Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemoselective seleno-click amidation in kinetic target-guided synthesis†
Lili
Huang‡
a,
Prakash T.
Parvatkar‡
 a,
Alicia
Wagner
a,
Sameer S.
Kulkarni
b and
Roman
Manetsch
a,
Alicia
Wagner
a,
Sameer S.
Kulkarni
b and
Roman
Manetsch
 *abcde
*abcde
aDepartment of Chemistry and Chemical Biology, Northeastern University, Boston, MA 02115, USA. E-mail: r.manetsch@northeastern.edu
bDepartment of Chemistry, University of South Florida, Tampa, FL 33620, USA
cDepartment of Pharmaceutical Sciences, Northeastern University, Boston, MA 02115, USA
dCenter for Drug Discovery, Northeastern University, Boston, MA 02115, USA
eBarnett Institute of Chemical and Biological Analysis, Northeastern University, Boston, MA 02115, USA
First published on 7th October 2024
Abstract
Capitalizing on our previous kinetic target-guided synthesis (KTGS) involving the sulfo-click reaction to form N-acylsulfonamide-linked inhibitors in the presence of the protein–protein interaction target Mcl-1, we herein report a seleno-click approach for amide-linked inhibitors of Mcl-1. The seleno-click reaction leverages the enhanced reactivity of selenocarboxylates, enabling the templated amidation with electron-rich azides, thereby expanding the scope of KTGS. The potential of this approach is demonstrated by generating N-benzyl-5-(4-isopropylthiophenol)-2-hydroxyl nicotinamide, a known Mcl-1 inhibitor featuring an amide, via KTGS at 37 °C against Mcl-1. Notably, the seleno-click strategy was also effective at 4 °C, offering a variant for thermally sensitive biological targets.
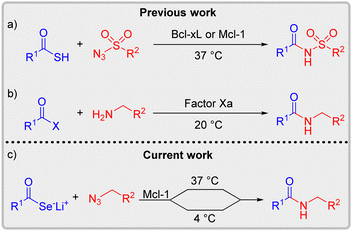
Since its inception over two decades ago, kinetic target-guided synthesis (KTGS) has emerged as a promising tool in the field of fragment-based lead discovery.1–6 This elegant approach involves the biological target itself, which facilitates a chemoselective reaction between two fragments bearing complementary functionalities, generating a bidentate ligand with higher affinity compared to its precursors. For example, in the pioneering work, Sharpless and co-workers demonstrated that the Huisgen 1,3-dipolar cycloaddition reaction between azides and alkynes in the presence of acetylcholine esterase resulted in the discovery of femtomolar inhibitors of acetylcholine esterase.7,8 Since this seminal report, several KTGS variants that rely on additional ligation reactions with bioorthogonal characteristics have been developed; examples include amidations (conventional9 or sulfo-click10,11), N- or S-alkylations,12,13 oxime formations,14 thio-Michael additions,15 epoxide ring openings,16 Ugi reactions,17 and Mannich reactions.18
Despite noteworthy advances, the sluggish reaction kinetics of the triazole-forming click reaction posed challenges in applying this method for non-enzymatic systems, specifically the seemingly ‘undruggable’ protein–protein interaction targets. To overcome this challenge, we successfully employed the sulfo-click reaction, reported by Williams and co-workers,19 to identify novel protein–protein interaction modulators (PPIMs) of the proteins of the Bcl-2 family.10,20,21 Benefiting from the superior reaction rates between thioacids and sulfonyl azides, this chemoselective reaction affords N-acylsulfonamides, thus providing a robust and highly efficient screening platform for rapid identification of PPIMs (Scheme 1a).20,21 Although promising, the reactivity profile of this transformation relies on electron-deficient sulfonyl azides and limits its KTGS application to the discovery of inhibitors containing an acylsulfonamide moiety. Therefore, we envisioned overcoming this constraint by replacing the thioacids with their more reactive chalcogen analogs, specifically the selenocarboxylates. We hypothesized that if the enhanced nucleophilicity of selenocarboxylates can compensate for the reduced electrophilicity of electron-rich azides compared to electron-poor sulfonyl azides, this approach could enable access to amides, further expanding the scope of KTGS. This would also contrast with the KTGS amidation approach by Rademann and coworkers (Scheme 1b),9 which depends on classical amidation conditions involving an activated carboxylic acid as an electrophile and a free amine as a nucleophile.
