and emission spectra of the fluorescent analogue of ENRO, namely BODIFLOXACIN, are shown in Fig. 4. The compound has an absorption maximum at 588 nm, ε = (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 750 ± 130) M−1 cm−1, and shows a fluorescence emission peak centered at 624 nm, in MeCN. The fluorescence quantum yield (Φem) was 0.73 ± 0.02. The selection of a BODIPY derivative for antibiotic labeling, instead of other commonly applied dansyl or cyanine dyes,14,35 was based on the high photostability and pH-insensibility of this fluorophore, which are two important properties for epi-fluorescence studies in competitive assays.
750 ± 130) M−1 cm−1, and shows a fluorescence emission peak centered at 624 nm, in MeCN. The fluorescence quantum yield (Φem) was 0.73 ± 0.02. The selection of a BODIPY derivative for antibiotic labeling, instead of other commonly applied dansyl or cyanine dyes,14,35 was based on the high photostability and pH-insensibility of this fluorophore, which are two important properties for epi-fluorescence studies in competitive assays.
Assay optimization and array characterization
Several parameters, including the incubation solvent, the amount of polymer and the concentration of BODIFLOXACIN have been evaluated to select the optimum conditions for the competitive assay. After incubation, the amount of ENRO and BODIFLOXACIN not retained by the MIP/NIP beads was monitored by HPLC-FLD (see ESI† for details). The optimized measurement conditions are shown in Table 1.| Parameter | Optimized value |
|---|---|
Solvent (MeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEPES 25 mM, pH 7.5, v/v) HEPES 25 mM, pH 7.5, v/v) |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 50 |
| [Polymer] (mg mL−1) | 0.25 |
| [BODIFLOXACIN] (nM) | 250 |
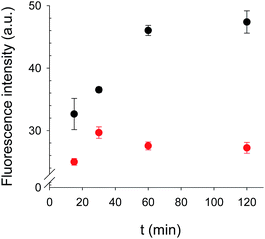
The binding kinetics of BODIFLOXACIN to the MIP microspheres in the presence and absence of a constant concentration of ENRO are depicted in Fig. 5. Incubations were carried out after depositing the microspheres onto the fiber-optic microwells. The signal response leveled off after 1 h incubation, which compares favorably with the analysis time of immunoassays reported in the literature for FQ detection36 and was selected for all subsequent competitive assays.
Competitive calibration curves were prepared by incubation of a freshly assembled MIP/NIP microarray in 1 mL of ENRO standards, with concentrations ranging from 0 to 1000 μM, prepared in 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (v/v) MeCN
50 (v/v) MeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
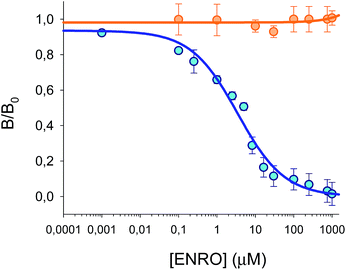
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEPES (25 mM, pH 7.5). As shown in Fig. 6, BODIFLOXACIN competes efficiently with ENRO for the selective binding sites in the MIP, as the labeled analogue was displaced from the imprinted beads in the presence of increasing concentrations of the antibiotic. The experimental data have been fitted to eqn (1) (n = 14, r2 = 0.98, blue points). The IC50 value is 3.5 μM and the LOD is 40 nM, calculated as the ENRO concentration for which BODIFLOXACIN binding to MIP was inhibited by 10%.37 The dynamic range (DR), i.e., the concentration range of ENRO that produces an inhibition of the signal between 20 and 80%, is between 290 nM and 21.5 μM. Furthermore, no competition was observed with the encoded NIP microspheres (n = 9, orange points), that were interrogated simultaneously with the MIP beads. This result confirms that nonspecific interactions are completely suppressed in the optimized assay conditions; therefore, it can be concluded that BODIFLOXACIN competes with ENRO for the specific binding sites in the MIP.
HEPES (25 mM, pH 7.5). As shown in Fig. 6, BODIFLOXACIN competes efficiently with ENRO for the selective binding sites in the MIP, as the labeled analogue was displaced from the imprinted beads in the presence of increasing concentrations of the antibiotic. The experimental data have been fitted to eqn (1) (n = 14, r2 = 0.98, blue points). The IC50 value is 3.5 μM and the LOD is 40 nM, calculated as the ENRO concentration for which BODIFLOXACIN binding to MIP was inhibited by 10%.37 The dynamic range (DR), i.e., the concentration range of ENRO that produces an inhibition of the signal between 20 and 80%, is between 290 nM and 21.5 μM. Furthermore, no competition was observed with the encoded NIP microspheres (n = 9, orange points), that were interrogated simultaneously with the MIP beads. This result confirms that nonspecific interactions are completely suppressed in the optimized assay conditions; therefore, it can be concluded that BODIFLOXACIN competes with ENRO for the specific binding sites in the MIP.
