Open Access Article
Open Access ArticleOxide nanowire microfluidics addressing previously-unattainable analytical methods for biomolecules towards liquid biopsy
Hiromi
Takahashi
a,
Yoshinobu
Baba
abc and
Takao
Yasui
 *abd
*abd
aDepartment of Biomolecular Engineering, Graduate School of Engineering, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8603, Japan
bInstitute of Nano-Life-Systems, Institutes of Innovation for Future Society, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8603, Japan
cInstitute of Quantum Life Science, National Institutes for Quantum Science and Technology, Anagawa 4-9-1, Inage-ku, Chiba 263-8555, Japan
dJapan Science and Technology Agency (JST), PRESTO, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan. E-mail: yasui@chembio.nagoya-u.ac.jp
First published on 26th November 2021
Abstract
Nanowire microfluidics using a combination of self-assembly and nanofabrication technologies is expected to be applied to various fields due to its unique properties. We have been working on the fabrication of nanowire microfluidic devices and the development of analytical methods for biomolecules using the unique phenomena generated by the devices. The results of our research are not just limited to the development of nanospace control with “targeted dimensions” in “targeted arrangements” with “targeted materials/surfaces” in “targeted spatial locations/structures” in microfluidic channels, but also cover a wide range of analytical methods for biomolecules (extraction, separation/isolation, and detection) that are impossible to achieve with conventional technologies. Specifically, we are working on the extraction technology “the cancer-related microRNA extraction method in urine,” the separation technology “the ultrafast and non-equilibrium separation method for biomolecules,” and the detection technology “the highly sensitive electrical measurement method.” These research studies are not just limited to the development of biomolecule analysis technology using nanotechnology, but are also opening up a new academic field in analytical chemistry that may lead to the discovery of new pretreatment, separation, and detection principles.
1. Introduction
Biomolecule analysis has greatly impacted the understanding of molecular mechanisms underlying human health and disease. The ability to analyze biomolecules is expected to provide a powerful tool for disease diagnosis based on molecular signatures.1 Because biomolecular analysis is largely dictated by the characteristics of biomolecules, an essential requirement for analysis is a sensitive method to overcome existing challenges and open new research fronts. Nanostructures fabricated by using self-assembly and nanofabrication technologies are expected to be applied to bioanalytical fields due to their unique properties and shapes.Breakthrough research in nanotechnology over the past decade has led to significant progress in biomolecular analysis with the emergence of engineered nanomaterials. Several types of nanomaterials, including zero-dimensional (0D) nanoparticles, one-dimensional (1D) nanowires, two-dimensional (2D) nanosheets, and three-dimensional (3D) nanoplates, were designed and developed to address challenges of materials and applications in traditional methods via their unique properties that lie in their nanoscale size.2–4 Among them, much effort has been made to explore and utilize the attractive properties of nanowires in a wide range of fields, including electronics, photonics, energy conversion, and storage, as well as biomedical applications, ever since Morales and Lieber reported a method to synthesize nanowires.5 The widespread adoption of nanowires has been made possible by the advancement of the chemical synthesis of nanowires, whereby unprecedented characteristics and properties could be obtained, thereby enhancing the effectiveness of applications.
In bioanalytical and biomedical applications, nanowires have come under the spotlight by virtue of their regime size compared with that of biomolecules and high surface area-to-volume ratio; both are the critical factors in achieving high levels of sensitivity.2,6,7 Several studies on the interfaces between nanowire devices and biological systems, notably by the Lieber group, have extensively explored a variety of applications over the past two decades. For example, a nanowire-based sensor was developed by the Lieber group in 2001 to be applied to biological and chemical systems.8 In that work, they demonstrated the possibility to exploit nanowires as biosensors by configuring them in a field-effect transistor (FET) system. The nanowires were modified with a receptor selective to a specific biomolecule; the receptors and biomolecules were 3-aminopropyltriethoxysilane (APTES), biotin, and calmodulin, which responded to changes in pH, streptavidin, and calcium ions, respectively. Since then, nanowires have been widely explored in biosensing applications. In another report by the Lieber group in 2005, they demonstrated a nanowire FET as a real-time multiplexed device for early-stage detection of cancer9 by modifying the surfaces of the nanowires with antibodies to detect four different cancer protein markers in serum samples. In 2012, Huang et al.10 developed a biosensor based on the earlier nanowire FET system,8,9 in which the nanowires were functionalized with glutaraldehyde to immobilize prostate-specific antigen, as a demonstration device to detect PSA biomarkers at the femtogram level in desalted human serum samples. These early investigations opened opportunities in the field of nanosensors for bioanalytical and biomedical applications, and led to the dramatic growth of research on nanowire-based sensors that continues even now.6,11–15
Besides nanowire-based sensors, there are several examples using nanowires as bioanalytical and biomedical tools that are due to the capabilities of nanowires to directly interface with biomolecules. For instance, the first cell-penetrating nanowire array for delivering genes was reported by the Yang group in 2007,16 and it showed that the nanowires could be used to deliver genes into living mammalian stem cells which were cultured on nanowires and survived up to a week. Later in 2009, Wang et al.17 demonstrated that 3D nanostructures coated with specific antibodies were able to capture circulating tumor cells (CTCs) with higher efficiency than a flat surface due to the high surface area-to-volume ratio of the nanowires. Based on a similar concept, previous studies have also reported nanowire arrays for isolating CD4+ T lymphocyte cells, a cancer biomarker, from heterogeneous cell mixtures by functionalizing nanowire surfaces with a specific antibody.18,19 More recently, the Cho group has developed a conductive polymer nanowire platform that is effective for capture and release based on a simple electrochemical method for biomolecules, such as cells,20 nucleic acids,21 and exosomes.22 Numerous studies on nanowires have been undertaken for interfacing with several biomolecules, including CTCs,23–32 nucleic acids,33–38 and exosomes,39–42 for a wide range of applications. These nanowire platforms showed improved biomolecule capture efficiency because of the nanowires, allowing for high sensitivity and specific recognition.
In the past decade there has been tremendous progress in the synthesis of nanowires, which results in new characteristics and properties, offering innovative applications. For example, Liu et al.43 demonstrated bendable polycrystalline nanowires that enable capture of bacteria in the bloodstream with higher efficiency than traditional nanowires due to their increased anti-shearing force toward bacteria. Ren et al.44 proposed a nanowire fabrication method based on a block copolymer-directed orthogonal assembly to construct arrays of 3D multilayer-crossed stacked metal oxide semiconducting nanowires, which can be subsequently doped with metal oxide, depending on a particular application. The nanowire arrays offer many target–receptor interfaces and improved molecule diffusion and good charge transport, which can benefit application in resistors, biosensors, and electrocatalysts (Table 1).
