Green synthesis of metal–organic framework loaded dexamethasone on wood aerogels for enhanced cranial bone regeneration
Zheng-Yang
Chen
ab,
Rui-Deng
Wang
ab,
Shi-Long
Su
ab,
You-Liang
Hao
*ab and
Fang
Zhou
 *ab
*ab
aDepartment of Orthopaedics, Peking University Third Hospital, Beijing 100191, China. E-mail: zhouf@bjmu.edu.cn; haoyl@bjmu.edu.cn
bEngineering Research Center of Bone and Joint Precision Medicine, Peking University Third Hospital, Beijing 100191, China
First published on 21st September 2023
Abstract
Bone defects have attracted increasing attention in clinical settings. To date, there have been no effective methods to repair defective bones. Balsa wood aerogels are considered as an excellent source of chemicals for chemical modification to facilitate the in situ immobilization of zeolitic imidazolate framework-8. Furthermore, dexamethasone has received considerable attention for bone tissue engineering. In this study, for the first time, a simple but effective one-pot method for developing a novel zeolitic imidazolate framework-8 with different concentrations of dexamethasone was developed. These findings illustrate that the novel scaffold has a significant positive impact on osteogenic differentiation in vitro and repairs defects in vivo, suggesting that it can be used in bone tissue engineering.
Introduction
Bone defects of various sizes induced by inflammation, trauma, and tumors have attracted increasing attention in clinical settings.1 Small defects rely on self-restorative abilities for restoration. However, the repair of large defects is beyond the critical restorative ability of the body and results in nonunion or delayed nonunion. Autologous grafting and allogeneic grafting are common treatments used in clinical practice2,3 including the use of a decalcified bone matrix4 and titanium alloys.5 Over the years, osteogenic, autologous, and allogenic grafting methods have been considered as the “gold standard” for treating larger defects because of their excellent osteogenic, osteoinductive, and osteoconductive properties. However, apart from their limited supply, infection transmission on grafting sites, secondary surgery, and immunological rejection, they are also expensive for patients.6,7 Various bioactive materials have been developed as substitutes for autografts.8–11 However, osteogenesis induced by these materials is poor and cannot satisfy the requirements of patients, particularly those with osteoporosis.12,13 Bone tissue engineering offers promising avenues for designing and fabricating innovative graft substitutes for substantial bone defects.14–16 Recent studies have explored various scaffolds to address bone defect challenges. For instance, scaffolds that regulate osteoimmunity have been shown to enhance bone regeneration.17,18 Moreover, biofunctionalized composite scaffolds create a favorable metabolic microenvironment that promotes both osteoconduction and angiogenesis.19,20Aerogel-based bioactive materials are novel and excellent alternatives to bone regeneration. The unique three-dimensional (3D) cross-linked structure of aerogels can partially simulate the natural meshwork of cancellous bone. In addition, apart from their low-density and high-porosity properties, aerogels have a high surface area that mimics the extracellular matrix.21,22 Furthermore, they can improve cellular functions (adhesion, migration, proliferation, and differentiation) during interaction and are regarded as a supporting template for bone regeneration.23 Many aerogel-based bioactive materials have been developed to promote osteogenesis. Recently, Zhang et al. prepared a gradient fibrous aerogel combined with a chemokine peptide. The aerogel enhanced rat bone marrow mesenchymal stem cell (rBMSC) migration and repaired the defect area.24 Additionally, Mo et al. developed a flexible SiO2 nanofiber aerogel with superior stretchability and demonstrated that it could induce bone reconstruction in both osteogenesis and angiogenesis in a rat calvarial defect model.25 The disadvantages of the aforementioned aerogels include their high production cost and fragility.26 Hence, seeking alternative substitutes that possess easy accessibility and superior mechanical features is essential. Wood is commonly used in most industries, and its advantages include its renewable nature, non-toxicity, and low-cost.27 Wood cell-walls are composed of cellulose (40–50%), hemicellulose (10–30%), and lignin (20–30%).28 Balsa wood aerogels (WAs) have been obtained by chemical treatment of natural wood (NW). Hemicellulose and lignin were removed from NW after soaking in chemical solutions, preserving the cellulose and resulting in a hierarchical structure in WA.29 WA has a fragile resistance feature because of its unique hierarchical structure.30 Furthermore, it is an excellent source of chemicals that are regarded as active sites for chemical modification to facilitate the functionalization of WA with polymers or inorganic substances.31
Metal–organic frameworks (MOFs) have been the focus of considerable interest among the scientific community because of their high and large surface areas; they have been considered as alternative multifunctional scaffolds for bone tissue engineering.32–34 Zeolitic imidazolate framework-8 (ZIF-8) is one of the most extensively studied MOFs.35,36 The reaction of sodium hydroxide can firmly immobilize ZIF-8 on WA, providing more sites for in situ immobilization.37 Furthermore, the immobilization of ZIF-8 on WA provides an optional method for the development of high-performance materials for water separation38 and energy fields.39 However, studies on the use of ZIF-8 for bone regeneration are limited. Dexamethasone (DEX) has received considerable attention for incorporation into scaffolds for tissue engineering owing to its high chemical stability.40 It can induce osteogenic differentiation of rBMSCs in vitro and promote the expression of osteogenesis-related genes.41
In this study, a simple but effective one-pot method was first described in the development of WA scaffolds with different concentrations of DEX. Low-density and high-porosity WA was selected as the template, and ZIF-8 was firmly immobilized on the lumen surface via H-bonding and electrostatic interactions.42 The DEX-releasing profile was measured in vitro, and the results indicated that it had a longer release profile, which could enhance the effects of DEX on cells. Furthermore, in vitro results illustrated that osteogenic differentiation was strongly promoted by the higher release of 3-DEX@ZIF-8 WA. Furthermore, the rat calvarial defect model was established by micro-computed tomography (micro-CT) and histological analysis to assess the bone regeneration ability in vivo and verify in vitro osteogenic results. Interestingly, novel DEX@ZIF-8 WAs showed significantly positive impacts on the osteogenic function of rBMSCs in vitro and the repair of bone defects in vivo.