Herein, we report for the first time the use of the seleno-click reaction in KTGS (Scheme 1c). The power of this strategy was showcased through the Mcl-1-templated assembly of a known Mcl-1 inhibitor featuring an amide bond. Impressively, the superior reactivity of selenocarboxylates enabled KTGS screening even at 4 °C, presenting tantalizing prospects of performing these reactions in complex biological systems, including live cells.
To explore the potential of a seleno-click amidation in KTGS, we chose an anti-apoptotic protein Mcl-1, from the Bcl-2 family, as the biological target. The Bcl-2 protein family is an important target for cancer therapy as it plays a central role in regulating apoptosis (programmed cell death). Previously, the Manetsch laboratory demonstrated that the Bcl-xL10,20,22 and Mcl-1-mediated21 assembly of N-acylsulfonamides using the sulfo-click amidation between in situ generated thiocarboxylic acids and electron-deficient sulfonyl azides at 37 °C is well suited for KTGS. The vast majority of KTGS approaches, including our original sulfo-click approach, are limited by the insufficient sensitivity of the single quadrupole mass spectrometer (LC-MS-SIM). This limitation requires incubation of samples containing the protein target and only two complementary reacting fragments at temperatures of 20 °C or 37 °C.10 In contrast, using a triple quadrupole mass spectrometer in a dynamic multiple reaction monitoring mode (LC-MS/MS-dMRM), we demonstrated that acylsulfonamide formation could be reliably determined in incubation samples containing the protein target and reactive fragments, giving rise to roughly 200 acylsulfonamide combinations in one single well.11,21 This increased sensitivity dramatically enhances the detection and quantification of low-abundance compounds, thereby improving the KTGS approach overall.
Selenium, despite sharing some chemical properties with sulfur, stands out due to its larger size and greater polarizability. This unique characteristic makes selenium more nucleophilic than sulfur. For instance, selenophenolate was found to react with methyl iodide approximately 7 times faster than thiophenolate in SN2 displacement reactions, highlighting the intriguing role of selenium in chemical reactions.23 Similarly, studies have shown that selenocarboxylic acids facilitate the formation of amides from azides more readily than thiocarboxylic acids.24,25 The seleno-click amidation is also characterized by a high degree of chemoselectivity, produces no undesired side products, and operates effectively under mild reaction conditions.24,26–28 In addition, most aromatic selenocarboxylate salts can be safely stored at −17 °C for at least one month in an oxygen-free environment,29 providing a practical approach for KTGS. In our current study, we performed the seleno-click amidation of an in situ generated selenocarboxylate with an electron-rich azide, with Mcl-1 serving as a template, at incubation temperatures of 37 °C and 4 °C, respectively (Scheme 1c).