| Materials | Applications | Diameter, length, and surface area-to-volume ratioa | Ref. | Note |
|---|---|---|---|---|
| a If the diameter or length was not given in the cited reference(s), these parameters were measured from the electron microscope images. | ||||
| ZnO | Extracellular vesicle separation | ∼100 nm, ∼2 μm, ∼40 μm−1 | 39, 86 and 87 | Dissolve under alkaline conditions such as blood plasma51 |
| Urinary miRNA extraction | ∼100 nm, ∼2 μm, ∼40 μm−1 | 55 and 56 | ||
| Cell lysis and DNA extraction | ∼75 nm, ∼1 μm, ∼54 μm−1 | 38 | ||
| Circulating tumor cell separation | ∼200 nm, ∼1 μm, ∼21 μm−1 | 23 | ||
| Fluorescence-based DNA detection | ∼90 nm, ∼1.4 μm, ∼45 μm−1 | 97 | ||
| ZnO/ZnO | ssDNA extraction | ∼100 nm, ∼1.7 μm, ∼41 μm−1 | 49 | |
| ZnO/Al2O3 | ssDNA and RNA extraction | ∼100 nm, ∼1.7 μm, ∼41 μm−1 | 49 and 51 | Stable under alkaline conditions51 |
| Extracellular vesicle extraction | ∼100 nm, ∼2 μm, ∼40 μm−1 | 55 | ||
| ZnO/SiO2 | ssDNA extraction | ∼100 nm, ∼1.7 μm, ∼41 μm−1 | 49 and 50 | |
| Bacterial extraction | ∼70 nm, ∼1.2 μm, ∼58 μm−1 | 64 | ||
| ZnO/TiO2 | Extracellular vesicle separation | ∼100 nm, ∼2 μm, ∼40 μm−1 | 86 | |
| SnO2/SiO2 | DNA separation | ∼30 nm, ∼2 μm, ∼134 μm−1 | 78–80 | |
| SiO2 | DNA separation | ∼500 nm, ∼4 μm, ∼8 μm−1 | 69 and 70 | |
| SnO2 | Bacterial extraction | ∼10 nm, ∼7 μm, ∼400 μm−1 | 63 | |
| ZnO/(Cu1−xZnx)O | Electrical-based DNA detection | ∼100 nm, ∼2 μm, ∼40 μm−1 | 99 | |
| WO3 | Electrical DNA detection | ∼400 nm, ∼5 μm, ∼10 μm−1 | 98 | |
| SiO2 | Circulating tumor cell separation | ∼300 nm, ∼4 μm, ∼14 μm−1 | 27 | |
| Electrical-based DNA detection | ∼20 nm, ∼4 μm, ∼200 μm−1 | 8, 12 and 35 | ||
| Magnetic | Extracellular vesicle separation | ∼36 nm, ∼2.3 μm, ∼112 μm−1 | 40–42 | |
| Polymer | Circulating tumor cell separation | ∼200 nm, ∼14 μm, ∼20 μm−1 | 22 | |
The advent of microfluidics45 has brought about a paradigm shift in bioanalytical and biomedical applications of nanowires, and led to the emergence of the concept of microfluidic nanowire platforms. Integration of microfluidic systems and nanowires provides a variety of advantages over conventional methods. First, a microfluidic system coupled with nanoscale dimension wires offers several advantages, including enhanced sensitivity, reduced sample and reagent consumption, and shortened processing time. Second, the structural and physical properties of nanowires can be readily tailored to a particular application. Third, the surfaces of the nanowires can be modified with biomaterials, including enzymes, antibodies, and nucleic acids, which promises diverse applications with enhanced specificity.
In this feature article, we provide a comprehensive review on bioanalytical methods using nanowire microfluidic devices that are not feasible with conventional techniques (Fig. 1). We developed nanospace control with “targeted dimensions” in “targeted arrangements” with “targeted materials/surfaces” in “targeted spatial locations/structures” in microfluidic channels (Fig. 2) and deployed them for precise control of the nanospace to develop innovative bioanalysis. Specifically, we developed the following technologies: extraction technologies for biomolecules that have been difficult to extract with conventional techniques, separation technologies of biomolecules employing different separation principles from conventional technology, and detection technologies for biomolecules that enable highly sensitive electrical measurements. These studies are not just limited to the development of biomolecule analysis technologies using nanotechnology, but they are also opening up a new academic field in analytical chemistry that may lead to the discovery of new pretreatment, separation, and detection principles.
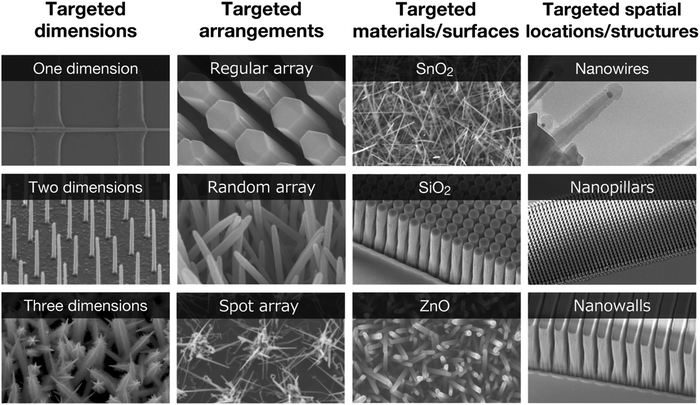 | ||
| Fig. 2 A concept of nanostructures with “targeted dimensions” in “targeted arrangements” with “targeted materials/surfaces” in “targeted spatial locations/structures” in microfluidic channels. | ||
2. Nanowire-based extraction
The extraction of nucleic acids is the initial and essential step in preparing them for downstream processes, a step that provides insights into their molecular signatures, which is the key to developing strategies for molecular diagnostics. The total yield and quality of biomolecules have a significant impact on the overall analytical performance. However, the existing technologies to extract biomolecules suffer from low extraction efficiency due to the low sensitivity of their extraction platforms. Therefore, there is a need to develop strategies that ensure the quality and maximum yield of nucleic acids, maximizing the effectiveness of analysis. Our work has focused on developing nanowire devices capable of extracting nucleic acids from different sources, including biological fluids, extracellular vesicles, and bacteria.2.1. DNA and RNA extraction from biological fluids
Tremendous progress over the past decade has been made in unraveling the clinical significance of circulating nucleic acids, including cell-free DNA, mRNA, and miRNA, as biomarkers and their role in diseases, to accelerate the growing field of nucleic acids for molecular diagnostics.46–49 Accessing molecular biomarkers in biological fluids, such as blood and urine, has been hampered by a lack of sensitive devices to isolate low levels of biomolecules in the presence of high levels of contaminants in the fluids. To overcome this challenge, we demonstrated that ZnO/Al2O3 core/shell nanowires can capture single-stranded DNA (ssDNA) with high efficiency, presumably due to the attraction of the phosphate backbones and the nucleobases of nucleic acids to nanowire surfaces.50 These findings suggest that ZnO/Al2O3 core/shell nanowires can be utilized as a nanostructure platform for capturing ssDNAs.Since the biocompatibility of nanostructures with biological fluids is essential for biomedical applications, in our recent work, we enhanced this biocompatibility by developing annealed ZnO/Al2O3 core/shell nanowires as a platform for capturing RNA molecules (Fig. 3).51 We demonstrated that our devices are biocompatible with blood plasma samples and enable the capture of RNA molecules with high efficiency. This work represents a feasibility study to develop nanowire devices for capturing nucleic acids. More in-depth studies, such as those regarding the interaction mechanism between nucleic acids and nanowire surfaces, would promote the direct isolation of circulating nucleic acids or even sequence-specific nucleic acids from biological samples.