Experimental section
Materials
Balsa wood was obtained from Tianjin Longmao Science and Technology Co., Ltd (China). Sodium chlorite (NaClO2, 80%) and acetic acid (CH3COOH, 99%) were obtained from Shanghai Chemical Reagent Co. Ltd (China). Zinc nitrate hexahydrate (Zn(NO3)2·6H2O, 97%) and 2-methylimidazole (C4H6N2, 98%) were supplied by Shanghai Energy Chemical Reagent Co., Ltd (China). Methanol (CH3OH, 99.9%) was supplied by Tianjin Fu Chen Chemical Reagent Co. Ltd (China). Dexamethasone (C22H29FO5, 97%) was purchased from Sigma-Aldrich. All chemical reagents and solvents used in this study were used without further purification.Fabrication of WAs
Balsa WAs were obtained from previous literature by removing lignin and hemicellulose from the cell wall of NW.43,44 NW cylinders were briefly immersed in 2 wt% NaClO2 solution for 18 h at 80 °C and refreshed every 6 h. Furthermore, acetic acid was added dropwise to the above NaClO2 solution until pH reached 4.6. Additionally, NW was treated with 8 wt% NaOH solution for 12 h at 100 °C. Finally, the NW was rinsed with DI water to remove residual chemicals and freeze-dried overnight to obtain WAs.Fabrication of the ZIF-8 WA and DEX@ZIF-8 WA
The WA was immersed in DI water for 24 h and then vacuumed for 2 h. For the preparation of the ZIF-8 WA, the WA immersed in Zn(NO3)2·6H2O (0.595 g, 0.002 mol) solution containing methanol (20 g) and DI water (3 g) was vacuum impregnated for 24 h at room temperature to ensure sufficient ion exchange from Na+ to Zn2+. A 2-MeIm (3.28 g, 0.04 mol) solution containing methanol (20 g) and DI water (3 g) was subsequently added dropwise to the Zn(NO3)2·6H2O solution under stirring for 24 h at 500 rpm. The WA scaffolds were then cleaned with methanol three times to remove the unreacted precursors, followed by freeze-drying to obtain the ZIF-8 WA.The DEX@ZIF-8 WA was prepared by adding DEX solution and 2-MeIm (3.28 g, 0.04 mol) solution containing methanol (15 g) and DI water (3 g) to vacuum-impregnated Zn(NO3)2·6H2O (0.595 g, 0.002 mol) and WA solution containing methanol (20 g) and DI water (3 g). The DEX masses in methanol (5 g) were 19, 29, and 39 mg, and the corresponding names were 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA, respectively. The subsequent steps were similar to those of the ZIF-8 WA.
WA characterization
The chemical structures of the WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA were characterized using attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR, PerkinElmer Frontier, USA) in the scanning range of 4000–600 cm−1 at a resolution of 2 cm−1. X-ray diffraction (XRD) patterns of the WA scaffolds were recorded using a D8 ADVANCE (Bruker, Germany) with a Cu-Kα radiation source, which was scanned in the 2θ Bragg angle range of 5–900 at a rate of 20 min−1. N2 adsorption/desorption isotherms were measured using a Micromeritics ASAP 2460 instrument at 77 K to determine the specific surface area (SSA) and pore size distribution (PSD). The scaffolds were placed at 120 °C overnight under a vacuum before testing. Thermal stability was measured under a N2 atmosphere using a Q5000IR TG analyzer (TA Instruments, USA) at temperatures ranging from 40 to 800 °C. The morphologies of all WA scaffolds were investigated by FE-SEM (JSM-7900F, JEOL, Japan).DEX release in vitro
The WA was punched into rectangular slices. The in vitro DEX release of 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA was performed in a PBS solution (pH = 7.4). Each slice was immersed in 5 mL of PBS solution and gently rotated (100 rpm) at room temperature. The release profile of DEX was measured using a UV/vis spectrophotometer at 242 nm for 0.5, 1, 2, 4, 7, 10, 13, 16, 19, 22, 25, 28, and 31 d. Each slice of WA was soaked in 5 mL of 6 M hydrochloric acid and gently rotated (100 rpm) to quantify the total loading amount of DEX at room temperature for 5 d. Finally, the collected solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The total loading amount of DEX was measured using the absorbance of the supernatant. The percentage (%) of DEX released from 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA was calculated by amount of released DEX/total loading amount of DEX. The releasing curve of all scaffolds was examined for five trials.
000 rpm for 10 min. The total loading amount of DEX was measured using the absorbance of the supernatant. The percentage (%) of DEX released from 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA was calculated by amount of released DEX/total loading amount of DEX. The releasing curve of all scaffolds was examined for five trials.
In vitro cytocompatibility of WA
Cylindrical scaffolds with the WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA were sterilized by Co60-irradiation (20 kGy; 6 h) for disinfection purposes prior to testing. Scaffolds containing 2 × 104/mL rBMSCs were placed in a 48-well culture plate.Cell proliferation and cell viability
A live/dead staining kit (Solarbio, Beijing, China) was used to observe cell viability using calcein-AM/propidium iodide (PI) under fluorescence staining. After culturing for 1 d and 3 d, the WA with cells was washed three times with PBS, and 250 μL of working solution was added to each WA scaffold based on the manufacturer's recommendations. Furthermore, the working solution was removed and observed under a fluorescence microscope (Olympus, Japan) after incubation at 37 °C for 15 min. Viable cells were stained green, whereas dead cells were stained red. ImageJ was used to quantify live and dead cells. The study was performed in triplicate.Proliferation was measured using cell counting kit-8 (CCK-8; Solarbio, Beijing, China). After 1 d and 3 d, the culture medium was removed, and the scaffolds (diameter: 4 mm, height: 2 mm) were transferred to a new 96-well culture plate. Then, 250 μL of the CCK-8 solution was added to each well. After 1 h, the optical density (OD) was measured at 450 nm with an enzyme-labeled meter (Semerce Aerospace Instruments, Multiskan FC, USA). The study was conducted in the sixth stage.
Cell morphology
WA scaffolds with cells were cleaned three times with PBS and fixed with 4% paraformaldehyde for 30 min. The fixed WA was then cleaned three times with PBS, and 0.2% Triton X-100 was added. After 15 min, 1% BSA was used as a nonspecific binding block for 2 h. Subsequently, primary antibodies: vinculin (ab129002, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Abcam) in PBS with 5% BSA was added to the fixed WA at 4 °C for 12 h. Furthermore, it was washed with PBS, and secondary antibodies of Alexa-Fluor 488 (ab150077, 1
200; Abcam) in PBS with 5% BSA was added to the fixed WA at 4 °C for 12 h. Furthermore, it was washed with PBS, and secondary antibodies of Alexa-Fluor 488 (ab150077, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500; Abcam) were added. After washing, F-actin and nuclei were stained with an FITC-Phalloidin kit (Solarbio, Beijing, China) and DAPI (Solarbio, Beijing, China), respectively, and analyzed by laser confocal microscopy (Leica TCS SP5, Germany). The study was performed in triplicate.
500; Abcam) were added. After washing, F-actin and nuclei were stained with an FITC-Phalloidin kit (Solarbio, Beijing, China) and DAPI (Solarbio, Beijing, China), respectively, and analyzed by laser confocal microscopy (Leica TCS SP5, Germany). The study was performed in triplicate.
