Portable all-in-one microfluidic system for CRISPR–Cas13a-based fully integrated multiplexed nucleic acid detection†
Ya
Zhang‡
 ab,
Yue
Guo‡
cd,
Guozhen
Liu
ab,
Shiqi
Zhou
ab,
Rouyu
Su
ab,
Qian
Ma
ab,
Yiyue
Ge
cd,
Yan-qing
Lu
*a,
Lunbiao
Cui
*cd and
Guanghui
Wang
ab,
Yue
Guo‡
cd,
Guozhen
Liu
ab,
Shiqi
Zhou
ab,
Rouyu
Su
ab,
Qian
Ma
ab,
Yiyue
Ge
cd,
Yan-qing
Lu
*a,
Lunbiao
Cui
*cd and
Guanghui
Wang
 *ab
*ab
aCollege of Engineering and Applied Sciences, Nanjing University, Jiangsu 210093, China. E-mail: yqlu@nju.edu.cn; wangguanghui@nju.edu.cn
bKey Laboratory of Intelligent Optical Sensing and Integration of the Ministry of Education, Nanjing University, Jiangsu 210093, China
cNHC Key Laboratory of Enteric Pathogenic Microbiology, Jiangsu Provincial Medical Key Laboratory of Pathogenic Microbiology in Emerging Major Infectious Diseases, Jiangsu 210009, China
dJiangsu Province Engineering Research Center of Health Emergency, Jiangsu Provincial Center for Disease Control and Prevention, Jiangsu 210009, China. E-mail: lbcui@jscdc.cn
First published on 31st May 2024
Abstract
Point-of-care testing of “sample in, answer out” is urgently needed for communicable diseases. Recently, rapid nucleic acid tests for infectious diseases have been developed for use in resource-limited areas, but they require types of equipment in central laboratories and are poorly integrated. In this work, a portable centrifugal microfluidic testing system is developed, integrated with magnetic bead-based nucleic acid extraction, recombinase-assisted amplification and CRISPR–Cas13a detection. The system, with the advantage of its power-supplied active rotating chip and highly programable flow control through integrated addressable active thermally-triggered wax valves, has a rapid turnaround time within 45 min, requiring only one user step. All reagents are preloaded into the chip and can be automatically released. By exploiting a multichannel chip, it is capable of simultaneously detecting 10 infectious viruses with limits of detection of 1 copy per reaction and 5 copies per reaction in plasmid samples and mock plasma samples, respectively. The system was used to analyse clinical plasma samples with good consistency compared to laboratory-based molecular testing. Moreover, the generalizability of our device is reported by successfully testing nasopharyngeal swabs and whole blood samples. The portable device does not require the operation of professional technicians, making it an excellent assay for on-site testing.
Introduction
The global impact of communicable diseases is still the leading cause of human death.1 Infectious diseases have become a major public health problem that not only threatens life and health but also causes massive economic burdens.2–4 In the past, the world has been swept by several major public health emergencies caused by infectious disease agents, such as the Ebola virus, Dengue virus, and Zika virus.5–7 Rapid and accurate detection methods are the key to controlling the spread of infectious diseases and timely treatment.8,9 However, infections with these pathogens demonstrate similar clinical symptoms.10 Therefore, a rapid and comprehensive method for the detection of multiple pathogens is urgently needed to monitor the differential diagnosis or coinfection.Laboratory detection methods have been widely used in the diagnosis of infectious diseases, such as immunoassay, microbial culture and nucleic acid detection.11–13 However, these methods usually require specially trained technicians, expensive equipment, and complicated operation steps.14,15 Point-of-care testing involves performing a rapid test in resource-limited settings.16,17 Compared to laboratory instruments, point-of-care (POC) devices are low-cost, portable and user-friendly, especially in resource-limited areas with underdeveloped healthcare systems.18–20
In recent years, advances in microfluidics and biological detection technologies have provided solutions to improve point-of-care testing (POCT) for infectious disease diagnosis.21–23 Isothermal amplification methods are promising assays for POCT, such as recombinase polymerase amplification (RPA), recombinase-aided amplification (RAA) and the application of loop-mediated isothermal amplification (LAMP), but the high nonspecificity of the results leads to unavoidable difficulty. Clustered regularly interspaced short palindromic repeat (CRISPR)-based technology and the application of Pf Ago protein improve the sensitivity and specificity of nucleic acid detection.24–26 Various detection methods, such as colorimetric LAMP, absorbance, fluorescence and lateral flow, have been applied to POCT.27–30 Moreover, the CRISPR/Cas system combined with isothermal amplification technology has been developed with the ability to rapidly and highly sensitively identify pathogens.31–34 However, the specific detection of a single target has difficulty handling the cocirculation of multiple pathogens from infectious diseases requiring differential diagnosis.