 | ||
| Fig. 3 (a) Annealed ZnO/Al2O3 core/shell nanowires were developed as a platform for capturing RNA molecules.51 (b) Characterization of the annealed ZnO/Al2O3 core/shell nanowires by scanning electron microscopy (SEM; first row left), transmission electron microscopy (TEM; first row right and second row left), X-ray diffraction (second row right), and TEM-energy dispersive X-ray spectroscopy (third and fourth rows).51 | ||
2.2. MicroRNA extraction from extracellular vesicles (EVs) in biological fluids
A number of findings now point to the properties of EV-encapsulated miRNAs being involved in cell communication and epigenetic regulation,52,53 leading them to emerge as a new potential source for liquid biopsy. Use of urine-based liquid biopsies serves as a truly non-invasive approach to access biomolecular information for molecular diagnostics. However, genomic analysis of miRNAs is restricted in sensitivity due to their limited abundance in urine.54 To address this challenge, we proposed a nanowire-based methodology for collecting urine miRNAs including EV-encapsulated miRNAs and EV-free miRNAs,55 which facilitated the first direct extraction of miRNA from EVs in urine samples (Fig. 4). In this work, ZnO nanowires were grown from buried nanowires embedded in a PDMS substrate and bonded with the microchannel in which there was a PDMS herringbone-structure. We demonstrated that this device collected EVs at high efficiency and massively extracted various miRNAs that unveiled up to 1000 types of miRNA sequences. The effectiveness of the device is the result of the nanowire properties, the mechanical stability of the nanowires that are firmly anchored into the substrate and unaffected during buffer flow, and the electrostatic collection that enables negatively charged EVs to be collected with higher efficiency than by conventional methods. The results we obtained in this work have led us to employ this methodology further to extract miRNA from EVs in urine samples.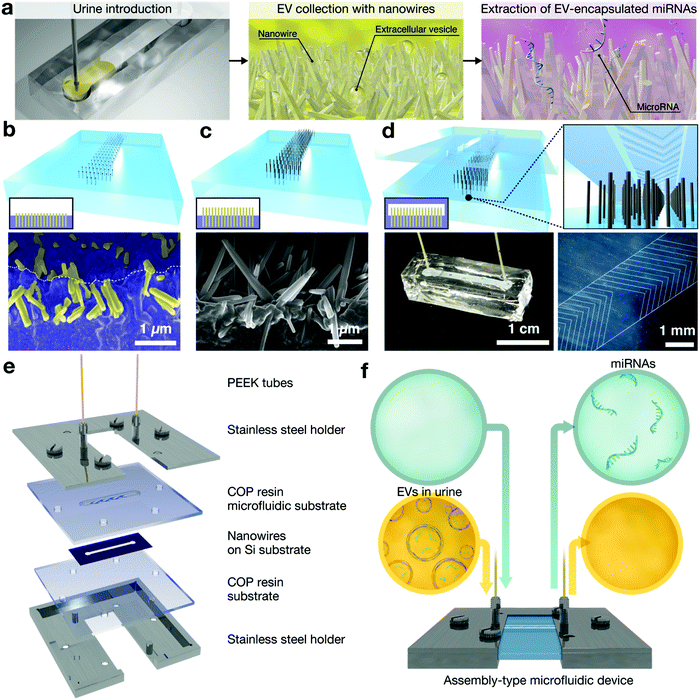 | ||
| Fig. 4 (a) Schematic of miRNA extraction in urine.55,56 (b–d) Schematic and SEM images for nanowire microfluidics to extract miRNAs from urine.55 (e) Exploded view of the assembly-type of nanowire microfluidics consisting of PEEK tubes, stainless steel holders, a COP resin microfluidic substrate, nanowires on a Si substrate, and a COP resin substrate.56 (f) Schematic of miRNA extraction from urine using the assembly-type of nanowire microfluidics.56 | ||
In one of our most recent papers, we developed a sterilizable and mass-producible nanowire-based device to help in meeting the long-term goal of achieving liquid biopsy using urinary miRNAs.56 An assembly-type microfluidic nanowire device was fabricated by two processes: first, ZnO nanowire scaffolds were fabricated on a silicon substrate, and second, ZnO nanowire scaffolds were assembled in an assembly-type microfluidic device (Fig. 4). We showed the effectiveness of our device to extract miRNAs from 1 mL of urine samples, and it provided a significantly greater variety and quantity of miRNA sequences compared to conventional methods. In addition, by identifying the origin of urinary miRNAs in patients with central nervous system (CNS) tumors, we confirmed that many CNS tumor-derived miRNAs could be identified in urine. These findings will surely impact the development of a diagnostic model based on urinary miRNA expression.
2.3. DNA extraction from bacteria
Bacteria are essential biomolecules that store genetic information both in single chromosomes, with the size of double-stranded DNA ranging from 600 kb to 9.5 Mb, and in plasmids, which play key roles in microorganisms.57 Therefore, this has resulted in several studies whose purposes were investigating antibiotic resistance from bacterial plasmids,58 molecular diagnostics of pathogens,59 and microbial genome sequencing from single microbial cells.60,61 The genetic information contained inside bacteria must be isolated via the lysis method based on enzymatic, thermal, chemical, or mechanical treatment.62 However, these treatments have many disadvantages, including ineffective lysis caused by insufficient thermal diffusion or reagent diffusion, and high costs and complex setups. To solve these challenges, we previously reported our concept of nanowire-mediated microbial cell lysis, which enables extraction of DNAs from microbial cells (Fig. 5).63,64 This concept relies on the biocompatibility, thinness, and flexibility of SnO2 nanowires, and on enhancing the absorption of bacteria by entanglements. Its mechanism involves mechanical force that stretches the cell membrane, thereby rupturing cells. In addition, we showed the versatility of a device that allows its integration with nucleic acid amplification assay, as the loop-mediated isothermal amplification method, to amplify target genes isothermally. The device realizing our concept has the advantages of being simple and rapid, and using fewer samples, all of which are very important as a microfluidic analytical platform.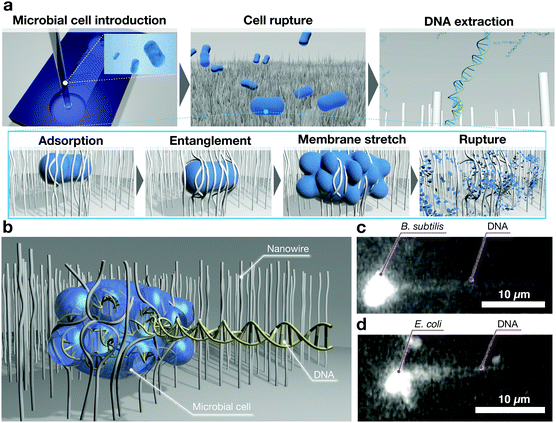 | ||
| Fig. 5 (a) Schematic of DNA extraction in microbial cells. The adsorbed cells on nanowires are ruptured by nanowire-entanglement and membrane stretching.63 (b–d) Schematic diagram of the moment of DNA extraction via nanowire-entanglement, and snapshots of fluorescence images of DNA extraction from B. subtilis and E. coli.63 | ||
3. Nanowire-based separation
In biomolecular analysis, separating biomolecules from a complex mixture is a crucial step in understanding the underlying mechanism of the functions in specific biomolecules. In general, the most widely used biomolecule separation method is electrophoresis,65 whereby biomolecules can be separated from each other under an electric field based on the charge-to-mass ratio. However, electrophoresis consumes time, labor, reagents, and sample resources. To overcome these limitations, nanostructure-based electrophoresis separation has been introduced due to its capability to manipulate fluid samples at the nanoscale, leading to reduced consumptions of time, labor, reagents, and sample resources. We have advanced the development of separation methods using nanostructures, such as nanopillars and nanowalls, for applications in combination with nanowires. Moreover, we have demonstrated the surface modification of nanowires with a specific peptide to separate biomolecules, including EVs and proteins, based on immunoaffinity, thereby promoting the separation efficiency.3.1. DNA separation using nanopillars and nanowalls
Toward improved performance of DNA separation using nanostructures, we proposed that a nanopillar array and a nanowall array were good candidates that affect the transport of DNA molecules.66–74 Since nonequilibrium transport is considered to have the potential to overcome the intrinsic trade-off between the separation speed and resolution of DNA separations, we demonstrated that the nonequilibrium transport of DNA molecules could be achieved by arranging a nanopillar square array, and we achieved a faster separation without any loss of resolution (Fig. 6).69 One major drawback of previous studies was the need for fluorescence labeling,75 which can affect the biomolecular characteristics. In our recent work, we aimed at label-free monitoring of biomolecular separation using the intensity variation of a laser diffracted using a nanopillar array,70 and we demonstrated that our proposed nanopillars yielded label-free monitoring and separating of DNA molecules in a nonequilibrium manner. In addition to the nanopillar array, we also proposed a nanowall array for DNA separations, methylation identification, and label-free detections (Fig. 6).71–74 We reported separation of a mixture of DNA fragments (48.5 kbp and 1 kbp fragments) within 30 s by applying an electric voltage, showing that larger DNA fragments migrate faster than shorter ones, and we confirmed that label-free detection based on the intensity variation of the laser diffracted by the nanowall array was achieved. These studies are not just limited to the development of DNA separation and detection technologies using nanotechnology, but are also opening up a new area of analytical chemistry that includes the discovery of new separation and detection principles.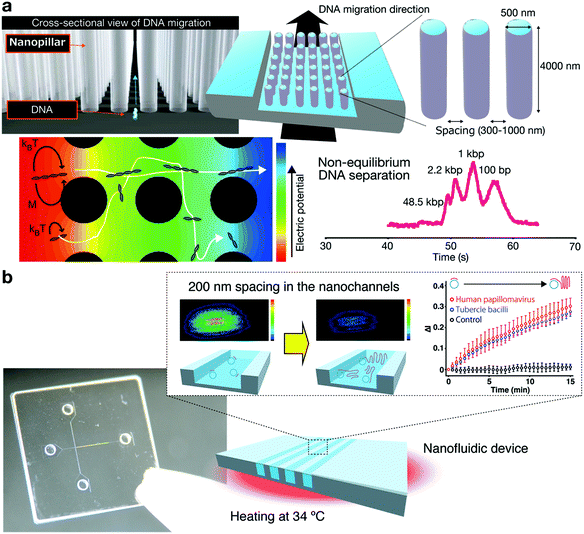 | ||