In vitro osteogenic differentiation of WA
To investigate the osteogenic differentiation of WA, 2 × 104/mL of rBMSCs were seeded on WA scaffolds in a 48-well culture plate. The medium was removed after 24 h and replaced with an osteogenic differentiation medium containing 10 mM β-glycerol phosphate and 50 μg mL−1 vitamin C. The osteogenic induction medium was refreshed every 3 d.Alkaline phosphatase (ALP) staining
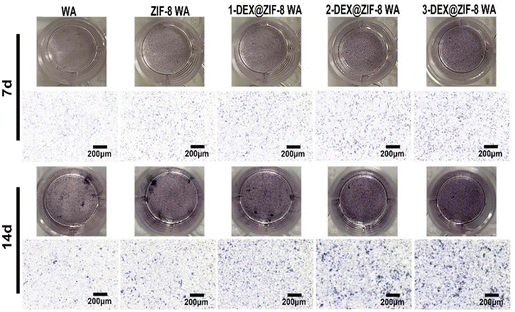
WA scaffolds were immersed in a complete medium and incubated at 37 °C for 48 h at a ratio of 0.2 g mL−1 to prepare the extracts. Then, the extracts were filtered through a 0.22 μm filter for sterilization. For ALP staining, the cells were washed three times with PBS and fixed with 4% paraformaldehyde for 30 min after 7 d and 14 d. The fixed WA was then washed three times with PBS and stained using a BCIP/NBT ALP Kit (Solarbio, Beijing, China). The study was performed in triplicate.Immunostaining
After 7 d of osteogenic differentiation, the cells were washed three times with PBS and fixed for 30 min with 4% paraformaldehyde. The fixed WA was then cleaned three times with PBS, and 0.2% Triton X-100 was added. After 15 min, 1% BSA was used as a non-specific binding block for 2 h. Subsequently, the fixed WA was incubated overnight at 4 °C with primary antibodies: RUNX2 (ab240329, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Abcam) and Col 1α (ab215969, 1
200; Abcam) and Col 1α (ab215969, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Abcam) in PBS with 5% BSA. After washing with PBS, the WA was incubated at room temperature with Alexa Fluor 488 secondary antibodies (ab150077, 1
200; Abcam) in PBS with 5% BSA. After washing with PBS, the WA was incubated at room temperature with Alexa Fluor 488 secondary antibodies (ab150077, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500; Abcam) against RUNX2 and Col 1α. Additionally, the nuclei were stained with DAPI and analyzed using LSCM. The study was performed in triplicate.
500; Abcam) against RUNX2 and Col 1α. Additionally, the nuclei were stained with DAPI and analyzed using LSCM. The study was performed in triplicate.
Quantitative real-time PCR (RT-PCR) analysis
Osteogenesis-related genes were analyzed by RT-PCR (ALP, RUNX2, OPN, and Col 1α). After 7 d of osteogenic differentiation, the cells were cleaned three times with PBS, and total RNA was extracted from cells using an RNA extraction kit (Invitrogen, US) according to the manufacturer's recommendations. The extracted RNA was reverse transcribed to cDNA using a reverse transcription kit (Takara, Japan) according to the manufacturer's recommendations. RT-PCR was performed using a 7500 RT-PCR system (Applied Biosystems, USA). The mRNA levels of the aforementioned osteogenesis-related gene were measured and normalized to the endogenous control GAPDH. The cell culture in osteogenic differentiation medium without the WA served as the control. The study was performed in triplicate. The primers and probes that were used are listed in Table 1.| Forward primer | Reverse primer |
|---|---|
| 5′-CCTAGACACAAGCACTCCCACTA-3′ | 5′-GTCAGTCAGGTTGTTCCGATTC-3′ |
| 5′-GTGTCACTGCGCTGAAGAGG-3′ | 5′-GACCAACCGAGTCATTTAAGGC-3′ |
| 5′-GCCGAGGTGATAGTGTGGTT-3′ | 5′-TGAGGTGATGTCCTCGTCTG-3′ |
| 5′-GAGGGCCAAGACGAAGACATC-3′ | 5′-CAGATCACGTCATCGCACAAC-3′ |
| 5′-ACAACTTTGGTATCGTGGAAGG-3′ | 5′-GCCATCACGCCACAGTTTC-3′ |
In vivo animal experiments
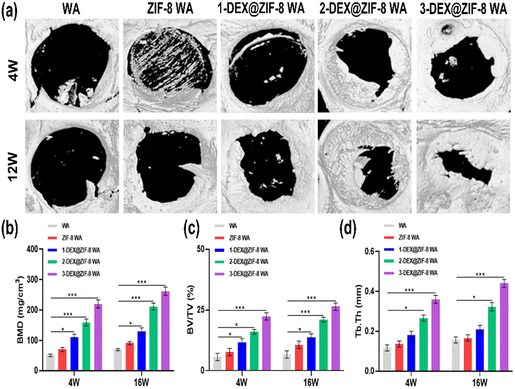
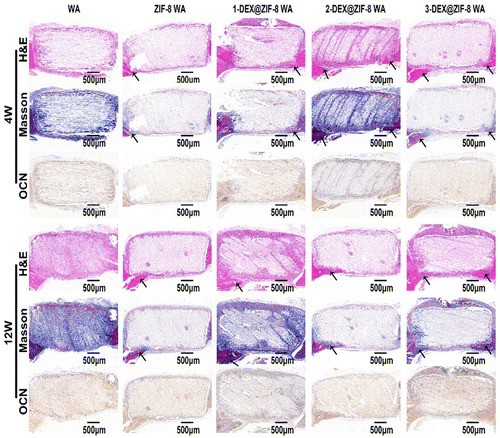
All animal studies were approved by the Peking University Biomedical Ethics Committee. All animal procedures were performed in accordance with the guidelines for care and use of Laboratory Animals of Peking University Third Hospital in China (Approval number: LA2019018). Female Sprague-Dawley rats were obtained from the Animal Research Center of the Peking University Health Science Center. 10% chloral hydrate (0.3 mL/100 g) was injected intraperitoneally according to the weight of the rats. Iodophor was used to sterilize the head of the rats after anesthesia. A longitudinal incision of approximately 1 cm was made in the middle of the scalp to expose the skull. A low-speed grinding drill was used to obtain a 5 mm critical-sized calvarial defect on the skull. WA scaffolds (diameter: 5 mm; height: 2 mm) were placed in the defect area. Rats were sacrificed at 4 W and 12 W. Skulls were obtained and fixed with 4% paraformaldehyde for subsequent experiments.Skulls were obtained and analyzed using micro-CT (Skyscan1176). After micro-CT observation, the skulls were decalcified with 10% EDTA for several weeks and embedded in paraffin. Hematoxylin and eosin (H&E) and Masson staining were performed to evaluate osteogenic ability. Finally, OCN was stained after deparaffinization. Images were obtained using a microscope.