The centrifugal microfluidic system can provide uniform actuation on the whole chip, which can facilitate multiple detections through the reasonable design of microchannels. Researchers have taken full advantage of centrifugal microfluidics to achieve multiplex detection of pathogens by combining the CRISPR/Cas system with centrifugal microfluidics.35 However, sample extraction still needs to be performed outside of the chip, which increases the complexity of manual manipulation and limits its application for onsite cases.36 More critically, the transfer of extraction products can potentially lead to nonnegligible contamination. Therefore, isothermal amplification technology combined with the CRISPR/Cas system is a powerful tool for portable platforms for nucleic acid diagnosis but is still limited by the integration of a full process including nucleic acid extraction, amplification and detection. Furthermore, most of the detection procedures for infectious diseases need to use complex samples that are dense biofluids, such as whole blood, plasma and mucus. Therefore, many simple extraction methods without target enrichment are not applicable.37–39
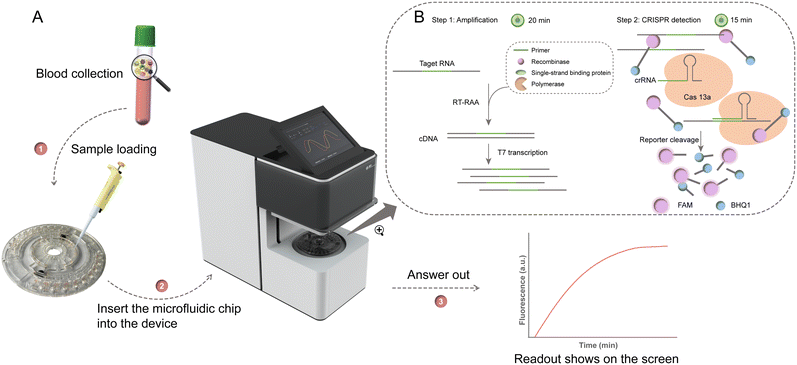
To address the on-site detection of “sample in, answer out,” we propose a portable centrifugal microfluidic testing (POCMT) system and a centrifugal microfluidic chip for the all-in-one-chip detection of 10 kinds of infectious pathogens with the integration of nucleic acid extraction from plasma samples. Within our POCMT system, the power-supplied active rotating chip enables the on-chip programmable control of the addressable thermally-triggered paraffin wax valves for flow switching and the release of prestored reagents. It is also equipped with an on-chip magnetic state switching structure for nucleic acid extraction. With the miniaturized fluorescence detection module, the air chamber temperature control unit and the embedded operating system, it becomes an all-in-one platform for on-site application. This assay combines nucleic acid extraction with magnetic beads (MBs) and RAA with the CRISPR–Cas13a system in a centrifugal microfluidic POC test. After one step of sample loading, ten targets can be simultaneously detected with the POCMT system, including Japanese encephalitis virus (JEV), yellow fever virus (YEV), West Nile virus (WNV), Ebola virus (EBOV), Chikungunya virus (CHIKV), Crimean-Congo haemorrhagic fever virus (CCHFV), Dengue virus (DENV), Rift valley fever virus (RVFV), Zika virus (ZIKV) and Marburg virus (MARV). The POCMT system has a rapid detection period as short as 45 min (nucleic acid extraction: 10 min, RAA: 20 min, CRISPR/Cas13a: 15 min). By utilizing an all-in-one chip, the limit of multiplex detection is reduced to at least 1 copy per reaction with nucleic acid templates and to 5 copies per reaction in mock plasma samples. To validate the system, the sensitivity and accuracy of the system were determined by using simulated clinical blood samples, with results compared with laboratory-based testing. Using multiplexed microfluidic chips, the entire assay process can be completed in an automated and independent manner. These features render the platform for POCT of infectious diseases, especially in resource-limited settings. Benefiting from the versatility of the chip and the flexibility of reagent replacement, the POCMT system can be easily updated and expanded to detect various diseases.
Materials and methods
Microfluidic system
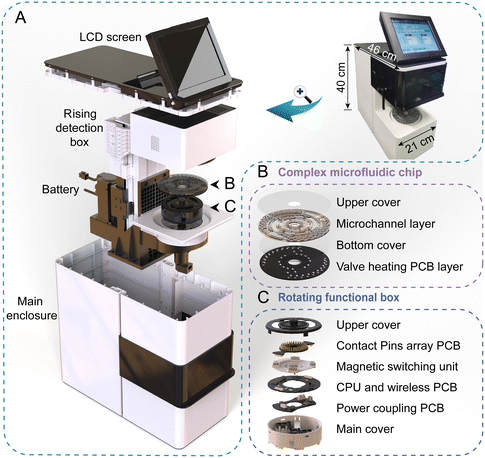
The assembled prototype of our integrated, portable, “sample in, answer out” POCMT system is shown in Fig. 1A, with a size of 21 cm (L) × 46 cm (W) × 40 cm (H) and mass of 12.5 kg. The devices were equipped with high precision servo motors for rotating control and an air heating unit for temperature control. It also had a rechargeable lithium polymer battery (6000 mAh) for the on-site case without a power supply. Our complex chip was housed on a rotating functional box with electric contact by using a spring-loaded pin array. Specifically, the chip is composed of four layers, including an upper chip cover, a microchannel layer with a carved double side channel and an embedded paraffin wax valve, a bottom chip cover and a buckled printed circuit board (PCB) layer for valve heating (with bottom solder pad) (Fig. 1B). Inside this rotating functional box, there were a contactless coupling coil module for wireless power supply, a CPU and a ZigBee communication