| Fig. 6 (a) Non-equilibrium DNA separation using nanopillar arrays.69 (b) Label-free detection of DNA amplification in a nanofluidic device.73 | ||
In electrophoretic separation, it is necessary to consider not only the separation mechanism, but also the various factors involved in the separation, such as electroosmotic flow (EOF) and preconcentration. We investigated the effect of electroosmotic flow and the possibility of preconcentration in arrays using nanopillars. We reported the first quantitative investigation and formulation of EOF mobility in microchannels with nanopillar array structures76 and demonstrated that the charge density of the nanopillar region affected the suppression of EOF, while the arrangement of nanopillars had no effect on it. These findings contributed to the development of a nanopillar array chip for DNA manipulation and separation. Moreover, to overcome the challenges for detecting low concentrations of target biomolecules in samples, an on-line stacking technique was proposed.77 The on-line stacking technique offers the capability to concentrate biomolecules into a smaller volume, and in this work, we developed a nanopillar array chip integrated with the on-line stacking for DNA separation, which allowed for faster separation, higher sensitivity and higher resolution compared to performance reported in previous studies. These results offer useful information for electrophoretic separation using not only nanopillars but also other nanostructures, such as nanowires.
3.2. DNA separation using nanowires
Despite the successful applications of nanopillar arrays and nanowall arrays for DNA separation, the fabrication process has remained technically challenging to obtain the desirable size range and form 3D designed spatial nanostructures. Nanowire arrays, therefore, have emerged as an alternative nanostructure due to the easy fabrication control for the desired size and spatial pattern. In addition, the thin structure of the nanowires offers a benefit for DNA separation, since it can elongate the DNA molecules effectively with minimal bending deformation. Among our early attempts to explore nanowire arrays, we developed a SnO2 nanowire spot-array for DNA manipulation and separation.78 This was the first time that a small structure with a diameter of about 10 nm had been fabricated at a targeted location inside a microchannel. We demonstrated that the nanowire spot-array could elongate long T4 DNA molecules via unique entanglement events. In addition, we found that the retardation of the electrophoretic mobility of DNA molecules caused DNA separation due to the frequent collisions and quick relaxation caused by the nanowire spot-array structure. The investigation presented in this work has provided the fundamentals for our development of nanowires for DNA manipulation and separation.To improve the resolution for DNA separation with the device, we have developed 3D nanowire arrays for ultra-fast separation of biomolecules.79–81 In our latest work, by advancing the self-assembling formation technology of nanowires, we fabricated branching nanowire structures and developed the world's first 3D nanowire arrays that can be spatially controlled to a level of a few nanometers at targeted locations in microchannels. The proposed methodology for controlling the pore size of the 3D nanowire arrays was varying the number of nanowire growth cycles (Fig. 7),81 which allows us to obtain nanowires sized as thin as the hydrodynamic radius of protein and RNA molecules.80 Using this technique, we have achieved fast separation of not only DNA but also RNA and proteins with molecular weights smaller than DNA within a few seconds to a few tens of seconds.
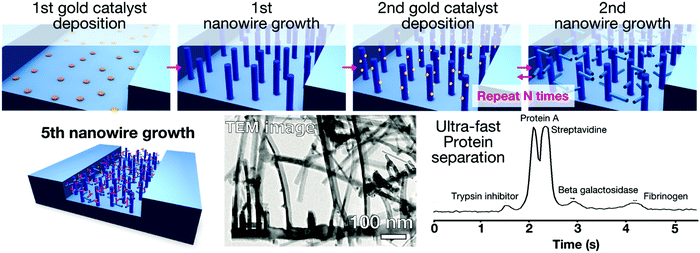 | ||
| Fig. 7 The 3D nanowire array for ultra-fast separation of biomolecules.79 The 3D nanowire array was fabricated by repeating catalyst deposition on the targeted special location and nanowire growth from the deposited catalyst. | ||
3.3. EV separation using nanowires
To take advantage of the full potential of EVs as biomarkers, methodologies that not only separate special subsets of EVs but also profile encapsulated miRNAs and membrane proteins of EVs are required to identify the correlation between the captured EV subsets and profiled EV-encapsulated miRNAs and EV membrane proteins. Current methodologies for EV separation are mainly divided into three groups:82 density-based separation using ultracentrifugation;83 size-based separation using size exclusion chromatography; and immunoaffinity-based separation (for specific membrane proteins, e.g., CD9, CD63, and CD81). As emerging alternatives, microfluidic-based platforms (density-, immunoaffinity-, and size-based separations)84,85 have been reported. The EV subsets obtained through density-, size-, and immunoaffinity-based separations have been extensively studied, but fewer efforts have been devoted to investigating the correlation between EV subsets obtained through surface charge-based separation and encapsulated miRNAs or expressed membrane proteins. We introduced a nanowire-based separation concept that could collect EVs based on nanowire surface potentials; the analysis targets were urinary microRNAs55,56 and expressed membrane proteins.86The feasibility of charge-based separation of EVs on nanowires was demonstrated by using different oxide materials86 (Fig. 8). As different oxide materials, we deposited TiO2 and SiO2 on ZnO nanowires to form different surface potentials of the nanowires in the same pH range. Around pH = 7, the ZnO (bare) nanowires had a positively charged surface, and the ZnO/TiO2 (core/shell) and ZnO/SiO2 (core/shell) nanowires had a negatively charged one. All types of nanowires could separate EVs, and the ZnO (bare) nanowires showed the highest separation efficiency among the three types of nanowires. Although the positively charged surfaces of the nanowires showed the highest separation efficiency, the negatively charged surfaces of the nanowires could also separate EVs. The ZnO (bare) nanowires have become our first choice for charge-based separation of EVs from the standpoint of separation efficiency.86
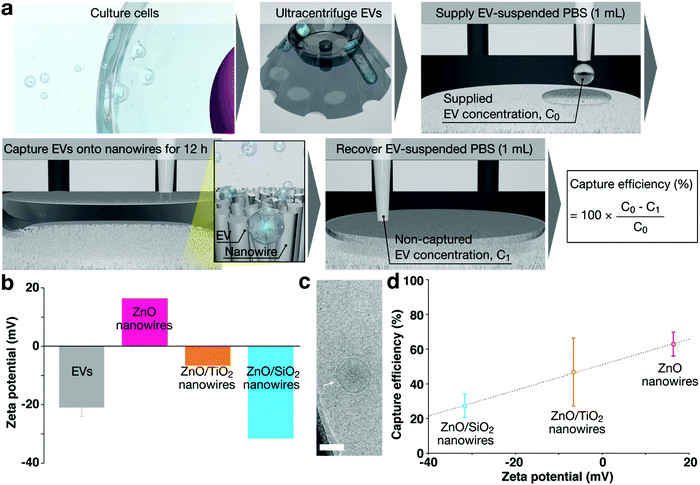 | ||
| Fig. 8 (a) Schematic showing steps for surface charge-based separation of EVs.86 (b) Zeta potentials of the EVs in PBS, and ZnO (bare), ZnO/TiO2 (core/shell) and ZnO/SiO2 (core/shell) nanowires in water.86 (c) A cryo-TEM image of the EVs; scale bar, 100 nm. The white arrow indicates the EVs.86 (d) Capture efficiency of EVs using different material nanowires (pink, ZnO (bare); orange, ZnO/TiO2 (core/shell); cyan, ZnO/SiO2 (core/shell)).86 | ||
Charge-based separation combined with immunoaffinity-based separation is expected to be used for enhanced specific separation of EVs with specific biomarkers. We recently reported peptide-functionalized ZnO nanowires in which the nanowires were functionalized with the bifunctional peptide specific to CD9 cancer-related EVs.87 Whereas the surface modification with antibodies could capture EVs, our methodology achieved functionalizing with the peptide without cross-linking in the immobilization step, and it provided simple, bio-friendly, and surface-specific functionalization strategies. This peptide-functionalized ZnO nanowire device provided capture and release of EVs under neutral salt conditions with higher efficiency than conventional methods.