Statistical analysis
Data are presented as the mean ± standard deviation. Data were analyzed using Tukey's multiple-comparison post hoc test using SPSS software (Version 22); *p < 0.05, **p < 0.01, and ***p < 0.001 were considered statistically significant.Results and discussion
Fabrication and characterization of WA scaffolds
The procedure to fabricate WA and DEX@ZIF-8 WA scaffolds is outlined in Scheme 1. The Balsa NW, chosen for its low density and enriched porosity, is ideal for in situ ZIF-8 immobilization.45 The NW was shaped into a cylinder along its growth direction to maintain its anisotropic structure. For the preparation of DEX@ZIF-8 WA, the NW underwent pretreatment with a mix of NaClO2 and acetic acid, enabling the strong oxidizing hypochlorite (ClO2) to remove lignin. This elimination of lignin enlarges the lumen area, promoting better attachment of metal ions and enhancing ZIF-8 immobilization.46 Subsequent treatment with a NaOH solution ensured efficient exchange between carboxyl (COO−) and sodium ions (Na+).47 As a result of lignin removal by NaClO2, the initial pale-yellow hue of the NW transformed into a white WA. This WA was then submerged in a Zn(NO3)2·6H2O solution, facilitating ion exchange from Na+ to Zn2+ through vacuum impregnation and offering more sites for in situ immobilization.42 The zinc ion (Zn2+) anchored itself onto the lumen surfaces of the WA, attaching to COO− after permeating the wood cell membrane.48 Finally, DEX@ZIF-8 became firmly affixed to the WA through electrostatic forces and hydrogen bonds, post its reaction with 2-MeIm and DEX.49 The fabrication of ZIF-8 WA followed a similar route, excluding the addition of DEX.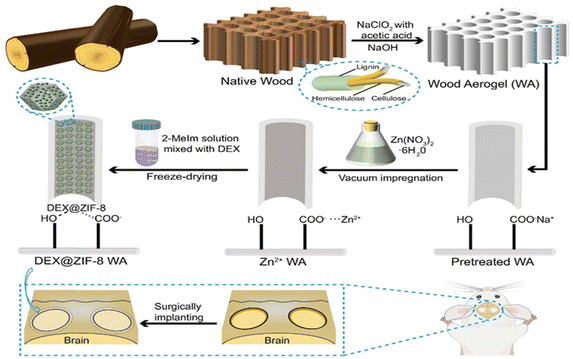 | ||
| Scheme 1 Schematic description of the fabrication process of WA from NW and the in situ immobilization of DEX@ZIF-8 on WA. | ||
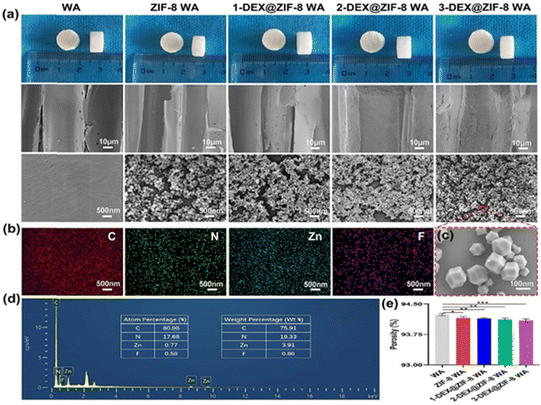
Fig. 1a shows the inner morphology and structure of the WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA. The SEM images of WAs indicated that the lumen surfaces were smooth, which provided a potential binding site for Zn ions. Many small MOF nanoparticles were observed after the MOF was immobilized in situ, and the lumen surfaces changed from smooth to rough. The lumen surfaces of 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA remained rough after the addition of DEX, and these MOF nanoparticles could be identified without obvious agglomeration. The DEX-loaded MOF nanoparticles were regular and rhombohedral dodecahedra at a higher resolution, indicating that the introduction of DEX did not influence the morphology of ZIF-8. Notably, the MOF nanoparticles in all WA scaffolds were firmly immobilized on the lumen surface, demonstrating the superior MOF-WA interface. The EDS mapping images and EDS spectra of the 3-DEX@ZIF-8 WA (Fig. 1b and d) illustrate the uniform distribution of C, N, Zn, and F on the lumen surfaces, confirming the successful introduction of DEX into ZIF-8 on WA channels. As shown in Fig. 1c, DEX@ZIF-8 nanoparticles are similar in size, monodispersed, uniformly shaped, and exhibit a rhombohedral dodecahedral structure. Fig. 1e shows that all WA scaffolds exhibited high porosity, while promoting the proliferation, differentiation, and migration of rBMSCs.
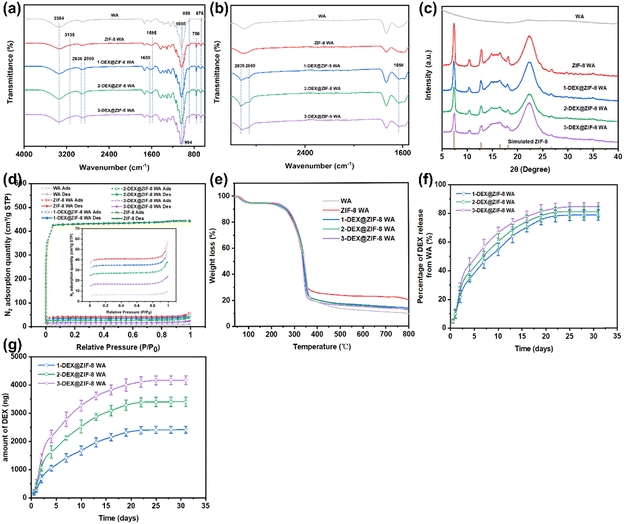
ATR-FTIR was used to further confirm the chemical structures of WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA (Fig. 2a and b). The characteristic peaks at 898 and 676 cm−1 corresponded to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending of WA. Furthermore, the peaks at 3384 and 1058 cm−1 were related to O–H stretching and C–O stretching, respectively.50 In this study, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2- DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA's characteristic peaks at 1585 cm−1 correspond to the stretching vibration of C
C bending of WA. Furthermore, the peaks at 3384 and 1058 cm−1 were related to O–H stretching and C–O stretching, respectively.50 In this study, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2- DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA's characteristic peaks at 1585 cm−1 correspond to the stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, as studied in previous literature.51,52 The two peaks at 750 (N–H) and 994 (C–N) cm−1 were ascribed to the out-of-plane and in-plane bending vibrations of the imidazolate ring.53–55 In addition, the peak at 3135 cm−1 was attributed to the stretching of aromatic C–H.56,57 ZIF-8 successfully immobilizes in-situ on the surface of WA. After adding DEX, some peaks related to DEX appeared; for example, one peak located at 1650 cm−1 was attributed to C