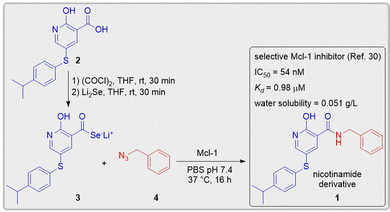
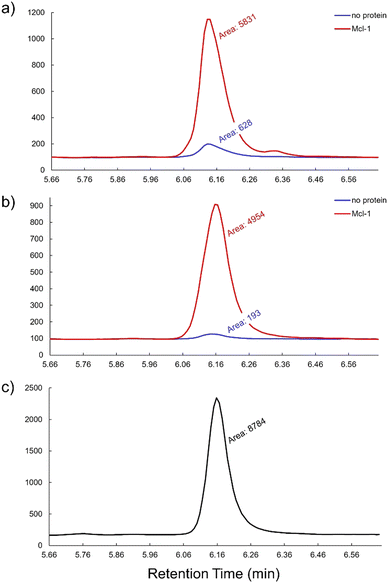
Zhang et al.30 recently discovered a potent Mcl-1 inhibitor, N-benzyl-2-hydroxy-5-((4-isopropylphenyl)thio)nicotinamide 1, through a fragment-based lead discovery approach. Nicotinamide derivative 1 effectively binds to the Mcl-1 protein with a Kd value of 0.98 μM as determined by isothermal titration calorimetry and exhibits an IC50 value of 54 nM. We envisioned that this nicotinamide derivative 1, featuring a suitably placed amide moiety, could serve as a perfect candidate to validate the potential of the seleno-click amidation with an electron-rich azide in KTGS. The carboxylic acid precursor 2, required for synthesizing selenocarboxylate fragment 3in situ, was prepared using literature procedures.30 Since the compounds containing a hydroxynicotine core demonstrated a high affinity for Mcl-1,30 the selenocarboxylate fragment 3 that encompasses this core was crucial for the Mcl-1 templated KTGS experiments. The other fragment, benzyl azide 4, was purchased from a commercial source. Next, Mcl-1-templated seleno-click amidations were conducted at 37 °C using the in situ generated selenocarboxylate 3 with the benzyl azide 4 (Scheme 2). The findings from these KTGS experiments were meticulously examined via LC-MS/MS and compared with the chemically synthesized nicotinamide derivative 1. Dynamic multiple reaction monitoring (dMRM) was used for the analysis of nicotinamide derivative 1 because of its high sensitivity, specificity, and reproducibility. The parent ion (m/z = 379 [M + H]+) of the nicotinamide derivative 1 and its two distinct fragmentation product ions (m/z = 272 for acylium ion fragment and m/z = 91 for benzyl carbonium ion fragment) were monitored. The results revealed a ∼9-fold increase in the formation of the amide-linked nicotinamide derivative 1 in the templated reaction (with Mcl-1 protein) compared to the background reaction (without protein), demonstrating the successful application of the seleno-click amidation in KTGS (Fig. 1a). To eliminate any non-specific templating effect, we conducted a control experiment using in situ formed benzoic acid selenocarboxylate 6 (a fragment with no known affinity for Mcl-1) and benzyl azide 4, both with and without Mcl-1 (see ESI,† Section S3.4 and Fig. S1). The absence of the Mcl-1 templating effect with benzoic acid selenocarboxylate 6 confirms that the specific templating effect is indeed due to the affinity of selenocarboxylate 3 to Mcl-1.
Due to the superior reactivity of selenocarboxylates, we decided to also perform the seleno-click amidation at 4 °C. Performing KTGS at a lower temperature offers significant advantages, such as maintaining the stability of biological targets for extended periods during the screening process. This is particularly advantageous for biological targets that are unstable at room temperature (23 °C) or body temperature (37 °C). It is also worth noting that certain proteins may attain a different conformation at lower temperatures, which would be inaccessible under standard KTGS conditions, offering unprecedented opportunities. In addition, reducing the temperature helps to decrease background reactions, thus enhancing the templation effect.
Notably, the seleno-click amidation at 4 °C led to the formation of nicotinamide derivative 1, confirmed through LC-MS/MS-dMRM and comparison to an authentic sample of nicotinamide derivative 1. As expected, more pronounced templation was observed at 4 °C for the Mcl-1-mediated assembly of the amide-linked nicotinamide derivative 1 (Fig. 1b). Our findings reveal that the seleno-click amidation can be successfully performed even at 4 °C, resulting in a remarkable ∼26-fold increase in amplification, compared to a ∼9-fold increase at 37 °C. This translates to an approximately 3-fold improvement in the KTGS process at 4 °C, highlighting the benefits of conducting the screening at a lower temperature. This successful application of seleno-click amidation in KTGS, using less reactive electron-rich azide at both 37 °C and 4 °C, demonstrates its potential to expand the chemical and biological space for identifying new modulators targeting various biological entities. This research, which holds the potential to inspire new approaches for developing novel modulators of other challenging biological targets, has profound practical implications for future research and development.
In summary, we have demonstrated that the seleno-click amidation can be applied to KTGS for the templated assembly of ligands featuring amides. In this proof-of-concept study, nicotinamide derivative 1, a previously reported Mcl-1 inhibitor,30 was successfully generated from the corresponding reactive fragments in the presence of Mcl-1, wherein the templation effect was analyzed and confirmed using a triple quadrupole mass spectrometer in dMRM mode. Our findings suggest that the significantly improved nucleophilicity of in situ generated selenocarboxylates, compared to their sulfur counterparts, can be harnessed to facilitate an amidation using less reactive electron-rich azides, making it an attractive bioorthogonal reaction for applications in the KTGS field. Furthermore, our research showcases the first example of successful implementation of KTGS at 4 °C, wherein most biological targets remain stable for extended periods, reducing the risk of protein denaturation. Additionally, KTGS at 4 °C exhibits a better amplification than corresponding incubations at 37 °C, further strengthening the potential impact of seleno-click amidations in KTGS. We are currently synthesizing a library of selenocarboxylates to react with various electron-rich and electron-deficient azides to discover novel modulators of protein–protein interactions using our previously developed multi-fragment KTGS approach.11,21
The experiments were proposed, designed, and supervised by PTP and RM. LH conducted the chemical synthesis experiments, while LH, PTP, and SSK conducted the KTGS experiments. AW and PTP were responsible for the LC-MS/MS analysis. PTP, RM, and SSK wrote the manuscript with assistance from all authors, and all authors approved the final version.
The authors express gratitude for the staffing and instrumentation support provided by the Northeastern University NMR Core Facility. This work utilized an NMR spectrometer purchased with funding from a National Institutes of Health SIG grant (S10OD032452).
Data availability
The data supporting this article have been included in the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- P. T. Parvatkar and R. Manetsch, Med. Chem. Res., 2024, 33, 1307–1314 CrossRef CAS.
- P. T. Parvatkar, A. Wagner and R. Manetsch, Trends Chem., 2023, 5, 657–671 CrossRef CAS.
- X. D. Hu and R. Manetsch, Chem. Soc. Rev., 2010, 39, 1316–1324 RSC.
- A. Lossouarn, P. Y. Renard and C. Sabot, Bioconjugate Chem., 2021, 32, 63–72 CrossRef CAS PubMed.
- D. Bosc, V. Camberlein, R. Gealageas, O. Castillo-Aguilera, B. Deprez and R. Deprez-Poulain, J. Med. Chem., 2020, 63, 3817–3833 CrossRef CAS.
- D. Bosc, J. Jakhlal, B. Deprez and R. Deprez-Poulain, Future Med. Chem., 2016, 8, 381–404 CrossRef CAS PubMed.
- W. G. Lewis, L. G. Green, F. Grynszpan, Z. Radic, P. R. Carlier, P. Taylor, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 1053–1057 CrossRef CAS.
- R. Manetsch, A. Krasinski, Z. Radic, J. Raushel, P. Taylor, K. B. Sharpless and H. C. Kolb, J. Am. Chem. Soc., 2004, 126, 12809–12818 CrossRef CAS PubMed.
- M. Jaegle, T. Steinmetzer and J. Rademann, Angew. Chem., Int. Ed., 2017, 56, 3718–3722 CrossRef CAS PubMed.
- X. D. Hu, J. Z. Sun, H. G. Wang and R. Manetsch, J. Am. Chem. Soc., 2008, 130, 13820–13821 CrossRef CAS PubMed.
- M. Kassu, P. T. Parvatkar, J. Milanes, N. P. Monaghan, C. Kim, M. Dowgiallo, Y. Z. Zhao, A. H. Asakawa, L. L. Huang, A. Wagner, B. Miller, K. Carter, K. F. Barrett, L. M. Tillery, L. K. Barrett, I. Q. Phan, S. Subramanian, P. J. Myler, W. C. Van Voorhis, J. W. Leahy, C. A. Rice, D. E. Kyle, J. Morris and R. Manetsch, ACS Infect. Dis., 2023, 9, 2190–2201 CrossRef CAS.
- J. Inglese and S. J. Benkovic, Tetrahedron, 1991, 47, 2351–2364 CrossRef CAS.
- R. Nguyen and I. Huc, Angew. Chem., Int. Ed., 2001, 40, 1774–1776 CrossRef CAS.
- P. Parvatkar, N. Kato, M. Uesugi, S. Sato and J. Ohkanda, J. Am. Chem. Soc., 2015, 137, 15624–15627 CrossRef CAS PubMed.
- E. Oueis, F. Nachon, C. Sabot and P. Y. Renard, Chem. Commun., 2014, 50, 2043–2045 RSC.
- T. Maki, A. Kawamura, N. Kato and J. Ohkanda, Mol. BioSyst., 2013, 9, 940–943 RSC.
- F. Mancini, M. Y. Unver, W. A. M. Elgaher, V. R. Jumde, A. Alhayek, P. Lukat, J. Herrmann, M. D. Witte, M. Köck, W. Blankenfeldt, R. Müller and A. K. H. Hirsch, Chem. – Eur. J., 2020, 26, 14585–14593 CrossRef CAS PubMed.
- E. L. Wong, E. Nawrotzky, C. Arkona, B. G. Kim, S. Beligny, X. N. Wang, S. Wagner, M. Lisurek, D. Carstanjen and J. Rademann, Nat. Commun., 2019, 10, 66 CrossRef CAS PubMed.
- N. Shangguan, S. Katukojvala, R. Greenburg and L. J. Williams, J. Am. Chem. Soc., 2003, 125, 7754–7755 CrossRef CAS.
- S. S. Kulkarni, X. D. Hu, K. Doi, H. G. Wang and R. Manetsch, ACS Chem. Biol., 2011, 6, 724–732 CrossRef CAS PubMed.
- K. Nacheva, S. S. Kulkarni, M. Kassu, D. Flanigan, A. Monastyrskyi, I. D. Iyamu, K. Doi, M. Barber, N. Namelikonda, J. D. Tipton, P. Parvatkar, H. G. Wang and R. Manetsch, J. Med. Chem., 2023, 66, 5196–5207 CrossRef CAS.
- N. K. Namelikonda and R. Manetsch, Chem. Commun., 2012, 48, 1526–1528 RSC.
- X. H. Wu and L. Q. Hu, J. Org. Chem., 2007, 72, 765–774 CrossRef CAS PubMed.
- L. Silva, R. F. Affeldt and D. S. Lüdtke, J. Org. Chem., 2016, 81, 5464–5473 CrossRef CAS PubMed.
- L. A. Wessjohann, A. Schneider, M. Abbas and W. Brandt, Biol. Chem., 2007, 388, 997–1006 CrossRef CAS PubMed.
- X. H. Wu and L. Q. Hu, Tetrahedron Lett., 2005, 46, 8401–8405 CrossRef CAS.
- L. Silva, A. R. Rosário, B. M. Machado and D. S. Lüdtke, Tetrahedron, 2021, 79, 131834 CrossRef CAS.
- X. H. Wu and L. Q. Hu, J. Org. Chem., 2007, 72, 765–774 CrossRef CAS PubMed.
- O. Niyomura, K. Tani and S. Kato, Heteroat. Chem., 1999, 10, 373–379 CrossRef CAS.
- Z. C. Zhang, C. W. Liu, X. Q. Li, T. Song, Z. Y. Wu, X. M. Liang, Y. Zhao, X. Y. Shen and H. B. Chen, Eur. J. Med. Chem., 2013, 60, 410–420 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: General information, synthetic procedure, compound characterization data, KTGS experimental procedures, and LC-MS/MS details. See DOI: https://doi.org/10.1039/d4cc04491f |
| ‡ Equally contributed. |
| This journal is © The Royal Society of Chemistry 2024 |