3.4. Protein separation using nanowires
As alternatives to electrophoresis-based separation and charge-based separation, strategies based on molecular recognition have received attention as they may target specific molecules, enhancing the specificity and sensitivity of the device. As such, we proposed surface-modified nanowires for biomolecular recognition that would allow the separation of target biomolecules.88 In this work, we modified nanowires with thiolated 2-methacryloyloxyethyl phosphorylcholine (MPC-SH) to capture the target protein, by allowing for the reduction of nonspecific absorption. The surface-modified nanowires developed here recognize biomolecules on the surfaces of the nanowires, which can be used in nanowire-embedded biosensors for label-free detection (described in Section 4).4. Nanowire-based detection
The detection of biomolecules is a critical capability for the effective diagnosis and management of cancers and infectious diseases. Molecular biomarkers are unique disease indicators that have long been recognized for their effectiveness in providing genetic information from the very early stage until the terminal stage of a disease and in leading a move toward personalized treatment and medicine. Nevertheless, the detection of biomolecules is largely dictated by their characteristics that are rarely present in biological samples. Thus, an essential requirement is a highly sensitive method that can detect low levels of biomolecular analytes. Through the convergence of engineering and life science, the past decades have been witnessing significant progress in advanced biological sensors,89–96 in a wide range of areas, including genomics, proteomics, biomedical diagnostics, and drug discovery. The regime of molecular sizes mostly lies in the nanometer scale, necessitating that their detection capability be nanoscale. In this regard, nanowire-based sensors have been long recognized as suitable biosensors due to their comparable dimensions with respect to Debye length and due to their good sensitivity and short response time.4.1. Fluorescence detection on nanowires
For fluorescence detection, we proposed the idea of in situ detection of DNAs and RNAs captured on nanowires. Previous success had led us to further develop a nanowire device integrated with a microheater that was capable of in situ heating.97 We utilized the device for in situ annealing ssDNA to serinol nucleic acid with a molecular beacon (SNA-MB) probe. In this work, we exploited the properties of nanowires to capture ssDNAs. We demonstrated that our device realized in situ annealed SNA-MB probes by which it subsequently detected target ssDNAs. We envision that this work will serve as a fundamental study on the development of nanowire-based DNA detection.4.2. Electrical detection on nanowires
We also reported a strategy to enhance the electrical molecular sensing of nanowires for aldehydes; we used nonanal, a biomarker for sensing.98 The strategy was based on the surface treatment with a strong acid (sulfuric acid) of WO3 nanowires, in which the catalytic activity of the nanowires was promoted. Our results revealed that the surface treatment improved both the oxidization of nonanal and the desorption of its products (nonanoic acid) from the surfaces. Moreover, we confirmed the long-term stability of the nanowires with a sensitivity of 4 orders of magnitude.Besides this device development study, another study of our recent studies demonstrated the impact of surface cation composition on the adsorption and chemical transformation of carbonyl compounds.99 We demonstrated that the suppressed adsorption of nonanal was affected by the surface-exposed Cu2+ and the surface Cu/Zn ratio of ZnO/(Cu1−xZnx)O heterostructure nanowires (Fig. 9). This behavior was interpreted to mean that the inactive surface Cu2+ site suppresses the aldol condensation reaction, preventing the adsorption of molecules on the Zn2+ site. The results we obtained promote understanding of the surface cation composition of nanoscale metal oxides, facilitating the future development of molecular sensing applications.
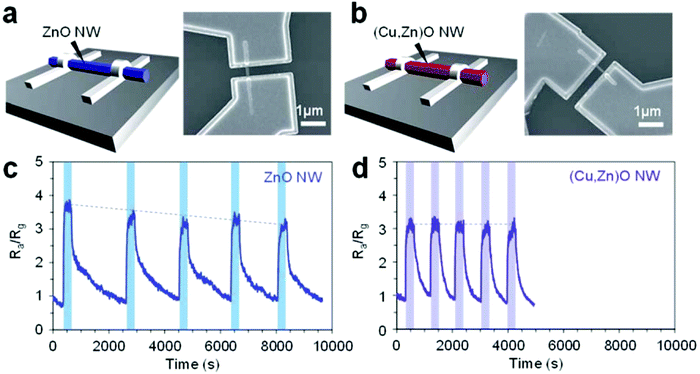 | ||
| Fig. 9 (a and b) Schematic and SEM images of single nanowire sensor devices using the (a) ZnO nanowire and (b) (Cu,Zn)O nanowire.99 (c and d) Five successive sensor responses to nonanal using the ZnO nanowire device and (Cu,Zn)O nanowire device, respectively.99 | ||
We proposed a method to simplify fabrication of a single-nanowire device.100–102 In this report, a single nanowire was obtained by the synthesis of a sparse array of ZnO nanowires. The nanowires were grown through hydrothermal synthesis with the addition of polyethylenimine (PEI) and NH3 to achieve spatially long nanowires. This nanowire device showed ∼100 (Iuv/Idark) sensitivity on exposure to UV irradiation. Further studies related to this work should be able to promote the development of a single nanowire device for biomolecule detection.