N, as studied in previous literature.51,52 The two peaks at 750 (N–H) and 994 (C–N) cm−1 were ascribed to the out-of-plane and in-plane bending vibrations of the imidazolate ring.53–55 In addition, the peak at 3135 cm−1 was attributed to the stretching of aromatic C–H.56,57 ZIF-8 successfully immobilizes in-situ on the surface of WA. After adding DEX, some peaks related to DEX appeared; for example, one peak located at 1650 cm−1 was attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, while the other peaks at 2936 cm−1 and 2850 cm−1 were assigned to saturated C–H bonds.58,59 These characteristic peaks confirmed the successful loading of DEX onto WA. As shown in Fig. 2c, the XRD pattern of all WA scaffolds revealed a diffraction peak at a Bragg angle of 22.3° associated with cellulose. In addition, some characteristic peaks belonging to ZIF-8 were clearly observed at Bragg angles of 7.30, 10.40, 12.80, 14.70, and 18.10, corresponding to the (110), (200), (211), (220), and (222) crystal of ZIF-8, respectively. Moreover, it demonstrated the successful in situ immobilization of ZIF-8 on ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA. The positions of the diffraction peaks and their intensities in the WA scaffolds loaded with ZIF-8 confirmed the presence of ZIF-8 in-situ immobilization. The crystal structure of ZIF-8 was not changed when the DEX was incorporated. However, the crystallinities of 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA decreased with an increase in the DEX concentration. The decreasing intensity values for 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA indicate that DEX was successfully encapsulated in the ZIF-8 pores. However, according to previous studies, the decreasing crystallinities might be related to the framework and pore cages that hinder X-ray penetrate, leading to the intensity value.60,61
O stretching, while the other peaks at 2936 cm−1 and 2850 cm−1 were assigned to saturated C–H bonds.58,59 These characteristic peaks confirmed the successful loading of DEX onto WA. As shown in Fig. 2c, the XRD pattern of all WA scaffolds revealed a diffraction peak at a Bragg angle of 22.3° associated with cellulose. In addition, some characteristic peaks belonging to ZIF-8 were clearly observed at Bragg angles of 7.30, 10.40, 12.80, 14.70, and 18.10, corresponding to the (110), (200), (211), (220), and (222) crystal of ZIF-8, respectively. Moreover, it demonstrated the successful in situ immobilization of ZIF-8 on ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA. The positions of the diffraction peaks and their intensities in the WA scaffolds loaded with ZIF-8 confirmed the presence of ZIF-8 in-situ immobilization. The crystal structure of ZIF-8 was not changed when the DEX was incorporated. However, the crystallinities of 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA decreased with an increase in the DEX concentration. The decreasing intensity values for 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA indicate that DEX was successfully encapsulated in the ZIF-8 pores. However, according to previous studies, the decreasing crystallinities might be related to the framework and pore cages that hinder X-ray penetrate, leading to the intensity value.60,61
BET-specific tests were performed to investigate the aperture properties of ZIF-8, WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA. The N2 adsorption–desorption isotherms and PSD curves are shown in Fig. 2d. ZIF-8, the ZIF-8 WA, the 1-DEX@ZIF-8 WA, the 2-DEX@ZIF-8 WA, and the 3-DEX@ZIF-8 WA exhibited type I N2 isotherms, and no distinct hysteresis loop was observed, demonstrating the existence of micropores. However, the WA had a small hysteresis loop with a high pressure (0.8–1 P/P0), caused by the mesoporous structure.62,63 Furthermore, the N2 adsorption–desorption capacity also decreased with a DEX loading compared to that of the ZIF-8 WA, indicating the encapsulation of DEX into ZIF-8. In addition, the PSD curves of ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA show a mesoporous structure with a uniform distribution of 7–30 nm diameters. Apart from being controlled, with a continued and steady release of DEX, the outstanding features of mesopores also promoted the adhesion, proliferation, and differentiation of rBMSCs. Such a pore distribution might have resulted from the higher pyrolysis temperature, and was better for adsorption, as the larger pores could transfer adsorbate and then rapidly adsorb in smaller pores by van der Waals forces.64
Thermogravimetric analysis was beneficial for understanding the components of the scaffold. The decomposition behavior of WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA under a N2 atmosphere is shown in Fig. 2e. All scaffolds exhibited similar weight loss trends. The TG curves of all scaffolds could be separated into three sections based on the weight loss, 40–100 °C, 100–360 °C, and 360–800 °C. The primary weight loss percentage in WA was 6%, which was ascribed to the evaporation of the adsorbed water. The second was 74.46% owing to the breakdown of cellulose. Finally, the weight loss at 360–800 °C was caused by the breakdown of residual lignin. After ZIF-8 loading, the ZIF-8 WA exhibited a large residual weight loss percentage, which is related to the stability of ZIF-8 at high temperatures. Upon increasing the DEX concentration, the TG curves did not change significantly for the 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA, demonstrating that the structure of ZIF-8 was not affected by different DEX concentrations.
Fabrication and characterization of WA scaffolds