There are only a few examples in the literature of MIP-based optical methods for FQ detection, with none of the measuring schemes being compatible with a multiplexing approach, as described in this work. Moreover, except for the work of Xuan-Anh Ton et al.38 that reported the analysis of FQs using a solution of MIP nanoparticles and direct fluorescence polarization measurements, the detection limit achieved with the microarray is lower than for other competitive fluoroimmuno-like assays, using either structurally related or unrelated labeled analogues.39,40
The inter-day assay reproducibility was evaluated in terms of the relative standard deviation (RSD in %) for a fixed amount of BODIFLOXACIN measured in five independent tests (B0) performed during different months and at different temperatures (17–28 °C). The average RSD ranged from 3% to 8%. These values are comparable, or even better, than those reported in other studies using microsphere-based platforms with on a variety of bio-recognition elements, assay formats, and target analytes.41 In addition, improved reproducibility was observed when compared to other immunoassays for the detection of FQs described in the literature.42
Cross reactivity
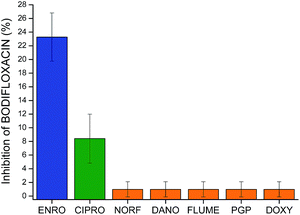
The potential cross-reactivity between MIP microspheres and other interfering substances was also evaluated by testing the microarrays with single antimicrobial standards. The responses caused by cross-reactivity, expressed as % Inhibition of BODIFLOXACIN, were less than 1% (see Fig. 7) for NORF, DANO, FLUME, PGP, and DOXY, while the inhibition obtained for CIPRO was 8%. Responses from NIP microspheres were negligible. These results confirm the excellent selectivity of the MIP based microarray for the template molecule and this lack of cross-reactivity represents a major improvement over many commercially available fluoroquinolone assays.41Sheep serum sample
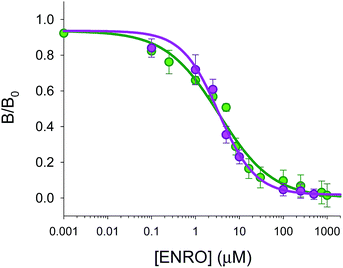
The optimized fiber-optic MIP array has been applied to the analysis of ENRO in sheep serum samples. In order to investigate the presence of matrix effects, sheep's serum samples not treated with antibiotics were diluted with water and spiked with increasing concentrations of ENRO, ranging from 0 to 500 μM and analysed using the microarray. Fig. 8 shows the comparison of the dose-calibration curves obtained in serum and in water. The results suggest that matrix effects for a 6-fold dilution of serum were negligible. No statistically significant differences were observed. In fact, the calibration plots overlap in water IC50 = 3.5 μM (±0.8 μM) compared to serum IC50 = 3.4 μM (±0.6 μM), as well as in the confidence intervals of 95% and 98%, respectively.Three samples of sheep serum were collected 5 min after drug administration, treated, and then measured using the array platform and HPLC-FLD for validation purposes. The results obtained using the MIP assay were 20 μM (±2 μM) of ENRO and 19.9 μM (±0.2 μM) of ENRO using the HPLC-FLD system. These results confirmed that no statistical differences in the variances and averages values at any confidence level were observed demonstrating the excellent performance of the fiber-optic array using MIP microspheres for ENRO analysis in serum samples.
Conclusions
In this paper, we described a new fiber-optic microarray based on MIP microspheres for the antibiotic ENRO. Exhaustive optimizations have been performed for all aspects of the assay including imprinted and non-imprinted microspheres synthesis and encoding, synthesis and characterization of a novel fluorescently-labeled ENRO, image processing, data analysis, and applications to real samples. Our platform, based on the combination of a microarray of optical fiber bundles with MIP microspheres and epi-fluorescence imaging detection, has not only shown the ability to improve upon the LODs previously described in the literature for this type of assay, but has also demonstrated its potential in multiplexed analysis. The basic assay design is flexible and should be applicable with other MIP microspheres selective to other target molecules. In addition, the platform can be applied to the analysis of real serum samples, can be used in the presence of organic solvents, and is both more reproducible and faster than other similar bioassays reported previously.Acknowledgements
Sergio Carrasco thanks the Ministry of Education, Culture and Sport for a doctoral grant (FPU). The authors gratefully acknowledge financial support from MINECO (CTQ2012-37573-C02-02). We thank Dr Alberto Benito-Peña and Dr Sonia Rubio from Faculty of Veterinary at Alfonso X “el Sabio” University (UAX) for their veterinary assistance and the donation of the sheep's serum blood samples. We also thank Stephanie M. Schubert and Thy L. Vu for insightful discussions during the preparation of this manuscript.Notes and references
- National Research Council, Inspired by Biology: From Molecules to Materials to Machines, The National Academies Press, Washington, DC, 2008 Search PubMed.
- W. Gunter and L. Junqiu, Acc. Chem. Res., 2012, 45, 239–247 CrossRef PubMed.
- B. Sellergren, Molecularly imprinted polymers. Man-made mimics of antibodies and their applications in analytical chemistry, Elsevier, Amsterdam, 2001 Search PubMed.
- L. Chen, S. Xuab and J. Lia, Chem. Soc. Rev., 2011, 40, 2922–2942 RSC.
- E. Benito-Peña, M. C. Moreno-Bondi, S. Aparicio, G. Orellana, J. Cederfur and M. Kempe, Anal. Chem., 2006, 78, 2019–2027 CrossRef PubMed.
- Handbook of Molecular Imprinting. Advanced Sensor Applications, ed. L. Woo and T. Kunitake, Pan Stanford Publishing, Singapore, 2013 Search PubMed.
- N. Perez-Moral and A. G. Mayes, in Molecular Imprinting of Polymers, ed. S. Piletsky and A. Turner, Landes Bioscience, Georgetown, 2006 Search PubMed.
- E. Rodriguez, F. Navarro-Villoslada, E. Benito-Peña, M. D. Marazuela and M. C. Moreno-Bondi, Anal. Chem., 2011, 83, 2046–2055 CrossRef CAS PubMed.
- Emerging Contaminants from Industrial and Municipal Waste. Ocurrence, Analysis and Effects, ed. D. Barceló and M. Petrovic, Springer-Verlag, Berlin-Heidelberg, 2008 Search PubMed.
- (a) Water Framework Directive (2000/60/EC), European Union, Brussels, 2000; (b) Directive (2008/105/EC), European Union, Brussels, 2008.
- L. Okerman, H. Noppe, V. Cornet and L. De Zutter, Food Addit. Contam., 2007, 24, 252–257 CrossRef CAS PubMed.
- W. Jiang, Z. Wang, R. C. Beier, H. Jiang, Y. Wu and J. Shen, Anal. Chem., 2013, 85, 1995–1999 CrossRef CAS PubMed.
- J. Zdunek, E. Benito-Peña, A. Linares, A. Falcimaigne-Cordin, G. Orellana, K. Haupt and M. C. Moreno-Bondi, Chem.–Eur. J., 2013, 19, 10209–10216 CrossRef PubMed.
- A. B. Descalzo, C. Somoza, M. C. Moreno-Bondi and G. Orellana, Anal. Chem., 2013, 85, 5316–5320 CrossRef CAS PubMed.
- S. Nie, W. H. Henley, S. E. Miller, H. Zhang, K. M. Mayer, P. J. Dennis, J. P. Alarie, Y. Wu, F. G. Oppenheim, F. F. Little, A. Z. Uluer, P. Wang, J. M. Ramsey and D. R. Walt, Lab Chip, 2014, 14, 1087–1098 RSC.
- K. Yoshimatsu, K. Reimhult, A. Krozer, K. Mosbach, K. Sode and L. Ye, Anal. Chim. Acta, 2007, 584, 112–121 CrossRef CAS PubMed.
- M. Manesse, A. F. Phillips, C. N. LaFratta, M. A. Palacios, R. B. Hayman and D. R. Walt, Lab Chip, 2013, 13, 2153–2160 RSC.
- H. Lin and K. S. Suslick, J. Am. Chem. Soc., 2010, 132, 15519–15521 CrossRef CAS PubMed.
- N. L. Rosi and C. A. Mirkin, Chem. Rev., 2005, 105, 1547–1562 CrossRef CAS PubMed.
- D. M. Rissin and D. R. Walt, Nano Lett., 2006, 6, 520–523 CrossRef CAS PubMed.
- D. M. Rissin and D. R. Walt, J. Am. Chem. Soc., 2006, 128, 6286–6287 CrossRef CAS PubMed.
- P. Pantano and D. R. Walt, Chem. Mater., 1996, 8, 2832–2835 CrossRef CAS.
- C. N. LaFratta and D. R. Walt, Chem. Rev., 2008, 108, 614–637 CrossRef CAS PubMed.
- G. Orellana, A. M. Gómez-Carneros, C. de Dios, A. A. García-Martínez and M. C. Moreno-Bondi, Anal. Chem., 1995, 67, 2231–2238 CrossRef CAS.
- T. M. Blicharz, W. L. Siqueira, E. J. Helmerhorst, F. G. Oppenheim, P. J. Wexler, F. F. Little and D. R. Walt, Anal. Chem., 2009, 81, 2106–2114 CrossRef CAS PubMed.
- R. F. Masseyeff, W. H. Albert and N. A. Staines, Methods of Immunological Analysis, Wiley-VCH, New York, 1st edn, 1992, pp. 655–671 Search PubMed.
- A. Rahal, A. Kumar, A. H. Ahmad, J. K. Malik and V. Ahuja, J. Vet. Pharmacol. Ther., 2006, 29, 321–324 CrossRef CAS PubMed.
- R. H. Wiley and J. I. Jin, J. Macromol. Sci., Chem., 1968, 6, 1097–1104 CrossRef.
- J. Brandrup, E. H. Immergut and E. A. Grulke, Polymer Handbook, Wiley-Interscience, New York, 4th edn, 1999, vol. VII, p. 507 Search PubMed.
- S. Nie, E. Benito-Peña, H. Zhang, Y. Wu and D. R. Walt, Anal. Chem., 2013, 85, 9272–9280 CrossRef CAS PubMed.
- D. G. Pinacho, F. F. Sánchez-Baeza and M. P. Marco, Anal. Chem., 2012, 84, 4527–4534 CrossRef CAS PubMed.
- G. T. Hermanson, Bioconjugate Techniques, Elsevier, Oxford, UK, Second edn, 2008 Search PubMed.
- G. Höfle, W. Steglich and H. Vorbrüggen, Angew. Chem., Int. Ed. Engl., 1978, 17, 569–583 CrossRef.
- B. Neises and W. Steglich, Angew. Chem., Int. Ed. Engl., 1978, 17, 522–524 CrossRef.
- C. S. Chen, W. N. U. Chen, M. Zhou, S. Arttamangkul and R. P. Haugland, J. Biochem. Biophys. Methods, 2000, 42, 137–151 CrossRef CAS.
- X. Tao, M. Chen, H. Jiang, J. Shen, Z. Wang, X. Wang, X. Wu and K. Wen, Anal. Bioanal. Chem., 2013, 405, 7477–7484 CrossRef CAS PubMed.
- M. P. Marco, S. Gee and B. D. Hammock, Trends Anal. Chem., 1995, 14, 341–350 CAS.
- T. Xuan-Anh, V. Acha, K. Haupt and B. T. S. Bui, Biosens. Bioelectron., 2012, 36, 22–28 CrossRef PubMed.
- K. Haupt, A. G. Mayes and K. Mosbach, Anal. Chem., 1998, 70, 3936–3939 CrossRef CAS.
- K. Haupt, React. Funct. Polym., 1999, 41, 125–131 CrossRef CAS.
- M. F. Elshal and J. P. McCoy, Methods, 2006, 38, 317–323 CrossRef CAS PubMed.
- G. Tsekenis, G. Z. Garifallou, F. Davis, P. A. Millner, D. G. Pinacho, F. Sanchez-Baeza, M. P. Marco, T. D. Gibson and S. P. J. Higson, Anal. Chem., 2008, 80, 9233–9239 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of: (1) synthesis of BODIFLOXACIN, (2) purification of BODIFLOXACIN by semi-preparative HPLC and spectroscopic characterization, (3) chromatographic conditions for the separation of fluoroquinolones and other antibiotics, (4) binding of BODIFLOXACIN and ENRO to the encoded polymer microspheres and, (5) image acquisition and data analysis. Also included: Scheme 1, Tables S1 and S2, Fig. S1–S5. See DOI: 10.1039/c5sc00115c |
| This journal is © The Royal Society of Chemistry 2015 |