5. Conclusions
In summary, we have brought innovations to bioanalytical methods using nanowires. We are proud that our pioneering research results based on these original ideas will contribute to the development of interdisciplinary research and the creation of a new discipline within analytical chemistry. Currently, fierce research and development is being conducted worldwide on extraction, separation, and detection technologies for disease-related biomolecules. In this context, we have developed nanowire devices that are extremely effective for the analysis of various biomolecules. In light of the fact that clinically meaningful information about diseases can be obtained from body fluid samples, which until a few decades ago had been the stuff of science fiction, we believe that further improvement and commercialization of various nanowire devices will lead to the realization of liquid biopsy for various diseases, including cancer, using biomolecules in body fluids as diagnostic markers.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This review was supported by PRESTO (JPMJPR19H9), Japan Science and Technology Agency (JST), the Strategic International Research Cooperative Program (eASIA, JPMJSC19E3), JST, the JSPS Grant-in-Aid for Scientific Research (B) 21H01960, the JSPS Grant-in-Aid for Scientific Research (S) 18H05243, the JSPS Grant-in-Aid for Exploratory Research 20K21124, and the JSPS Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems”.References
- R. Mayeux, NeuroRx, 2004, 1, 182–188 CrossRef PubMed.
- Y. N. Xia, P. D. Yang, Y. G. Sun, Y. Y. Wu, B. Mayers, B. Gates, Y. D. Yin, F. Kim and Y. Q. Yan, Adv. Mater., 2003, 15, 353–389 CrossRef CAS.
- A. Zhang and C. M. Lieber, Chem. Rev., 2016, 116, 215–257 CrossRef CAS PubMed.
- J. Hu, T. W. Odom and C. M. Lieber, Acc. Chem. Res., 1999, 32, 435–445 CrossRef CAS.
- A. M. Morales and C. M. Lieber, Science, 1998, 279, 208–211 CrossRef CAS PubMed.
- B. Tian and C. M. Lieber, Chem. Rev., 2019, 119, 9136–9152 CrossRef CAS PubMed.
- G. Zhou, L. Wang, X. Wang, Y. Yu and A. Mutzke, Vacuum, 2019, 166, 88–96 CrossRef CAS.
- Y. Cui, Q. Q. Wei, H. K. Park and C. M. Lieber, Science, 2001, 293, 1289–1292 CrossRef CAS PubMed.
- G. F. Zheng, F. Patolsky, Y. Cui, W. U. Wang and C. M. Lieber, Nat. Biotechnol., 2005, 23, 1294–1301 CrossRef CAS PubMed.
- Y.-W. Huang, C.-S. Wu, C.-K. Chuang, S.-T. Pang, T.-M. Pan, Y.-S. Yang and F.-H. Ko, Anal. Chem., 2013, 85, 7912–7918 CrossRef CAS PubMed.
- K. Dawson and A. O’Riordan, J. Phys.: Conf. Ser., 2011, 307, 012004 CrossRef.
- R. Smith, S. M. Geary and A. K. Salem, ACS Appl. Nano Mater., 2020, 3, 8522–8536 CrossRef CAS.
- E. Garnett, L. Mai and P. Yang, Chem. Rev., 2019, 119, 8955–8957 CrossRef CAS PubMed.
- S. Rahong, T. Yasui, N. Kaji and Y. Baba, Lab Chip, 2016, 16, 1126–1138 RSC.
- H. Zeng, G. Zhang, K. Nagashima, T. Takahashi, T. Hosomi and T. Yanagida, Chemosensors, 2021, 9, 41 CrossRef CAS.
- W. Kim, J. K. Ng, M. E. Kunitake, B. R. Conklin and P. D. Yang, J. Am. Chem. Soc., 2007, 129, 7228–7229 CrossRef CAS PubMed.
- S. Wang, H. Wang, J. Jiao, K. J. Chen, G. E. Owens, K. Kamei, J. Sun, D. J. Sherman, C. P. Behrenbruch, H. Wu and H. R. Tseng, Angew. Chem., Int. Ed., 2009, 48, 8970–8973 CrossRef CAS PubMed.
- D.-J. Kim, J.-K. Seol, M.-R. Lee, J.-H. Hyung, G.-S. Kim, T. Ohgai and S.-K. Lee, Appl. Phys. Lett., 2012, 100, 163703 CrossRef.
- S. T. Kim, D.-J. Kim, T.-J. Kim, D.-W. Seo, T.-H. Kim, S.-Y. Lee, K. Kim, K.-M. Lee and S.-K. Lee, Nano Lett., 2010, 10, 2877–2883 CrossRef CAS PubMed.
- W. Y. Hong, S. H. Jeon, E. S. Lee and Y. Cho, Biomaterials, 2014, 35, 9573–9580 CrossRef CAS PubMed.
- S. Jeon, H. Lee, K. Bae, K. A. Yoon, E. S. Lee and Y. Cho, Theranostics, 2016, 6, 828–836 CrossRef CAS PubMed.
- J. Lim, M. Choi, H. Lee, J.-Y. Han and Y. Cho, Front. Chem., 2019, 6, 664 CrossRef PubMed.
- H. Cui, Q. Liu, R. Li, X. Wei, Y. Sun, Z. Wang, L. Zhang, X.-Z. Zhao, B. Hua and S.-S. Guo, Nanoscale, 2020, 12, 1455–1463 RSC.
- M. A. Doucey and S. Carrara, Trends Biotechnol., 2019, 37, 86–99 CrossRef CAS PubMed.
- Y. D. Ivanov, T. S. Romanova, K. A. Malsagova, T. O. Pleshakova and A. I. Archakov, Molecules, 2021, 26, 3734 CrossRef CAS PubMed.
- H. J. Yoon, M. Kozminsky and S. Nagrath, ACS Nano, 2014, 8, 1995–2017 CrossRef CAS PubMed.
- C. Wang, M. Ye, L. Cheng, R. Li, W. Zhu, Z. Shi, C. Fan, J. He, J. Liu and Z. Liu, Biomaterials, 2015, 54, 55–62 CrossRef CAS PubMed.
- S.-K. Lee, G.-S. Kim, Y. Wu, D.-J. Kim, Y. Lu, M. Kwak, L. Han, J.-H. Hyung, J.-K. Seol and C. Sander, Nano Lett., 2012, 12, 2697–2704 CrossRef CAS PubMed.
- M. Kwak, L. Han, J. J. Chen and R. Fan, Small, 2015, 11, 5600–5610 CrossRef CAS PubMed.
- W. Hong, S. Lee, H. J. Chang, E. S. Lee and Y. Cho, Biomaterials, 2016, 106, 78–86 CrossRef CAS PubMed.
- Q. Shen, H. Yang, C. Peng, H. Zhu, J. Mei, S. Huang, B. Chen, J. Liu, W. Wu and S. Cao, Int. J. Nanomed., 2019, 14, 205 CrossRef CAS PubMed.
- H. Lee, M. Choi, J. Lim, M. Jo, J.-Y. Han, T. M. Kim and Y. Cho, Theranostics, 2018, 8, 505 CrossRef CAS PubMed.
- R. Niepelt, U. C. Schröder, J. Sommerfeld, I. Slowik, B. Rudolph, R. Möller, B. Seise, A. Csaki, W. Fritzsche and C. Ronning, Nanoscale Res. Lett., 2011, 6, 1–7 CrossRef PubMed.
- W. C. Maki, N. N. Mishra, E. G. Cameron, B. Filanoski, S. K. Rastogi and G. K. Maki, Biosens. Bioelectron., 2008, 23, 780–787 CrossRef CAS PubMed.
- Z. Gao, A. Agarwal, A. D. Trigg, N. Singh, C. Fang, C.-H. Tung, Y. Fan, K. D. Buddharaju and J. Kong, Anal. Chem., 2007, 79, 3291–3297 CrossRef CAS PubMed.
- H. Lee, N. R. Hwang, S.-H. Hwang and Y. Cho, Biosens. Bioelectron., 2016, 86, 864–870 CrossRef CAS PubMed.
- Y. He, S. Su, T. Xu, Y. Zhong, J. A. Zapien, J. Li, C. Fan and S.-T. Lee, Nano Today, 2011, 6, 122–130 CrossRef CAS.
- H. Y. So, K. Lee, N. Murthy and A. P. Pisano, ACS Appl. Mater. Interfaces, 2014, 6, 20693–20699 CrossRef CAS PubMed.
- Z. Chen, S.-B. Cheng, P. Cao, Q.-F. Qiu, Y. Chen, M. Xie, Y. Xu and W.-H. Huang, Biosens. Bioelectron., 2018, 122, 211–216 CrossRef CAS PubMed.
- J. Lim, M. Choi, H. Lee, Y.-H. Kim, J.-Y. Han, E. S. Lee and Y. Cho, J. Nanobiotechnol., 2019, 17, 1–12 CrossRef PubMed.
- Z. Nemati, M. R. Zamani Kouhpanji, F. Zhou, R. Das, K. Makielski, J. Um, M.-H. Phan, A. Muela, M. Fdez-Gubieda and R. R. Franklin, Nanomaterials, 2020, 10, 1662 CrossRef CAS PubMed.
- Z. Nemati, J. Um, M. R. Zamani Kouhpanji, F. Zhou, T. Gage, D. Shore, K. Makielski, A. Donnelly and J. Alonso, ACS Appl. Nano Mater., 2020, 3, 2058–2069 CrossRef CAS.
- L. Liu, S. Chen, Z. Xue, Z. Zhang, X. Qiao, Z. Nie, D. Han, J. Wang and T. Wang, Nat. Commun., 2018, 9, 444 CrossRef PubMed.
- Y. Ren, Y. Zou, Y. Liu, X. Zhou, J. Ma, D. Zhao, G. Wei, Y. Ai, S. Xi and Y. Deng, Nat. Mater., 2020, 19, 203–211 CrossRef CAS PubMed.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- J. C. M. Wan, C. Massie, J. Garcia-Corbacho, F. Mouliere, J. D. Brenton, C. Caldas, S. Pacey, R. Baird and N. Rosenfeld, Nat. Rev. Cancer, 2017, 17, 223–238 CrossRef CAS PubMed.
- I. Amelio, R. Bertolo, P. Bove, O. C. Buonomo, E. Candi, M. Chiocchi, C. Cipriani, N. Di Daniele, C. Ganini, H. Juhl, A. Mauriello, C. Marani, J. Marshall, M. Montanaro, G. Palmieri, M. Piacentini, G. Sica, M. Tesauro, V. Rovella, G. Tisone, Y. Shi, Y. Wang and G. Melino, Cell Death Discovery, 2020, 6, 131 CrossRef CAS PubMed.
- H. Takahashi, T. Yasui and Y. Baba, Chem. Lett., 2021, 50, 1244–1253 CrossRef CAS.
- M. Musa, T. Yasui, K. Nagashima, M. Horiuchi, Z. T. Zhu, Q. L. Liu, T. Shimada, A. Arima, T. Yanagida and Y. Baba, Anal. Methods, 2021, 13, 337–344 RSC.
- M. Musa, T. Yasui, Z. Zhu, K. Nagashima, M. Ono, Q. Liu, H. Takahashi, T. Shimada, A. Arima, T. Yanagida and Y. Baba, Anal. Sci., 2021, 37, 1139–1145 CrossRef CAS PubMed.
- H. Takahashi, T. Yasui, A. Klamchuen, N. Khemasiri, T. Wuthikhun, P. Paisrisarn, K. Shinjo, Y. Kitano, K. Aoki, A. Natsume, S. Rahong and Y. Baba, Nanomaterials, 2021, 11, 1768 CrossRef CAS PubMed.
- A. M. Rusek, M. Abba, A. Eljaszewicz, M. Moniuszko, J. Niklinski and H. Allgayer, Mol. Cancer, 2015, 14, 1–10 CrossRef CAS PubMed.
- Z. Qian, Q. Shen, X. Yang, Y. Qiu and W. Zhang, BioMed Res. Int., 2015, 2015, 649161 Search PubMed.
- P. Paisrisarn, T. Yasui and Y. Baba, Anal. Sci., 2020, 36, 791–798 CrossRef CAS PubMed.
- T. Yasui, T. Yanagida, S. Ito, Y. Konakade, D. Takeshita, T. Naganawa, K. Nagashima, T. Shimada, N. Kaji, Y. Nakamura, I. A. Thiodorus, Y. He, S. Rahong, M. Kanai, H. Yukawa, T. Ochiya, T. Kawai and Y. Baba, Sci. Adv., 2017, 3, e1701133 CrossRef PubMed.
- Y. Kitano, K. Aoki, F. Ohka, S. Yamazaki, K. Motomura, K. Tanahashi, M. Hirano, T. Naganawa, M. Iida, Y. Shiraki, T. Nishikawa, H. Shimizu, J. Yamaguchi, S. Maeda, H. Suzuki, T. Wakabayashi, Y. Baba, T. Yasui and A. Natsume, ACS Appl. Mater. Interfaces, 2021, 13, 17316–17329 CrossRef CAS PubMed.
- A. Griswold, Nat. Education, 2008, 1, 57 Search PubMed.
- P. M. Bennett, Br. J. Pharmacol., 2008,(153 Suppl 1), S347–S357 CrossRef CAS PubMed.
- L. Váradi, J. L. Luo, D. E. Hibbs, J. D. Perry, R. J. Anderson, S. Orenga and P. W. Groundwater, Chem. Soc. Rev., 2017, 46, 4818–4832 RSC.
- M. C. Wilson, T. Mori, C. Ruckert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. E. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. N. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crusemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, Nature, 2014, 506, 58–62 CrossRef CAS PubMed.
- C. Rinke, P. Schwientek, A. Sczyrba, N. N. Ivanova, I. J. Anderson, J. F. Cheng, A. Darling, S. Malfatti, B. K. Swan, E. A. Gies, J. A. Dodsworth, B. P. Hedlund, G. Tsiamis, S. M. Sievert, W. T. Liu, J. A. Eisen, S. J. Hallam, N. C. Kyrpides, R. Stepanauskas, E. M. Rubin, P. Hugenholtz and T. Woyke, Nature, 2013, 499, 431–437 CrossRef CAS PubMed.
- M. Shehadul Islam, A. Aryasomayajula and P. R. Selvaganapathy, Micromachines, 2017, 8, 83 CrossRef.
- T. Yasui, T. Yanagida, T. Shimada, K. Otsuka, M. Takeuchi, K. Nagashima, S. Rahong, T. Naito, D. Takeshita, A. Yonese, R. Magofuku, Z. T. Zhu, N. Kaji, M. Kanai, T. Kawai and Y. Baba, ACS Nano, 2019, 13, 2262–2273 CAS.
- T. Shimada, T. Yasui, A. Yonese, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, Micromachines, 2020, 11, 610 CrossRef PubMed.
- T. B. L. Kist and M. Mandaji, Electrophoresis, 2004, 25, 3492–3497 CrossRef CAS PubMed.
- T. Yasui, N. Kaji and Y. Baba, Annu. Rev. Anal. Chem., 2013, 6, 83–96 CrossRef CAS PubMed.
- T. Yasui, Y. Inoue, T. Naito, Y. Okamoto, N. Kaji, M. Tokeshi and Y. Baba, Anal. Chem., 2012, 84, 9282–9286 CrossRef CAS PubMed.
- T. Yasui, S. Rahong, N. Kaji and Y. Baba, Isr. J. Chem., 2014, 54, 1556–1563 CrossRef CAS.
- T. Yasui, N. Kaji, R. Ogawa, S. Hashioka, M. Tokeshi, Y. Horiike and Y. Baba, Nano Lett., 2015, 15, 3445–3451 CrossRef CAS PubMed.
- T. Ajiri, T. Yasui, M. Maeki, A. Ishida, H. Tani, J. Nishii, Y. Baba and M. Tokeshi, ACS Appl. Nano Mater., 2020, 3, 8810–8816 CrossRef CAS.
- T. Yasui, N. Kaji, R. Ogawa, S. Hashioka, M. Tokeshi, Y. Horiike and Y. Baba, Anal. Chem., 2011, 83, 6635–6640 CrossRef CAS PubMed.
- X. Y. Sun, T. Yasui, T. Yanagida, N. Kaji, S. Rahong, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, Sci. Technol. Adv. Mater., 2016, 17, 644–649 CrossRef CAS PubMed.
- T. Yasui, K. Ogawa, N. Kaji, M. Nilsson, T. Ajiri, M. Tokeshi, Y. Horiike and Y. Baba, Sci. Rep., 2016, 6, 31642–31649 CrossRef CAS PubMed.
- T. Ajiri, T. Yasui, M. Maeki, A. Ishida, H. Tani, Y. Baba and M. Tokeshi, Sens. Actuators, B, 2017, 250, 39–43 CrossRef CAS.
- T. Yasui, K. Motoyama, N. Kaji, M. Tokeshi and Y. Baba, RSC Adv., 2013, 3, 3237–3240 RSC.
- T. Yasui, N. Kaji, M. R. Mohamadi, Y. Okamoto, M. Tokeshi, Y. Horiike and Y. Baba, ACS Nano, 2011, 5, 7775–7780 CrossRef CAS PubMed.
- T. Yasui, N. Kaji, Y. Okamoto, M. Tokeshi, Y. Horiike and Y. Baba, Microfluid. Nanofluid., 2013, 14, 961–967 CrossRef CAS.
- T. Yasui, S. Rahong, K. Motoyama, T. Yanagida, Q. Wu, N. Kaji, M. Kanai, K. Doi, K. Nagashima, M. Tokeshi, M. Taniguchi, S. Kawano, T. Kawai and Y. Baba, ACS Nano, 2013, 7, 3029–3035 CrossRef CAS PubMed.
- S. Rahong, T. Yasui, T. Yanagida, K. Nagashima, M. Kanai, G. Meng, Y. He, F. W. Zhuge, N. Kaji, T. Kawai and Y. Baba, Sci. Rep., 2015, 5, 10584 CrossRef CAS PubMed.
- S. Rahong, T. Yasui, T. Yanagida, K. Nagashima, M. Kanai, G. Meng, Y. He, F. W. Zhuge, N. Kaji, T. Kawai and Y. Baba, Anal. Sci., 2015, 31, 153–157 CrossRef CAS PubMed.
- S. Rahong, T. Yasui, T. Yanagida, K. Nagashima, M. Kanai, A. Klamchuen, G. Meng, Y. He, F. Zhuge, N. Kaji, T. Kawai and Y. Baba, Sci. Rep., 2014, 4, 5252–5259 CrossRef CAS PubMed.
- R. Szatanek, J. Baran, M. Siedlar and M. Baj-Krzyworzeka, Int. J. Mol. Med., 2015, 36, 11–17 CrossRef CAS PubMed.
- D. K. Jeppesen, A. M. Fenix, J. L. Franklin, J. N. Higginbotham, Q. Zhang, L. J. Zimmerman, D. C. Liebler, J. Ping, Q. Liu, R. Evans, W. H. Fissell, J. G. Patton, L. H. Rome, D. T. Burnette and R. J. Coffey, Cell, 2019, 177, 428–445 CrossRef CAS PubMed.
- J. C. Contreras-Naranjo, H. J. Wu and V. M. Ugaz, Lab Chip, 2017, 17, 3558–3577 RSC.
- Y. Hattori, T. Shimada, T. Yasui, N. Kaji and Y. Baba, Anal. Chem., 2019, 91, 6514–6521 CrossRef CAS PubMed.
- T. Yasui, P. Paisrisarn, T. Yanagida, Y. Konakade, Y. Nakamura, K. Nagashima, M. Musa, I. A. Thiodorus, H. Takahashi, T. Naganawa, T. Shimada, N. Kaji, T. Ochiya, T. Kawai and Y. Baba, Biosens. Bioelectron., 2021, 194, 113589 CrossRef CAS PubMed.
- T. Suwatthanarak, I. A. Thiodorus, M. Tanaka, T. Shimada, D. Takeshita, T. Yasui, Y. Baba and M. Okochi, Lab Chip, 2021, 21, 597–607 RSC.
- T. Shimada, T. Yasui, A. Yokoyama, T. Goda, M. Hara, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, Y. Miyahara, T. Kawai and Y. Baba, Lab Chip, 2018, 18, 3225–3229 RSC.
- T. Shimada, H. Yasaki, T. Yasui, N. Kaji and Y. Baba, IEEE Int. Conf. Nano, 2018, 240–243 Search PubMed.
- H. Yasaki, T. Shimada, T. Yasui, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, ACS Sens., 2018, 3, 574–579 CrossRef CAS PubMed.
- M. Sano, N. Kaji, A. C. Rowat, H. Yasaki, L. Shao, H. Odaka, T. Yasui, T. Higashiyama and Y. Baba, Anal. Chem., 2019, 91, 12890–12899 CrossRef CAS PubMed.
- T. Suzuki, N. Kaji, H. Yasaki, T. Yasui and Y. Baba, Anal. Chem., 2020, 92, 2483–2491 CrossRef CAS PubMed.
- T. Shimada, H. Yasaki, T. Yasui, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, Anal. Sci., 2018, 34, 1347–1349 CrossRef CAS PubMed.
- H. Yasaki, T. Yasui, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, Chem. Lett., 2018, 47, 350–353 CrossRef CAS.
- H. Yasaki, T. Yasui, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, J. Am. Chem. Soc., 2017, 139, 14137–14142 CrossRef CAS PubMed.
- H. Yasaki, T. Yasui, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, Sens. Actuators, B, 2018, 260, 746–752 CrossRef CAS.
- H. Takahashi, T. Yasui, H. Kashida, K. Makino, K. Shinjo, Q. Liu, T. Shimada, S. Rahong, N. Kaji, H. Asanuma and Y. Baba, Nanotechnology, 2021, 32, 255301 CrossRef CAS PubMed.
- G. Zhang, T. Hosomi, W. Mizukami, J. Liu, K. Nagashima, T. Takahashi, M. Kanai, T. Sugiyama, T. Yasui, Y. Aoki, Y. Baba, J. C. Ho and T. Yanagida, J. Mater. Chem. A, 2021, 9, 5815–5824 RSC.
- J. Liu, K. Nagashima, Y. Nagamatsu, T. Hosomi, H. Saito, C. Wang, W. Mizukami, G. Zhang, B. Samransuksamer, T. Takahashi, M. Kanai, T. Yasui, Y. Baba and T. Yanagida, Chem. Sci., 2021, 12, 5073–5081 RSC.
- Q. Liu, T. Yasui, K. Nagashima, T. Yanagida, M. Horiuchi, Z. Zhu, H. Takahashi, T. Shimada, A. Arima and Y. Baba, Anal. Sci., 2020, 36, 1125–1129 CrossRef CAS PubMed.
- Q. L. Liu, T. Yasui, K. Nagashima, T. Yanagida, M. Hara, M. Horiuchi, Z. T. Zhu, H. Takahashi, T. Shimada, A. Arima and Y. Baba, J. Phys. Chem. C, 2020, 124, 20563–20568 CrossRef CAS.
- Z. Zhu, T. Yasui, Q. Liu, K. Nagashima, T. Takahashi, T. Shimada, T. Yanagida and Y. Baba, Micromachines, 2021, 12, 642 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2021 |