The total loading amounts of DEX in the 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA are 3063.02 ± 118.67 ng, 4192.18 ± 123.34 ng, and 4922.52 ± 138.62 ng, respectively. The total loading amounts of DEX in the scaffold are 141 937 ± 5 499 ng, 19 4262 ± 5 715 ng, and 228 105 ± 6 423 ng. Furthermore, the percentage and accumulative release profiles show that DEX was released in a sustained manner and increased upon increasing the DEX feeding concentration, confirming that the majority of DEX was loaded inside the pores, while the rest was adsorbed on the surface of ZIF-8. Fig. 2f shows that the released percentages of the 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA are 78.89 ± 3.71%, 81.28 ± 3.72%, and 84.71 ± 2.78% after 35 d, respectively. The loaded DEX was completely released from the 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA after 22 d and 25 d, which reached 2415.21 ± 122.44, 3406.49 ± 165.17, and 4169.01 ± 155.11 (ng), respectively (Fig. 2g). Compared with other DEX carriers, such as mesoporous silica nanoparticles, the DEX@ZIF-8 WA had a longer release profile, which could enhance the effects of DEX on cells.65 This might be ascribed to the lumen surface in WA, which hinders DEX release to some extent, thereby protecting DEX@ZIF-8 nanoparticles from fast degradation and reducing side effects.Cytocompatibility of WA scaffolds
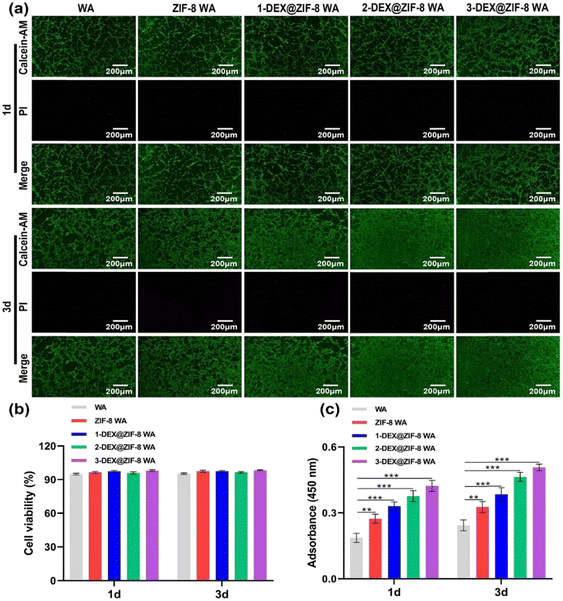
rBMSCs were co-cultured with the WA scaffolds to verify cytocompatibility. The live/dead staining kit and CCK-8 method were used to assess the influence of WA scaffolds on the cell viability and proliferation activity. Furthermore, they indicate that rBMSCs are more viable (stained green); only a few dead cells (stained red) can be seen on the WA surface under a fluorescence microscope (Fig. 3a) after being cultured for 1 d and 3 d. As shown in Fig. 3b, the cell viability in the WA, ZIF-8 WA, 1-DEX@ZIF-8 WA, 2-DEX@ZIF-8 WA, and 3-DEX@ZIF-8 WA was beyond 95% on 1 d and 3 d, demonstrating that the incorporation of ZIF-8 and DEX on the WA was safe for rBMSC growth. Additionally, the proliferation activity measured using CCK-8 was similar to live/dead staining results (Fig. 3c). On 1 d and 3 d, the rBMSC proliferation activity was higher in 3-DEX@ZIF-8 WA than in the other four scaffolds with an increase in DEX concentration (P < 0.05), indicating that more DEX could be released from ZIF-8, which may benefit the rBMSC proliferation process. Consequently, the incorporation of DEX into ZIF-8 and in-situ immobilization on the WA lumen surface at different concentrations did not influence the viability and proliferation of 3-DEX@ZIF-8 WA than in other scaffolds.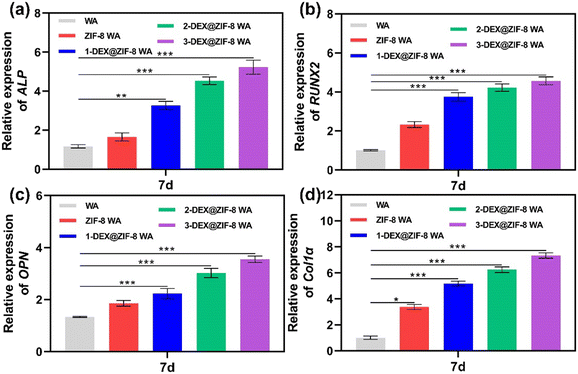 | ||
| Fig. 6 RT-PCR analysis of the osteogenic-associated gene expression of (a) ALP, (b) RUNX2, (c) OPN, and (d) Col1α in WA scaffolds after 7 d. | ||
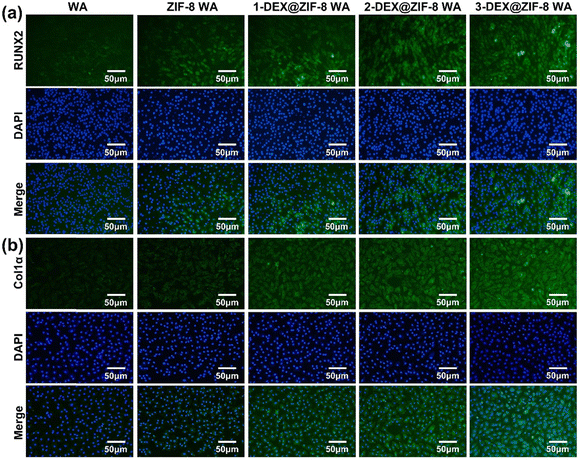 | ||
| Fig. 7 Effects of WA scaffolds on (a) RUNX2 and (b) Col1α expression after 7 d. Cell nuclei were labeled with blue, while RUNX2 and Col1α were green. | ||
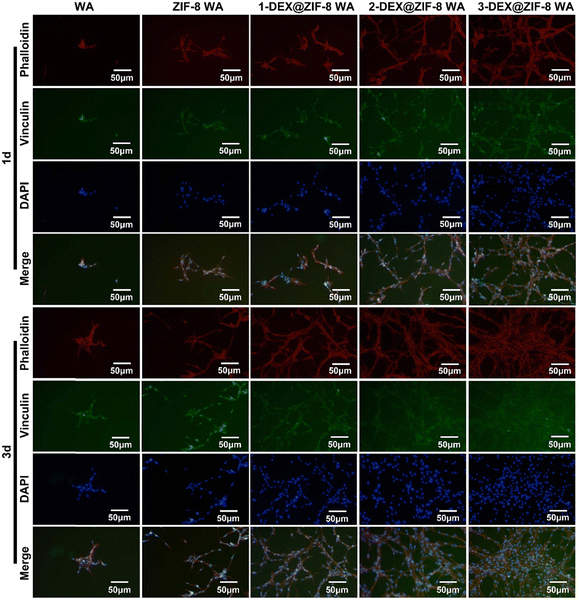
Both the RT-PCR and immunostaining results illustrated that the in situ immobilization of ZIF-8 loaded DEX on the WA promoted osteogenic induction. Compared with the WA, the 3-DEX@ZIF-8 WA significantly increased the expression of osteogenic-associated genes and proteins after osteogenic differentiation for 7 d. Additionally, the gene and protein levels of osteogenic-associated genes were in agreement with the ALP activity and ARS staining, which demonstrated that the in-situ immobilization of DEX on the WA could promote osteogenesis in vitro.
Conclusions
In this study, we designed the biological activity of WA by in situ immobilization of ZIF-8 loading of DEX on the lumen surface to create a scaffold for bone regeneration. This is the first study to prepare DEX@ZIF-8 WA and evaluate its osteogenic ability in vitro and in vivo. The obtained WA was thoroughly characterized using FTIR, XRD, BET, TGA, and FE-SEM to demonstrate the existence of DEX@ZIF-8. The DEX-releasing profile was measured in vitro, and the results indicated that it had a longer released profile in 3-DEX@ZIF-8 WA, related to the long-term regeneration of new bone. The cytocompatibility and effect on the proliferation and cell morphology of rBMSCs were measured in vitro, illustrating that adding DEX to the 3-DEX@ZIF-8 WA has a positive effect. The results of ALP staining, ALP activity, ARS staining, RT-PCR, and immunostaining experiments showed that the 3-DEX@ZIF-8 WA could significantly induce the osteogenic differentiation of rBMSCs in vitro by the long-term release of DEX. Based on the in vitro experiments, the high concentration of DEX in 3-DEX@ZIF-8 WA exhibited superior osteoinductive ability in the calvarial defect model after implantation for four and 12 W. Moreover, the results of H&E, Masson, and immunohistochemical staining of calvarial terminal sections confirmed that new bone formation was most frequently observed in 3-DEX@ZIF-8 WA. Interestingly, the novel DEX@ZIF-8 WA showed significant impacts on the osteogenic differentiation of rBMSCs in vitro and the repair of bone defects in vivo, thus possessing potential applications in bone regeneration.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81971160), the National Health and Family Planning Commission of China (No. A72500-20), and the Innovation and Transformation Fund of Peking University Third Hospital (BYSYZHKC2021106).References
- Z. Y. Chen, S. Gao, Y. W. Zhang, R. B. Zhou and F. Zhou, J. Mater. Chem. B, 2021, 9, 2594–2612 RSC.