module for receiving and executing instructions. It also had a magnetic switching structure for magnetic state control, which was important for washing of MBs in the on-chip nucleic acid extraction process. Above the chip, there was a motor-driven rising detection box containing 4 integrated optical detection modules for fluorescence excitation and measurement. Our portable device was operated with an embedded Android system, and the operator only needed to select an operational procedure file with all the detailed settings, including the reaction time, temperature, etc. The result and conclusion were displayed on the LCD screen. The operational procedure file is provided in the ESI.†Wireless power supply unit for the rotating functional box
In our system, the rotating functional box has a power supply and is used for on-board localized temperature control, magnet state switching and wireless communication. Beneath the box, there is a pair of flat coupling coils for a contactless power supply. Different from the method we proposed before,40 the flat coil method is much more stable and can provide more power. The transmitting coil and the receiving coil are attached on two PCBs, with the distance optimized at 8 mm. The lower stationary one is fixed on the platform and used for the modulation of transmitting electromagnetic waves, while the upper rotating one is rotating with the box, which stabilizes the voltage at 5 V with a power of approximately 20 W. In addition, a magnetic separator layer is used to isolate the electromagnetic wave radiation and reduce its interference with the electronics system. The design layout and schematic diagram of the rotating functional box can be found in the ESI.†Magnetic switching structure within the rotating functional box
To achieve the integration of nucleic acid extraction by MBs on the chip, we designed a two-state magnetic switching structure. Most importantly, in the washing of nucleic acid extraction process, magnet switching is needed to be carried out in a high-speed rotating state. Therefore, we could integrate this mechanical structure into the rotating functional box. The aggregation and dispersion of MBs was achieved by changing the magnet position through the magnetic switching structure. Further information on the design can be found in the ESI.†Optical structure within the rising detection box
To ensure the sensitivity of fluorescence detection, we used the coaxial optical path scheme (Fig. S4†). The fluorescence detection module was composed of a photodetector (S1133, Hamamatsu Photonics, Japan), a 488 nm semiconductor laser module model (HX548855D-AL, Zhuhai TengXing Optoelectronics Technology Co., Ltd., China), a fluorescence filter set (Suzhou Wendi Optoelectronics Technology Co., Ltd., China), a dichroscope (Suzhou Wendi Optoelectronics Technology Co., Ltd., China) and a biconvex lens (ST004, Nanyang BoFan Optics, China). The laser driving circuit could adjust the driving the current by pulse high-speed width modulation (PWM) to regulate the optical power of the semiconductor laser. In the assay, the optimal optical power of the laser was 20 mW. Further information on the design scheme of the optical path and hardware architecture of the control system can be found in the ESI.†Complex centrifugal microfluidics chip
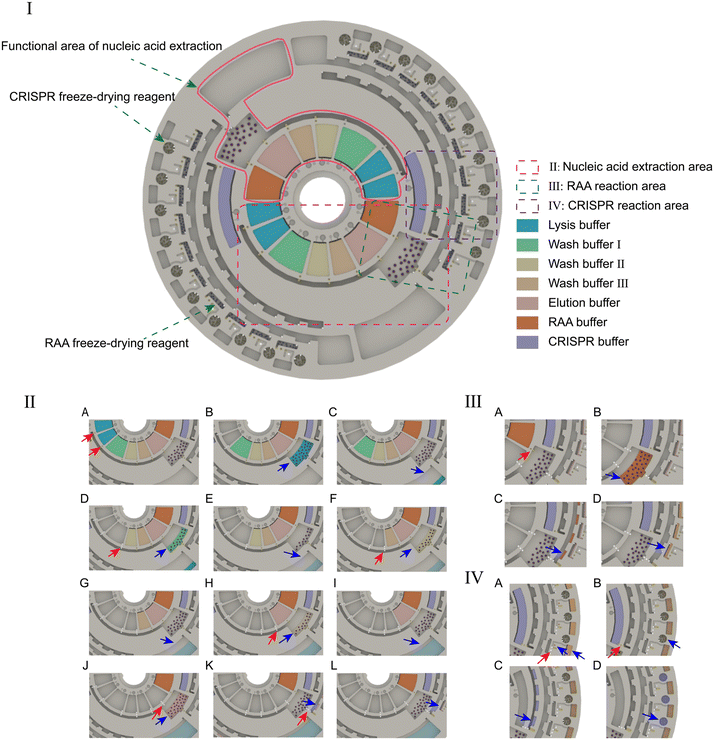
The complex centrifugal microfluidics chip has a CD-sized footprint with a 120 mm diameter. It is made of PC by the microinjection moulding method. The chip can simultaneously detect 2 samples, and each sample is divided into 10 parts for parallel detection of the infectious pathogens. According to different functions, the chip is divided into reagent storage, nucleic acid extraction, amplification and detection areas. A physical map of the POCMT chip is shown in Fig. S6.†Double side channel design of the chip
Our microchannel layer fabricated by injection moulding is delicately designed with double side channels. The upper side is carved with the main fluid channels, while the lower side has paraffin wax chambers, hydrophobic valves and crossing overpass channels. The channels of both sides are connected by vertical through-holes. In this way, the paraffin wax blocks are located at the bottom of the microchannel layer and very close to the MCH on the PCB layer, which can largely reduce the melting time of the paraffin wax valve. Furthermore, the double-sided channel design can also solve the problem of fluid channel crossing and greatly enhance the degree of integration of our chip. In our design for CRISPR detection, another CRISPR buffer need to be added into each detection chamber after amplification. A flow channel at the bottom is introduced to deliver this reagent and cross the main channel, similar to an underground passage.The switching structure for nucleic acid extraction
The nucleic acid extraction part of our chip is designed with a switching structure at the outlet of the lysis chamber; this structure is critical for the processes that require multiple transfers of liquids to different destinations. In our chip design, the switching structure has two branches, including a hydrophobic capillary valve and a paraffin wax valve. During the washing process, the paraffin wax valve is kept closed. After the lysis or washing step, we can simply increase the spinning speed to a higher value than the threshold of the hydrophobic capillary valve with the value set as 800 rpm; this will open the valve and release the waste liquid to one side. After the whole washing process is finished, we can open the paraffin wax valve to release the extracted product to the other side without any spinning threshold.Prestored reagents and controllable release
To reduce manual handling and attain all-in-one-chip operation, all reagents need to be pre-stored in the chip. Reagents include the liquid reagents and freeze-dried solid reagents. The reagent loading step can be carried out after the bottom cover bonding, paraffin wax valve injection and upper cover bonding. An injection hole and a vent hole at the upper cover for each reagent chamber are present. A barrier between both holes is designed to guide the injecting flow and prevent direct leakage from the inlet to the outlet. There are 14 pre-stored reagent chambers for the handling of 2 samples. After loading, the orderly release of reagents can be completely controlled by the paraffin wax valves, independent of rotational speed.Preparation of the clinical samples and spike-in samples
For on-site testing, 45 clinical blood samples from patients with suspected dengue were obtained from Jiangsu Provincial Center for Disease and Prevention and Control, and validation was conducted using quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). The blood samples were then shipped to Nanjing University and stored at −80 °C. For the spike-in samples, plasma samples from healthy donors (provided by Jiangsu Provincial Center for Disease and Prevention and Control, China) were mixed with RNA targets (provided by Sangon Biotech Inc., China). The plasma samples were maintained at −80 °C before nucleic acid extraction. The program was approved by the ethics committee of Jiangsu Provincial Center for Disease Control and Prevention (JSCDCLL2023[001]).Laboratory-based molecular assay
For laboratory-based molecular detection, nucleic acids were extracted by an automatic nucleic acid extraction instrument (Tianlong Technology Co., China). Real-time reverse transcription-polymerase chain reaction (RT-PCR) was carried out using a master mix reagent consisting of 2× One Step RT-PCR Buffer III, 1U Ex Taq Hot Start Version (TaKaRa Biotechnology, Kusatsu, Japan), 0.2 μL of PrimeScript RT Enzyme MX II, 2 μL of RNase free water, 0.2 μL of 10 μmol L−1 of each forward primer, 0.2 μL of 10 μmol L−1 of each reverse primer, and 0.2 μL of 10 μmol L−1 of each probe. The thermal cycling program included a reverse transcription step at 42 °C for 5 min, an enzyme activation step at 95 °C for 10 min, and 40 cycles at 95 °C for 5 s and 60 °C for 34 s. PCRs were performed on a QuantStudio 7 Pro system (Thermo Fisher Scientific, USA).Traditional CRISPR–Cas-based assay
We used commercial equipment as a reference method for the POCMT system. Nucleic acids were extracted by an automatic nucleic acid extraction system (Tianlong Technology Co., China). The RAA step was performed on a Thermo Veriti PCR system (Thermo Fisher Scientific, USA). The CRISPR–Cas13a system was performed on an F1620 nucleic acid amplification detector (Qitian Gene Biotechnology Co., Ltd., China).Results and discussion
Design and optimization of the POCMT system
The entire workflow of the detection system is shown in Fig. 2. Briefly, plasma samples are automatically tested in the portable device, and output fluorescence signals are monitored (Fig. 2A). Users only need to inject the sample into the chip, and the instrument does the following whole process. Firstly, a 200 μL plasma sample is added into the chip, then the detection is initiated by selecting its corresponding program. With the paraffin wax valve open in sequence, the reagents are released in turn (ESI† Movie S1). By switching the magnetic state, efficient nucleic acid washing and enrichment are realized. Through switching the structure, the extracted RNA product was released to RAA chambers. Then, the air heating unit is set to 39 °C for 20 min for RAA until the end of the assay. The miniaturized fluorescence detection module starts working during CRISPR detection for 15 min (Fig. 2B). The excited fluorescence signal is collected by a photodetector. The fluorescence acquisition data can be displayed in real time on the screen. The viral load of samples was evaluated by the fluorescence signal over a threshold readout when performed in the device. For samples that showed a positive result, the threshold time (Tt) is defined as the time required in the detection phase for the fluorescence signal to reach the threshold.Operational protocol for the microfluidic assay
Fig. 3 illustrates the operational protocol of the microfluidic chip. Two hundred microlitres of the plasma sample was added to the reaction chamber and mixed with the lysis buffer, proteinase K, acryl carrier and magnetic beads (MBs). The mixture was shaken for 6 min at room temperature to allow the MBs and nucleic acid to sufficiently bind. After that, the magnetic switching structure changed to a magnetic state for immobilization the MBs in the reaction chamber, and the liquid was released to the waste chamber (ESI† Movie S2). Then the MBs were washed sequentially with 500 μL of wash buffer I, wash buffer II, and wash buffer III, which were controlled by the paraffin wax valve. Finally, 500 μL of elution buffer was added to the reaction chamber. After changing to the non-magnetic state, a gentle shaking of 3 min was applied for the sufficiently elution of the nucleic acid on the MBs. Then, changing to the magnetic state and opening the wax valve, the template was released to the spiral channel for distribution and metering. The 12.5 μL of template and 12.5 μL of RAA buffer were measured and transferred to each RAA chamber. The total 25 μL RAA reaction mixture incubated at 39 °C for 20 min. After amplification, 2 μL of the RAA product was measured and transferred to the CRISPR chamber, and then mixed with the CRISPR buffer. The mixture was incubated at 39 °C for 15 min while collecting fluorescence. The operation of the microfluidic chip is shown in ESI† Movie S3.Optimizing the detection time and performance
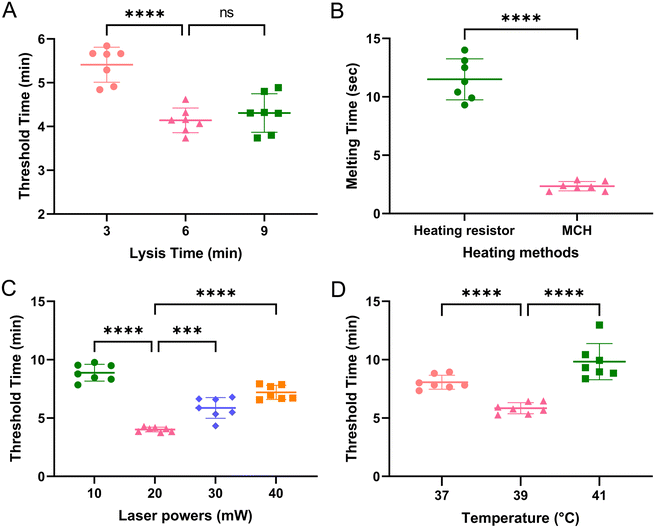
To improve the detection efficiency and shorten the turnaround time, we optimized four parameters. The nucleic acid extraction time provided by the instructions requires a long time. Fig. 4A shows the effect of lysis time on nucleic acid extraction efficiency. The lowest value of threshold time (Tt) indicated that 6 min was sufficient for cell lysis in the chip. The Tt value increased when increasing or decreasing the duration of lysis time. Based on this optimization, total RNA extraction could be rapidly accomplished in 10 min, which was approximately one-third the time required by standard laboratory protocols. To shorten the opening time of the paraffin valve, the heating efficiency of two kinds of heating elements were compared. The metal ceramic heater (MCH) is more efficient than the heating resistor (Fig. 4B). Only 2 s heating time was needed to melt the paraffin wax and open the valve. To improve the detection performance, we tested the effect of different laser powers on the threshold time. From Fig. 4C, the threshold time was the lowest at the laser power of 20 mW. At last, the temperature of the RAA reaction was also optimized. The optimal reaction temperature was 39 °C, and the Tt value increased at higher or lower temperatures (Fig. 4D).Specificity analysis
To assess the specificity of the detection system, a cross-reactivity test was performed. The reaction chambers were preloaded with reagents for ten viruses to verify specificity. All ten plasma samples representing viral infections of a single infectious disease showed positive reactions in the corresponding designed chambers. Other reaction chambers exhibited negative results. The positive signals were displayed as real-time curves, while negative responses did not reach the fluorescence threshold. From Fig. 5, the specificity of the microfluidic platform was reasonably good. The system exhibited no cross-reactivity between the ten viruses and exhibited excellent specificity.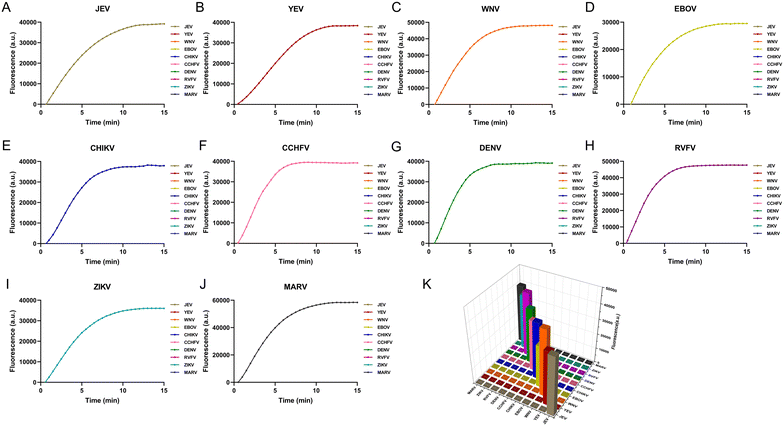 | ||
| Fig. 5 Specificity analysis for each virus. (A) JEV; (B) YEV; (C) WNV; (D) EBOV; (E) CHIKV; (F) CCHFV; (G) DENV; (H) RVFV; (I) ZIKV; (J) MARV. (K) Histogram overview of the specificity analysis. | ||
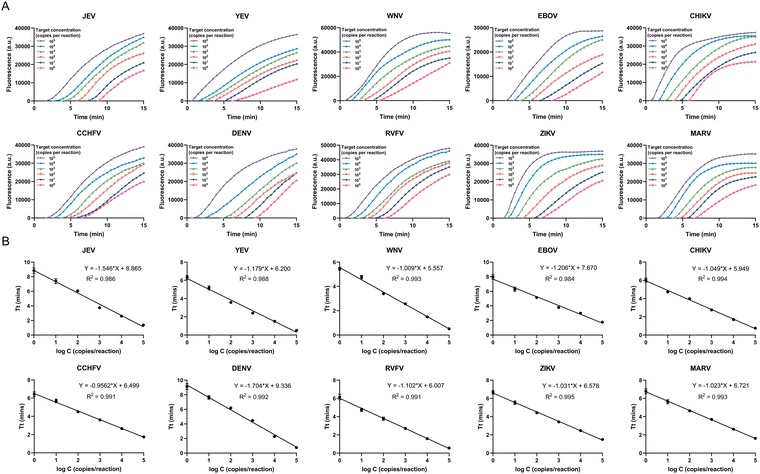
Sensitivity analysis
All plasmid samples, JEV, YEV, WNV, EBOV, CHIKV, CCHFV, DENV, RVFV, ZIKV and MARV, were diluted in a tenfold gradient with plasma samples. The platform remained detectable at dilution levels of 5 copies per mL for JEV, YEV, WNV, EBOV, CHIKV, CCHFV, DENV, RVFV, ZIKV and MARV (Fig. 6A). Our results showed a lower limit of detection (LOD) of 1 copy per reaction for the 10 viruses using the POCMT system, which was higher than that of the laboratory-based molecular assay (Fig. S8A†). The LOD of the laboratory-based molecular assay reached 10 copies per reaction (Fig. S8B†). The LOD of the POCMT system was consistent with the traditional CRISPR–Cas-based assay (Fig. S8C†).The relationship between the concentration and Tt value of the 10 detected viruses is shown in Fig. 6B. The Tt value was logarithmically related to the concentrations of the plasmid samples in the range from 105 to 1 copies per reaction for all 10 viruses with coefficients of determination (R2) as follows: R2 = 0.986 for JEV; R2 = 0.988 for YEV; R2 = 0.993 for WNV; R2 = 0.984 for EBOV; R2 = 0.994 for CHIKV; R2 = 0.991 for CCHFV; R2 = 0.992 for DENV; R2 = 0.991 for RVFV; R2 = 0.995 for ZIKV; R2 = 0.993 for MARV. These results indicated that the centrifugal microfluidic platform had the potential for quantitative detection. The system was suitable for rapid and accurate differential diagnosis of pathogens of infectious diseases.
Performance of the microfluidic platform in clinical specimens and spike-in samples
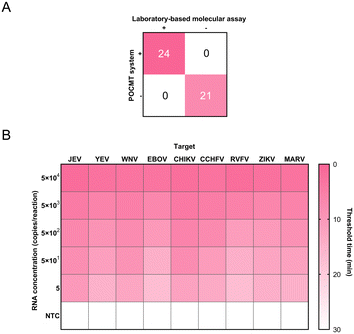
The practical potential of the device for detecting pathogens was evaluated further using clinical specimens. A total of 45 specimens were collected from suspected dengue patients: 21 were DENV-negative and 24 were DENV-positive (including samples with high and low DENV loads). Both the positive and negative coincidences were identical with that of the laboratory-based molecular assay (RT-PCR), as shown in Fig. 7A. They were also consistent with the results of the traditional CRISPR–Cas-based assay by using commercial equipment (Table S2†).For the detection of the spike-in samples, the device also showed excellent performance and could better distinguish negative and positive samples (Fig. 7B).
Application prospects
This study performed simultaneous and quantitative detection of ten pathogens in plasma samples. The feasibility of detecting other infectious agents was also successfully validated using this device, such as the Middle East respiratory syndrome coronavirus (MERS-CoV) in nasopharyngeal swabs (Fig. S9†) and Plasmodium in whole blood (Fig. S10†). No cross-reactivity was observed between the 12 infectious disease pathogens (Fig. S11†). These results indicated that the device could detect a variety of infectious disease pathogens with ideal sensitivity and specificity. During the global COVID-19 pandemic, medical devices for self-testing at home were urgently needed. The use this low-cost, automated and user-friendly device would be highly attractive for self-testing at home. Other types of assays, such as chemiluminescence immunoassays, are also suitable for this platform due to similar reaction conditions and operational steps, which greatly extends the applicability of POC testing.Conclusions
In the context of the SARS-CoV-2 outbreak, it has become critical to develop POC medical devices. We have developed an automated POC molecular detection platform that provides rapid, highly sensitive and specific screening for infectious pathogens.By integrating nucleic acid extraction with the magnetic switching structure, the detection mode of “sample in, answer out” was achieved. Based on the RAA and CRISPR technology, we designed a centrifugal microfluidic chip for the nucleic acid detection of infectious diseases. For the clinical sample results, our assay has a good concordance with laboratory methods and a significant reduction in testing time. The detection systems are relatively low-cost and have great commercial and practical potential as reliable diagnostic tools for controlling infectious disease outbreaks.
Author contributions
Ya Zhang: conceptualization, methodology, formal analysis, investigation, validation, visualization, and writing – original draft. Yue Guo: formal analysis, investigation, validation, and visualization. Guozhen Liu: formal analysis and investigation. Shiqi Zhou: investigation and writing – original draft. Rouyu Su: investigation and writing – original draft. Qian Ma: investigation. Yiyue Ge: supervision and funding acquisition. Yan-qing Lu: supervision and funding acquisition. Lunbiao Cui: writing – review & editing, supervision, and funding acquisition. Guanghui Wang: writing – review & editing, validation, supervision, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was sponsored by the National Natural Science Foundation of China (62375121, 61875083, 61535005), the Social Development Project of Jiangsu Province (BE2019761), the Natural Science Foundation of Jiangsu Province (BK20211373), the Key Scientific Research Project of Jiangsu Provincial Health Commission (ZD2021060), the Key Research and Development Program of Shandong Province (2020CXGC011304) and Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX24_0273).Notes and references
- World Health Organization, World Health Statistics 2021, World Health Organization, Geneva, 2021 Search PubMed.
- I. Ali and O. M. L. Alharbi, Sci. Total Environ., 2020, 728, 138861 CrossRef CAS PubMed
.
- W. Chen, H. Yu, F. Sun, A. Ornob, R. Brisbin, A. Ganguli, V. Vemuri, P. Strzebonski, G. Cui, K. J. Allen, S. A. Desai, W. Lin, D. M. Nash, D. L. Hirschberg, I. Brooks, R. Bashir and B. T. Cunningham, Anal. Chem., 2017, 89, 11219–11226 CrossRef CAS PubMed
.
- D. Musso, A. I. Ko and D. Baud, N. Engl. J. Med., 2019, 381, 1444–1457 CrossRef PubMed
.
- A. Shepherd, BMJ Clinical Research, 2019, 365, l4405 Search PubMed
.
- A. M. Ahmed, A. T. Mohammed, T. T. Vu, M. Khattab, M. F. Doheim, A. A. Mohamed, M. M. Abdelhamed, B. E. Shamandy, M. T. Dawod, W. A. Alesaei, M. A. Kassem, O. M. Mattar, C. Smith, K. Hirayama and N. T. Huy, Rev. Med. Virol., 2020, 30, e2093 CrossRef PubMed
.
- U. Bhardwaj, N. Pandey, M. Rastogi and S. K. Singh, Virology, 2021, 560, 86–95 CrossRef CAS PubMed
.
- N. J. Brendish, A. K. Malachira, L. Armstrong, R. Houghton, S. Aitken, E. Nyimbili, S. Ewings, P. J. Lillie and T. W. Clark, Lancet Respir. Med., 2017, 5, 401–411 CrossRef PubMed
.
- T. R. Kozel and A. R. Burnham-Marusich, J. Clin. Microbiol., 2017, 55, 2313–2320 CrossRef CAS PubMed
.
- O. Yaren, B. W. Alto, P. V. Gangodkar, S. R. Ranade, K. N. Patil, K. M. Bradley, Z. Yang, N. Phadke and S. A. Benner, BMC Infect. Dis., 2017, 17, 293 CrossRef PubMed
.
- S. G. Beal, N. Assarzadegan and K. H. Rand, Expert Rev. Mol. Diagn., 2016, 16, 323–341 CrossRef CAS PubMed
.
- P. J. Tighe, R. R. Ryder, I. Todd and L. C. Fairclough, Proteomics: Clin. Appl., 2015, 9, 406–422 CAS
.
- O. Pashchenko, T. Shelby, T. Banerjee and S. Santra, ACS Infect. Dis., 2018, 4, 1162–1178 CrossRef CAS PubMed
.
- M. Zarei, Biosens. Bioelectron., 2018, 106, 193–203 CrossRef CAS PubMed
.
- L. Bissonnette and M. G. Bergeron, Expert Rev. Mol. Diagn., 2017, 17, 471–494 CrossRef CAS PubMed
.
- Y. Xiao, S. Li, Z. Pang, C. Wan, L. Li, H. Yuan, X. Hong, W. Du, X. Feng, Y. Li, P. Chen and B. F. Liu, Biosens. Bioelectron., 2022, 206, 114130 CrossRef CAS PubMed
.
- P. K. Drain, E. P. Hyle, F. Noubary, K. A. Freedberg, D. Wilson, W. R. Bishai, W. Rodriguez and I. V. Bassett, Lancet Infect. Dis., 2014, 14, 239–249 CrossRef PubMed
.
- C. Dincer, R. Bruch, A. Kling, P. S. Dittrich and G. A. Urban, Trends Biotechnol., 2017, 35, 728–742 CrossRef CAS PubMed
.
- K. J. Land, D. I. Boeras, X. S. Chen, A. R. Ramsay and R. W. Peeling, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS PubMed
.
- J. M. Perkel, Nature, 2017, 545, 119–121 CrossRef CAS PubMed
.
- M. Zarei, TrAC, Trends Anal. Chem., 2017, 91, 26–41 CrossRef CAS
.
- C. S. Wood, M. R. Thomas, J. Budd, T. P. Mashamba-Thompson, K. Herbst, D. Pillay, R. W. Peeling, A. M. Johnson, R. A. McKendry and M. M. Stevens, Nature, 2019, 566, 467–474 CrossRef CAS PubMed
.
- P. Brangel, A. Sobarzo, C. Parolo, B. S. Miller, P. D. Howes, S. Gelkop, J. J. Lutwama, J. M. Dye, R. A. McKendry, L. Lobel and M. M. Stevens, ACS Nano, 2018, 12, 63–73 CrossRef CAS PubMed
.
- H. De Puig, R. A. Lee, D. Najjar, X. Tan, L. R. Soeknsen, N. M. Angenent-Mari, N. M. Donghia, N. E. Weckman, A. Ory, C. F. Ng, P. Q. Nguyen, A. S. Mao, T. C. Ferrante, G. Lansberry, H. Sallum, J. Niemi and J. J. Collins, Sci. Adv., 2021, 7, eabh2944 CrossRef PubMed
.
- G. Xun, S. T. Lane, V. A. Petrov, B. E. Pepa and H. Zhao, Nat. Commun., 2021, 12, 2905 CrossRef CAS PubMed
.
- Y. X. Zhang, Y. Song, Z. Y. Weng, J. Yang, L. Avery, K. D. Dieckhaus, R. B. C. Lai, X. Gao and Y. Zhang, Lab Chip, 2023, 23, 3862–3873 RSC
.
- M. A. Lalli, J. S. Langmade, X. Chen, C. C. Fronick, C. S. Sawyer, L. C. Burcea, M. N. Wilkinson, R. S. Fulton, M. Heinz, W. J. Buchser, R. D. Head, R. D. Mitra and J. Milbrandt, Clin. Chem., 2021, 67, 415–424 CrossRef PubMed
.
- X. Ge, T. Meng, X. Tan, Y. Wei, Z. Tao, Z. Yang, F. Song, P. Wang and Y. Wan, Biosens. Bioelectron., 2021, 189, 113350 CrossRef CAS PubMed
.
- J. Joung, A. Ladha, M. Saito, N. G. Kim, A. E. Woolley, M. Segel, R. P. J. Barretto, A. Ranu, R. K. Macrae, G. Faure, E. I. Ioannidi, R. N. Krajeski, R. Bruneau, M. W. Huang, X. G. Yu, J. Z. Li, B. D. Walker, D. T. Hung, A. L. Greninger, K. R. Jerome, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed
.
- S. Wang, N. Liu, L. Zheng, G. Cai and J. Lin, Lab Chip, 2020, 20, 2296–2305 RSC
.
- Y. Chen, Y. Mei, X. Zhao and X. Jiang, Anal. Chem., 2020, 92, 14846–14852 CrossRef CAS PubMed
.
- Y. Chen, N. Zong, F. Ye, Y. Mei, J. Qu and X. Jiang, Anal. Chem., 2022, 94, 9603–9609 CrossRef CAS PubMed
.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS PubMed
.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed
.
- J. Liu, H. Wang, L. Zhang, Y. Lu, X. Wang, M. Shen, N. Li, L. Feng, J. Jing, B. Cao, X. Zou, J. Cheng and Y. Xu, Small, 2022, 18, e2200854 CrossRef PubMed
.
- W. Liu, F. Yue and L. P. Lee, Acc. Chem. Res., 2021, 54, 4107–4119 CrossRef CAS PubMed
.
- M. M. Hassan, A. Ranzoni and M. A. Cooper, Biosens. Bioelectron., 2018, 99, 150–155 CrossRef CAS PubMed
.
- G. Czilwik, T. Messinger, O. Strohmeier, S. Wadle, F. Von Stetten, N. Paust, G. Roth, R. Zengerle, P. Saarinen, J. Niittymäki, K. McAllister, O. Sheils, J. O'Leary and D. Mark, Lab Chip, 2015, 15, 3749–3759 RSC
.
- R. Paul, E. Ostermann and Q. Wei, Biosens. Bioelectron., 2020, 169, 112592 CrossRef CAS PubMed
.
- G. H. Wang, H. P. Ho, Q. L. Chen, A. K. L. Yang, H. C. Kwok, S. Y. Wu, S. K. Kong, Y. W. Kwan and X. P. Zhang, Lab Chip, 2013, 13, 3698–3706 RSC
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00326h |
| ‡ These authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2024 |