- L. A. Vonk, G. Roel, J. Hernigou, C. Kaps and P. Hernigou, Int. J. Mol. Sci., 2021, 22 Search PubMed.
- C. Lakkireddy, S. K. Vishwakarma, A. Bardia, N. Raju, S. I. Ahmed, S. M. Fathima, S. Annamaneni and A. A. Khan, Tissue Cell, 2021, 73, 101631 CrossRef CAS PubMed.
- M. Zhang, J. P. Matinlinna, J. K. H. Tsoi, W. Liu, X. Cui, W. W. Lu and H. Pan, J. Orthop. Translat., 2020, 22, 26–33 CrossRef PubMed.
- F. A. Spitznagel, E. A. Bonfante, F. Vollmer and P. C. Gierthmuehlen, J. Prosthodont., 2022, 31, 136–146 CrossRef CAS PubMed.
- Z. Stopa, M. Siewert-Gutowska, K. Abed, D. Szubinska-Lelonkiewicz, A. Kaminski and P. Fiedor, Transplant. Proc., 2018, 50, 2199–2201 CrossRef CAS PubMed.
- H. Siadat, A. Khojasteh and E. Beyabanaki, Front. Dent., 2019, 16, 153–157 Search PubMed.
- X. Liu, L. Li, B. Gaihre, S. Park, Y. Li, A. Terzic, B. D. Elder and L. Lu, ACS Nano, 2022, 16, 2741–2755 CrossRef CAS PubMed.
- X. Li, B. Dai, J. Guo, Y. Zhu, J. Xu, S. Xu, Z. Yao, L. Chang, Y. Li, X. He, D. H. K. Chow, S. Zhang, H. Yao, W. Tong, T. Ngai and L. Qin, ACS Nano, 2022, 16, 18071–18089 CrossRef CAS PubMed.
- C. Wu, X. He, W. Weng, T. Zhang, D. Huang, K. Cheng and Z. Chen, Chem. Eng. J., 2022, 443, 136508 CrossRef CAS.
- X. Yu, X. Wang, D. Li, R. Sheng, Y. Qian, R. Zhu, X. Wang and K. Lin, Chem. Eng. J., 2022, 433, 132799 CrossRef CAS.
- J. Li, Y. Chen, N. Kawazoe and G. Chen, Nano Res., 2018, 11, 1247–1261 CrossRef CAS.
- E. B. Hunziker, L. Enggist, A. Kuffer, D. Buser and Y. Liu, Bone, 2012, 51, 98–106 CrossRef CAS PubMed.
- X. Wang, M. Zhang, T. Zhu, Q. Wei, G. Liu and J. Ding, Adv. Sci., 2023, 10 Search PubMed.
- X. Wan, Y. Zhao, Z. Li and L. Li, Exploration, 2022, 2, 20210029 CrossRef PubMed.
- Y. Peng, Y. Zhuang, Y. Zhang, J. Zuo and J. Ding, MedComm – Biomater. Appl., 2023, 2, e49, DOI:10.1002/mba2.49.
- J. Zhang, D. Tong, H. Song, R. Ruan, Y. Sun, Y. Lin, J. Wang, L. Hou, J. Dai, J. Ding and H. Yang, Adv. Mater., 2022, 34, 2202044 CrossRef CAS PubMed.
- Z. Liu, J. Zhang, C. Fu and J. Ding, Asian J. Pharm. Sci., 2023, 18, 100774, DOI:10.1016/j.ajps.2023.100774.
- Y. Peng, Y. Zhuang, Y. Liu, H. Le, D. Li, M. Zhang, K. Liu, Y. Zhang, J. Zuo and J. Ding, Exploration, 2023, 20210043, DOI:10.1002/exp.20210043.
- T. Zhu, M. Jiang, M. Zhang, L. Cui, X. Yang, X. Wang, G. Liu, J. Ding and X. Chen, Bioactive Mater., 2022, 9, 446–460 CrossRef CAS PubMed.
- C. Bonnans, J. Chou and Z. Werb, Nat. Rev. Mol. Cell Biol., 2014, 15, 786–801 CrossRef CAS PubMed.
- Y. Jiang, H. Zhang, J. Wang, Y. Liu, T. Luo and H. Hua, J. Hematol. Oncol., 2022, 15, 34 CrossRef PubMed.
- H. Maleki, L. Duraes, C. A. Garcia-Gonzalez, P. Del Gaudio, A. Portugal and M. Mahmoudi, Adv. Colloid Interface Sci., 2016, 236, 1–27 CrossRef CAS PubMed.
- L. Zhang, J. Fang, L. Fu, L. Chen, W. Dai, H. Huang, J. Wang, X. Zhang, Q. Cai and X. Yang, Chem. Eng. J., 2021, 422 Search PubMed.
- M. Liu, M. Shafiq, B. Sun, J. Wu, W. Wang, M. El-Newehy, H. El-Hamshary, Y. Morsi, O. Ali, A. U. R. Khan and X. Mo, Adv. Healthcare Mater., 2022, 11, e2200499 CrossRef PubMed.
- Y. G. Zhang, Y. J. Zhu, Z. C. Xiong, J. Wu and F. Chen, ACS Appl. Mater. Interfaces, 2018, 10, 13019–13027 CrossRef CAS PubMed.
- J. Sun, H. Guo, J. Ribera, C. Wu, K. Tu, M. Binelli, G. Panzarasa, F. Schwarze, Z. L. Wang and I. Burgert, ACS Nano, 2020, 14, 14665–14674 CrossRef CAS PubMed.
- Z. Wang, Y. He, L. Zhu, L. Zhang, B. Liu, Y. Zhang and T. Duan, Mater. Chem. Phys., 2021, 258, 123964 CrossRef CAS.
- L. A. Berglund and I. Burgert, Adv. Mater., 2018, 30, e1704285 CrossRef PubMed.
- Y. Li, E. Vasileva, I. Sychugov, S. Popov and L. Berglund, Adv. Opt. Mater., 2018, 6, 1800059 CrossRef.
- V. Merk, M. Chanana, N. Gierlinger, A. M. Hirt and I. Burgert, ACS Appl. Mater. Interfaces, 2014, 6, 9760–9767 CrossRef CAS PubMed.
- X. Ma, Y. Xiong, Y. Liu, J. Han, G. Duan, Y. Chen, S. He, C. Mei, S. Jiang and K. Zhang, Chem, 2022, 8, 2342–2361 CAS.
- C. Xu, Y. Kang, X. Dong, D. Jiang and M. Qi, Chin. Chem. Lett., 2023, 34, 107528, DOI:10.1016/j.cclet.2022.05.042.
- C. Xu, Y. Kang, S. Guan, X. Dong, D. Jiang and M. Qi, Chin. Chem. Lett., 2023, 34, 107825, DOI:10.1016/j.cclet.2022.107825.
- Z.-Y. Chen, S. Gao, R.-B. Zhou, R.-D. Wang and F. Zhou, Mater. Des., 2022, 217, 110627 CrossRef CAS.
- Z.-Y. Chen, R.-B. Zhou, R.-D. Wang, S.-L. Su and F. Zhou, Int. J. Biol. Macromol., 2023, 234, 123788, DOI:10.1016/j.ijbiomac.2023.123788.
- D. You, Y. Zhao, W. Yang, Q. Pan and J. Li, Chem. Res. Chin. Univ., 2021, 38, 186–191 CrossRef.
- Y. Li, Z. Lin, X. Wang, Z. Duan, P. Lu, S. Li, D. Ji, Z. Wang, G. Li, D. Yu and W. Liu, Sep. Purif. Technol., 2021, 270, 118794 CrossRef CAS.
- J. Sun, K. Tu, S. Büchele, S. M. Koch, Y. Ding, S. N. Ramakrishna, S. Stucki, H. Guo, C. Wu, T. Keplinger, J. Pérez-Ramírez, I. Burgert and G. Panzarasa, Matter, 2021, 4, 3049–3066 CrossRef CAS.
- E. K. Medlock Kakaley, B. R. Blackwell, M. C. Cardon, J. M. Conley, N. Evans, D. J. Feifarek, E. T. Furlong, S. T. Glassmeyer, L. E. Gray, Jr., P. C. Hartig, D. W. Kolpin, M. A. Mills, L. Rosenblum, D. L. Villeneuve and V. S. Wilson, Sci. Total Environ, 2020, 699, 134297 CrossRef CAS PubMed.
- S. B. Gasson, L. K. Dobson, L. Chow, S. Dow, C. A. Gregory and W. B. Saunders, Stem Cells Dev., 2021, 30, 214–226 CrossRef CAS PubMed.
- K. Tu, B. Puertolas, M. Adobes-Vidal, Y. Wang, J. Sun, J. Traber, I. Burgert, J. Perez-Ramirez and T. Keplinger, Adv. Sci., 2020, 7, 1902897 CrossRef CAS PubMed.
- J. Garemark, X. Yang, X. Sheng, O. Cheung, L. Sun, L. A. Berglund and Y. Li, ACS Nano, 2020, 14, 7111–7120 CrossRef CAS PubMed.
- M. B. Wu, C. Zhang, Y. Xie, S. Huang, C. Liu, J. Wu and Z. K. Xu, ACS Appl. Mater. Interfaces, 2021, 13, 51039–51047 CrossRef CAS PubMed.
- Z. Cheng, H. Guan, J. Meng and X. Wang, ACS Omega, 2020, 5, 14096–14103 CrossRef CAS PubMed.
- Z. Wang, F. Yin, X.-F. Zhang, T. Zheng and J. Yao, Sep. Purif. Technol., 2022, 293 Search PubMed.
- P. Song, X. Shen, X. He, K. Feng, L. Kong, Z. Ji, L. Zhai, G. Zhu and D. Zhang, Cellulose, 2018, 26, 1195–1208 CrossRef.
- X.-F. Zhang, Z. Wang, L. Song and J. Yao, Sep. Purif. Technol., 2021, 266, 118527 CrossRef CAS.
- M. Liu, Z. Zhao and W. Yu, Chem. Eng. J., 2020, 393, 124748 CrossRef CAS.
- G. Chen, S. He, G. Shi, Y. Ma, C. Ruan, X. Jin, Q. Chen, X. Liu, H. Dai, X. Chen and D. Huang, Chem. Eng. J., 2021, 423, 130184 CrossRef CAS.
- H. Fan, Q. Shi, H. Yan, S. Ji, J. Dong and G. Zhang, Angew. Chem., Int. Ed., 2014, 53, 5578–5582 CrossRef CAS PubMed.
- G. Liu, Z. Jiang, K. Cao, S. Nair, X. Cheng, J. Zhao, H. Gomaa, H. Wu and F. Pan, J. Membr. Sci., 2017, 523, 185–196 CrossRef CAS.
- Q. Li, S. Jiang, S. Ji, M. Ammar, Q. Zhang and J. Yan, J. Solid State Chem., 2015, 223, 65–72 CrossRef CAS.
- N. M. Mahmoodi, M. Oveisi, A. Taghizadeh and M. Taghizadeh, Carbohydr. Polym., 2020, 227, 115364 CrossRef CAS PubMed.
- L. B. Vaidya, S. S. Nadar and V. K. Rathod, Int. J. Biol. Macromol., 2020, 146, 678–686 CrossRef CAS PubMed.
- A. Karakecili, B. Topuz, S. Korpayev and M. Erdek, Mater. Sci. Eng., C, 2019, 105, 110098 CrossRef CAS PubMed.
- L. Ning, S. Liao, C. Dong, M. Zhang, W. Gu and X. Liu, ACS Appl. Mater. Interfaces, 2020, 12, 7198–7205 CrossRef CAS PubMed.
- M. Lian, B. Sun, Z. Qiao, K. Zhao, X. Zhou, Q. Zhang, D. Zou, C. He and X. Zhang, Colloids Surf., B, 2019, 176, 219–229 CrossRef CAS PubMed.
- M. Roozbahani, M. Kharaziha and R. Emadi, Int. J. Pharm., 2017, 518, 312–319 CrossRef CAS PubMed.
- X. Zhu, J. Gu, Y. Wang, B. Li, Y. Li, W. Zhao and J. Shi, Chem. Commun., 2014, 50, 8779–8782 RSC.
- J. Gu, M. Huang, J. Liu, Y. Li, W. Zhao and J. Shi, New J. Chem., 2012, 36 CAS.
- S. He, G. Chen, H. Xiao, G. Shi, C. Ruan, Y. Ma, H. Dai, B. Yuan, X. Chen and X. Yang, J. Colloid Interface Sci., 2021, 582, 90–101 CrossRef CAS PubMed.
- S. He, G. Shi, H. Xiao, G. Sun, Y. Shi, G. Chen, H. Dai, B. Yuan, X. Chen and X. Yang, Chem. Eng. J., 2021, 410, 128286 CrossRef CAS.
- P. Song, C. Chen, X. Shen, S. Zeng, S. Premlatha, Z. Ji, L. Zhai, A. Yuan and Q. Liu, J. Colloid Interface Sci., 2022, 620, 376–387 CrossRef CAS PubMed.
- Q. Gan, J. Zhu, Y. Yuan, H. Liu, J. Qian, Y. Li and C. Liu, J. Mater. Chem. B, 2015, 3, 2056–2066 RSC.
- J. M. Kalappurakkal, A. A. Anilkumar, C. Patra, T. S. van Zanten, M. P. Sheetz and S. Mayor, Cell, 2019, 177, 1738–1756 CrossRef CAS PubMed.
- L. Qiu, J. Cai, N. Zhang, L. Ma, F. Y. Fan and X. M. Li, Tissue Cell, 2022, 76, 101791 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |