Open Access Article
Open Access ArticleIntegration of semi-empirical MS/MS library with characteristic features for the annotation of novel amino acid-conjugated bile acids†
Yan
Ma
 *ab,
Yang
Cao
a,
Xiaocui
Song
a,
Weichen
Xu
c,
Zichen
Luo
c,
Jinjun
Shan
*ab,
Yang
Cao
a,
Xiaocui
Song
a,
Weichen
Xu
c,
Zichen
Luo
c,
Jinjun
Shan
 c and
Jingjie
Zhou
d
c and
Jingjie
Zhou
d
aNational Institute of Biological Sciences, Beijing, Beijing 102206, China. E-mail: mayan@nibs.ac.cn
bTsinghua Institute of Multidisciplinary Biomedical Research, Tsinghua University, Beijing 100084, China
cInstitute of Pediatrics, Jiangsu Key Laboratory of Pediatric Respiratory Disease, Medical Metabolomics Center, Nanjing University of Chinese Medicine, Nanjing 210023, China
dThe Affiliated Jiangyin Hospital of Nanjing University of Chinese Medicine, Jiangyin 214400, China
First published on 13th September 2023
Abstract
Recently, amino acids other than glycine and taurine were found to be conjugated with bile acids by the gut microbiome in mouse and human. As potential diagnostic markers for inflammatory bowel disease and farnesoid X receptor agonists, their physiological effects and mechanisms, however, remain to be elucidated. A tool for the rapid and comprehensive annotation of such new metabolites is required. Thus, we developed a semi-empirical MS/MS library for bile acids conjugated with 18 common amino acids, including alanine, arginine, asparagine, aspartate, glutamine, glutamate, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine. To investigate their fragmentation rules, these amino acids were chemically conjugated with lithocholic acid, deoxycholic acid, and cholic acid, and their accurate-mass MS/MS spectra were acquired. The common fragmentation patterns from the amino acid moieties were combined with 10 general bile acid skeletons to generate a semi-empirical MS/MS library of 180 structures. Software named BAFinder 2.0 was developed to combine the semi-empirical library in negative mode and the characteristic fragments in positive mode for automatic unknown identification. As a proof of concept, this workflow was applied to the LC-MS/MS analysis of the feces of human, beagle dogs, and rats. In total, 171 common amino acid-conjugated bile acids were annotated and 105 of them were confirmed with the retention times of synthesized compounds. To explore other potential bile acid conjugates, user-defined small molecules were in-silico conjugated with bile acids and searched in the fecal dataset. Four novel bile acid conjugates were discovered, including D-Ala-D-Ala, Lys(iso)-Gly, L-2-aminobutyric acid, and ornithine.
Introduction
Gut microbiota play a critical role in regulating the host physiology and pathophysiology. The approximately 1013–1014 bacteria in the human microbiome produce numerous metabolites, among which one of the most important classes is bile acids.1,2 Gut bacteria chemically modify the primary bile acids synthesized from the host liver and produce secondary bile acids through deconjugation, dehydroxylation, oxidation, and epimerization.3 These modified bile acids can act as signaling molecules to modulate host glucose and lipid metabolism and can affect the gut microbiota composition.4,5Bile acids have long been known to be conjugated with glycine and taurine by liver enzymes.6 Recently, other amino acids, such as leucine, phenylalanine, and tyrosine, were reported for the first time to be conjugated to bile acids by the gut microbiome in human and mouse.7 These novel metabolites agonized the farnesoid X receptor in vitro and were enriched in patients with inflammatory bowel disease.7 Since then, more bile acid conjugations have been discovered, covering almost all common amino acids.8–10Fig. 1 shows the simplified structure of an amino acid-conjugated bile acid. The structural diversities from both the steroid core and the amino acid moiety pose a challenge for unknown identification.
Liquid chromatography-mass spectrometry (LC-MS) has been widely applied in the analysis of amino acid-conjugated bile acids. For example, Zhu et al.11 developed a method that combined chemical derivatization and alternating dual-collision energy scanning mass spectrometry, and identified 17 bile acids conjugated with alanine, proline, leucine, and phenylalanine from mouse intestine contents and feces. Wang et al.12 established a polarity-switching multiple reaction monitoring (MRM) mass spectrometry method and screened 118 amino acid-conjugated bile acids, which was the most comprehensive profiling of such metabolites to our knowledge so far. However, most of these reported methods are targeted at only certain amino acid conjugations. Besides, additional expertise and time are often required for manual spectral interpretation.
To facilitate the automatic annotation of amino acid-conjugated bile acids in biological samples, a MS/MS library is needed. However, due to the lack of reference standard compounds, a comprehensive experimental library is not feasible. In silico MS/MS libraries generated from fragmentation rules and computational methods have been applied to the annotation of various compound classes, such as lipids13 and acyl-CoA.14 Therefore, we set out to expand the coverage of an experimental MS/MS library with the spectra predicted from the fragmentation rules of 18 common amino acid-conjugated bile acids in negative ESI mode. This semi-empirical library can be used with any generic spectra search engine (e.g., NIST MS Search), or with BAFinder, a software dedicated for the identification of bile acids.15 The updated BAFinder 2.0 software combined the characteristic fragments in the positive mode with the semi-empirical library in the negative mode to further increase the confidence of annotation. What's more, novel bile acid conjugates other than common amino acids could be annotated using a similar strategy.
Experimental
Materials
Alanine, arginine, asparagine, aspartate, cysteine, glutamine, glutamate, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, valine, and cholic acid (CA) were purchased from Sigma-Aldrich (St Louis, MO, USA). Lithocholic acid (LCA), deoxycholic acid (DCA), and N-ethoxycarbonyl-2-ethoxy-1,2-dihydroquinoline (EEDQ) were purchased from Aladdin (Shanghai, China). Other chemicals and reagents were obtained from Sigma-Aldrich (St Louis, MO, USA).Synthesis of amino acid-conjugated bile acids
The chemical synthesis of the amino acid-conjugated bile acids was modified from a published protocol.16 Briefly, 3 mg mL−1 bile acid and 100 mM EEDQ were prepared in 10 μL CH3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) t-BuOH (3
t-BuOH (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Next, a solution of 125 mM amino acid and 100 mM NaOH was prepared in 10 μL water. For cysteine and tyrosine, which are less soluble in water, the volume of solvent was increased to 100 μL. The two solutions were mixed and shaken at 80 °C for 2 h. When the reaction was done, the pH of the resulting solution was adjusted to 7 with 100 mM HCl. It was then diluted 100 times with 50% MeOH and 1 μL was injected in to the LC-MS system for analysis.
1, v/v). Next, a solution of 125 mM amino acid and 100 mM NaOH was prepared in 10 μL water. For cysteine and tyrosine, which are less soluble in water, the volume of solvent was increased to 100 μL. The two solutions were mixed and shaken at 80 °C for 2 h. When the reaction was done, the pH of the resulting solution was adjusted to 7 with 100 mM HCl. It was then diluted 100 times with 50% MeOH and 1 μL was injected in to the LC-MS system for analysis.
MS/MS fragmentation investigation
LC-MS/MS acquisition was performed using a Vanquish UHPLC system coupled to a Q Exactive HF-X mass spectrometer with an ESI source (Thermo Fisher Scientific, Bremen, Germany). The synthesized bile acids were analyzed using flow injection with isocratic condition of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 mobile phase A (7.5 mM ammonium acetate in water, adjusted to pH 4.35 using acetic acid): mobile phase B (acetonitrile). The flow rate was 0.3 mL min−1. MS spectra were acquired using the following ESI source settings: spray voltage 3.5 kV (positive mode) or 2.5 kV (negative mode), aux gas heater temperature 380 °C, capillary temperature 320 °C, sheath gas flow rate 30 units, aux gas flow gas 10 units. MS1 scan parameters included a resolution of 60
60 mobile phase A (7.5 mM ammonium acetate in water, adjusted to pH 4.35 using acetic acid): mobile phase B (acetonitrile). The flow rate was 0.3 mL min−1. MS spectra were acquired using the following ESI source settings: spray voltage 3.5 kV (positive mode) or 2.5 kV (negative mode), aux gas heater temperature 380 °C, capillary temperature 320 °C, sheath gas flow rate 30 units, aux gas flow gas 10 units. MS1 scan parameters included a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, AGC target of 3e6, and maximum injection time of 200 ms. MS/MS spectra were acquired in negative ESI mode with a normalized collision energy of 60 and positive ESI mode with a normalized collision energy of 45. Data-dependent MS2 (dd-MS2) acquisition employed a resolution of 30
000, AGC target of 3e6, and maximum injection time of 200 ms. MS/MS spectra were acquired in negative ESI mode with a normalized collision energy of 60 and positive ESI mode with a normalized collision energy of 45. Data-dependent MS2 (dd-MS2) acquisition employed a resolution of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, AGC target of 2e5, and maximum injection time of 100 ms.
000, AGC target of 2e5, and maximum injection time of 100 ms.
Raw data were converted to mgf format using ProteoWizard MSConvert17 software. MS/MS fragmentation patterns were then manually investigated with NIST MS Search software. For each amino acid conjugation, the common characteristic product ions from conjugated LCA, DCA, and CA were selected. The formulas of these fragments were annotated according to their m/z and chemical structures. Their theoretical m/z were then calculated using Molecular Weight Calculator (Tables S1 and S2†). For the negative mode data, the average relative intensities were also calculated and rounded to the nearest 5% for the semi-empirical library development (Table S1†).
Semi-empirical library and software development
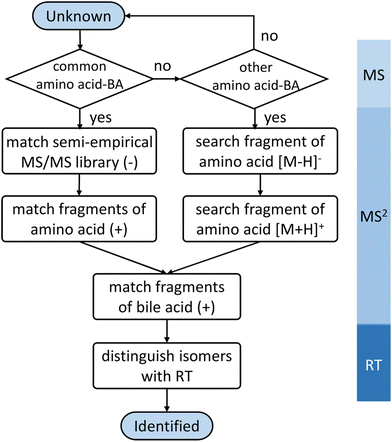
A semi-empirical MS/MS library in the negative mode was developed for the common amino acid-conjugated bile acids. Here, 18 common amino acids (cysteine excluded) were in silico coupled with 10 bile acid skeletons, including bile acids with one to four hydroxyl groups (1OH-BA, 2OH-BA, 3OH-BA, 4OH-BA) and oxidized forms (1O-BA, 1O1OH-BA, 2O-BA, 1O2OH-BA, 2O1OH-BA, 3O-BA). Their accurate masses and [M − H]− precursor m/z were calculated. An in-house Java (v 15.0.1) program was developed to combine the theoretical m/z of the precursors and characteristic product ions and export the semi-empirical MS/MS spectra in MSP format.BAFinder is a software tool for bile acid annotation from LC-MS/MS data.15 The previously released version 1.0 covered free bile acids and common conjugations, such as glycine, taurine, sulfate, and glucuronide. The software was updated to version 2.0 with an expanded scope for amino acid-conjugated bile acids other than glycine and taurine. The overall workflow is shown in Fig. 3. First, the unknown MS/MS spectra in negative mode were searched against the semi-empirical library, and a normalized dot product was calculated using eqn (1):
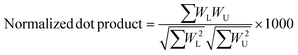 | (1) |
For other amino acid conjugations not included in the semi-empirical library, a new feature was added to BAFinder 2.0 software to allow the annotation of user-defined conjugations. The annotation of novel conjugations was based on the observation that most amino acid-conjugated bile acids generated fragments of amino acid [M − H]− and [M + H]+ in negative and positive mode, respectively. Therefore, matching of the calculated precursor m/z, presence of amino acid [M − H]− product ion in negative mode, as well as amino acid [M + H]+ and bile acids fragments in positive mode were used as the criteria for screening.
Cross-validation of the semi-empirical library using the synthesized compounds
The synthesized amino acid-conjugated bile acids were annotated using a semi-empirical library developed from other bile acid classes to test the methodology. First, the experimental spectra of the 18 amino acid-conjugated DCA and CA in the negative mode were used to develop a semi-empirical library. The experimental spectra of the amino acid-conjugated LCA were then searched against this library using NIST MSPepSearch software. The mass tolerances were 0.005 Da for the precursor and 0.01 Da for the product ions. This process was repeated for annotation of the amino acid-conjugated DCA and CA. The top hits and their dot-product scores were reported (Table S3†).Application to the fecal analysis of human, dog, and rat samples
Fecal samples from healthy human subjects were obtained from the Affiliated Jiangyin Hospital of Nanjing University of Chinese Medicine, Jiangyin, China. Feces of beagle dogs and SD rats were purchased from iPhase Biosciences, Beijing, China. Fecal samples were extracted using a published method with slight modifications.18 Briefly, 1 mL 5% ammonia in ethanol was added to 100 mg wet feces. The mixture was shaken for 15 min at 4 °C, sonicated for 15 min, and then centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 15 min. The supernatant was transferred to a new tube, dried by SpeedVac, and stored at −20 °C. On the day of the experiment, the fecal extracts were resuspended with 100 μL 50% methanol, centrifuged, and filtered through a 0.45 μm filter.
000g at 4 °C for 15 min. The supernatant was transferred to a new tube, dried by SpeedVac, and stored at −20 °C. On the day of the experiment, the fecal extracts were resuspended with 100 μL 50% methanol, centrifuged, and filtered through a 0.45 μm filter.
LC-MS/MS analysis of the fecal samples was performed with the same instrumental setup and mobile phases as above. An Acquity BEH C18 column (2.1 mm × 100 mm, 1.7 μm) (Waters, Milford, MA, USA) was used for the separation.19,20 The column temperature was 45 °C and flow rate was 0.45 mL min−1. The gradient program was 0.0–0.5 min (5% B), 0.5–1.0 min (5%–20% B), 1.0–2.0 min (20%–25% B), 2.0–5.5 min (25% B), 5.5–6.0 min (25%–30% B), 6.0–8.0 min (30% B), 8.0–9.0 min (30%–35% B), 9.0–17.0 min (35%–65% B), 17.0–18.0 min (65%–100% B), 18.0–19.0 min (100% B), 19.0–19.5 (100%–5% B) and 19.5–24.0 min (5% B). The injection volume was 5 μL.
Raw data were converted to mzML format using ProteoWizard MSConvert17 and then processed by XCMS.21 MS/MS spectra were converted to mgf files using RawConverter.22 The resulting peak table and alignment files, together with the mgf files, were imported into BAFinder 2.0 software. The m/z and RT tolerance was set to 0.005 Da and 0.05 min, respectively. The semi-empirical library of amino acid-conjugated bile acids in negative mode was added to the existing experimental library of bile acids. Spectral match hits with dot-product scores higher than 500 were reported. If the corresponding spectra in positive mode matched the characteristic fragments of amino acid and bile acid, a higher confidence level would be assigned to the annotation result.
To explore novel conjugates of bile acids, small molecules containing amino and carboxyl groups were extracted from the HMDB database23 using InstantJchem, ChemAxon (https://www.chemaxon.com). The following filters were applied to the unique formulas: (1) containing only C, H, O, N, P, S elements; (2) molecular weight less than 500 Da. Glycine, taurine, and other common amino acids in the semi-empirical library were excluded. Accurate masses of these metabolites were imported into BAFinder 2.0, and bile acids conjugated with these molecules were screened in the fecal dataset.
To validate the annotation result and distinguish isomers, some candidate structures were synthesized, and their retention times were compared with the features in biological samples. Specifically, to determine the isomer position of bile acids conjugated with C8H17N3O3, Boc-Lys-OH was reacted with glycocholic acid (GCA) and glycochenodeoxycholic acid (GCDCA) using the same method mentioned previously. To the dried conjugation products, 30 μL dichloromethane and 10 μL TFA pre-cooled at 0 °C were added to remove the Boc protecting groups. After the reaction was done (∼45 min), the mixture was evaporated and 200 μL saturated aqueous NaHCO3 solution was added. Synthesized Lys(iso)-GCA and Lys(iso)-GCDCA were loaded on Sep-Pak C18 cartridges (1 cc, 100 mg) and eluted with methanol for clean-up.
Results and discussion
Development of the semi-empirical MS/MS library in negative ESI mode
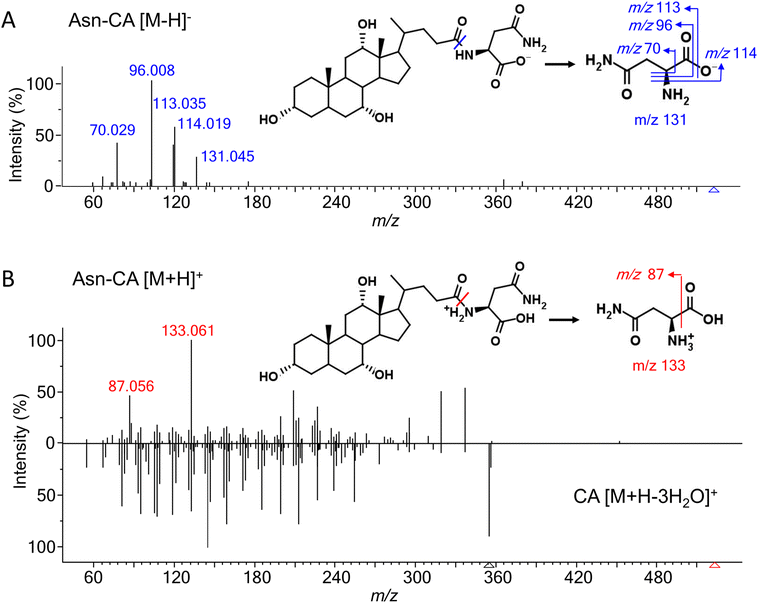
Most of the amino acids (except for glycine and taurine)-conjugated bile acids were not commercially available. To investigate their fragmentation patterns, 19 common amino acid (alanine, arginine, asparagine, aspartate, cysteine, glutamine, glutamate, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine)-conjugated lithocholic acid (LCA), deoxycholic acid (DCA), and cholic acid (CA) were synthesized and their experimental MS/MS spectra were acquired under negative and positive ESI modes. Glycine- and taurine-conjugated bile acids have been well-studied and thus were not discussed here. [M − H]− and [M + H]+ were the most abundant adducts in the negative and positive mode, respectively. [M + H–H2O]+ and other adducts with further water losses were also detected. For all the amino acids except arginine, histamine, and lysine, the MS intensity of [M − H]− were higher than for [M + H]+ (Fig. S1†). According to our previous experience with other bile acids,15 a normalized collision energy of 60 was applied in negative mode to enhance the characteristic product ions while minimizing general neutral losses, such as H2O and HCOOH. Table 1 shows the common product ions found in the amino acid-conjugated LCA, DCA, and CA. For alanine, leucine, isoleucine, lysine, methionine, and proline, the only product ions were [M − H]− of the amino acids themselves. Leucine and isoleucine conjugates could not be distinguished by their MS/MS spectra. For other amino acids, further fragmentations were observed, due to neutral losses such as NH3, CO2, NH3 + CO2, and H2O. Fig. 2A shows the MS/MS spectrum and proposed fragmentation pathway for a representative compound, asparagine-conjugated CA. Detailed information for the other conjugates is presented in Table S1.† Overall, these amino acid-conjugated bile acids showed similar fragmentation patterns to glycine and taurine conjugates, in which the characteristic amino acids product ions were dominant regardless of the bile acid species.24 The only exception was cysteine, whose fragmentation patterns were different when conjugating with LCA, DCA, and CA (Fig. S2†), indicating a bile-acid-dependent fragmentation process. Therefore, cysteine conjugates were excluded from the following semi-empirical library development.| Conjugate | Fragment ions | ||||||
|---|---|---|---|---|---|---|---|
| AA | AA-NH3 | AA-CO2 | AA-(NH3 + CO2) | AA-H2O | AA-C3H5NO2 | Others | |
| Ala | 88 | ||||||
| Leu/Ile | 130 | ||||||
| Lys | 145 | ||||||
| Met | 148 | ||||||
| Pro | 114 | ||||||
| Val | 116 | ||||||
| Arg | 131(AA-CH2N2) | ||||||
| Thr | 74(AA-C2H4O) | ||||||
| Ser | 104 | 74(AA-CH2O) | |||||
| Phe | 164 | 147 | 72(AA-C7H8) | ||||
| Asp | 115 | 88 | 71 | ||||
| Glu | 146 | 102 | 128 | 84(AA-CH2O3) | |||
| His | 154 | 137 | 110 | 93 | |||
| Asn | 131 | 114 | 70 | 113 | 96(AA-H2O + NH3) | ||
| Gln | 145 | 128 | 101 | 84 | 127 | 109(AA-H4O2) | |
| Trp | 203 | 159 | 142 | 116 | 74(AA-C9H7N), 72(AA-C9H9N) | ||
| Tyr | 180 | 163 | 119 | 93 | 107(AA-C2H3NO2), 74(AA-C7H6O), 72(AA-C7H8O) | ||
To expand the structural space of the experimental MS/MS library, a semi-empirical library was developed. Bile acids with various numbers of hydroxyl groups (1OH-BA, 2OH-BA, 3OH-BA, 4OH-BA) and oxidized forms (1O-BA, 1O1OH-BA, 2O-BA, 1O2OH-BA, 2O1OH-BA, 3O-BA) were combined with 18 amino acids to generate 180 conjugated structures. Here, positional and stereoisomers of the bile acids were not considered since they cannot be distinguished with current MS/MS information. The theoretical m/z of the [M − H]− precursors were combined with their corresponding characteristic product ions to generate a series of semi-empirical MS/MS spectra (Fig. S3†). The resulting library was exported in MSP format, which could be used as itself (e.g., with BAFinder15 or MS-DIAL25), or converted to other library formats, such as NIST MS library through lib2nist software.
To evaluate the performance of the semi-empirical library generated in this way, cross-validation was performed using the synthesized compounds. Specifically, a semi-empirical library was created from the experimental spectra of amino acid-conjugated DCA and CA, and the experimental spectra of amino acid-conjugated LCA were searched against this library. The same process was repeated to annotate the amino acid-conjugated DCA and CA. As shown in Table S3,† all the synthesized compounds were correctly identified using the semi-empirical library built with the other two bile acid classes, with an average dot-product score of 834.
Characteristic features in positive ESI mode and the workflow for compound annotation
In positive mode, a normalized collision energy of 45 yielded fragments from both amino acid and bile acid moieties. Fig. 2B shows the MS/MS spectrum of asparagine-conjugated CA in positive mode, compared with the spectrum of CA. The fragmentation patterns of the other amino acid conjugates were similar (Fig. S4†). Most fragments in the conjugated bile acids were also found in the free bile acids, indicating that these fragments originated from the bile acid part. The fragmentation pathway of selected bile acids was investigated in the atmospheric pressure chemical ionization (APCI) positive mode,26 yet it was challenging to predict the spectra for other bile acids due to the complex fragments that were not fully understood. Apart from these ions, characteristic product ions from amino acids were observed, including amino acid [M + H]+, [M + H–H2O–CO]+, and [M + H–NH3]+ (Table S2†), similar to the fragments of the individual amino acids.27To improve the confidence of the compound identification, an automatic workflow was developed using data from both the negative and positive mode (Fig. 3). Unknown features were first filtered and grouped according to the calculated precursor m/z of the amino acid-conjugated bile acids. After that, the MS/MS spectra in the negative mode were searched against the semi-empirical library, and hits with dot-product scores higher than 500 were reported. In addition, characteristic product ions of amino acids were searched in the MS/MS spectra in the positive mode (if available). If these amino acid-related fragments were found, the other fragments in the MS/MS spectra, which were supposed to come from bile acids, were searched against the experimental MS/MS library of free bile acids.15 A higher confidence level would be assigned to the annotation result if hits were found in both positive and negative mode. This workflow was integrated into a software tool named BAFinder 2.0, which was the updated version of BAFinder 1.0 software for common bile acid annotation.15
Annotation of the unknown amino acid-conjugated bile acids in the fecal samples
As an application example, fecal samples from human, beagle dogs, and SD rats were analyzed using LC-MS/MS in negative and positive mode. Unknown amino acid-conjugated bile acids were annotated using BAFinder 2.0 software. In total, 171 amino acid-conjugated bile acids were annotated in at least one type of sample (Table S4†). The annotation result included amino acid type and bile acid class. For example, Ala-2OH-BA meant an alanine (or its isomer)-conjugated bile acid with two hydroxyl groups. Further distinguishing the isomers required retention time information. Potential candidates for the annotated bile acids were synthesized through a single-step reaction, and 105 out of the 171 annotations were identified using the retention times (Table S4†). The number of validated annotations was mainly limited by the availability of bile acid reference standards to perform the synthesis. During the retention time comparison, uncommon amino acid conjugates, including β-alanine and D-glutamic acid, were discovered as isomers of alanine and L-glutamic acid, respectively. The experimental data of the synthesized compounds demonstrated that these isomers had similar fragmentation patterns but different retention times. D-Glutamic acid, along with other D-amino acids, could be produced by the gut microbiota,28 therefore it was not surprising that D-amino acids were involved in the microbial conjugation of bile acids.Fig. 4A shows the distribution of amino acid conjugates in the feces of the different species. In terms of the number of bile acids annotated, the top 5 most frequently observed conjugates included β-alanine/alanine, leucine/isoleucine, lysine, phenylalanine, and glutamic acid. Actually, more β-alanine-conjugated bile acids were identified compared to alanine. The proline conjugate was not detected in this study, probably due to the wider peak shape and the resulting lower peak height compared to other conjugates with the current LC method. The bile acids conjugated with amino acids also varied with different species (Fig. 4B). Bile acids with two hydroxyl groups (2OH-BA, e.g., DCA) contributed the most in the human fecal samples, while 3OH-BA (e.g., CA and β-MCA) was the major component in the rat feces. These patterns were consistent with the bile acids profiles of the two species.29 Compared to the other two, the dog feces contained more amino acid-conjugated oxidized bile acids, including 1O1OH-BA and 1O2OH-BA.
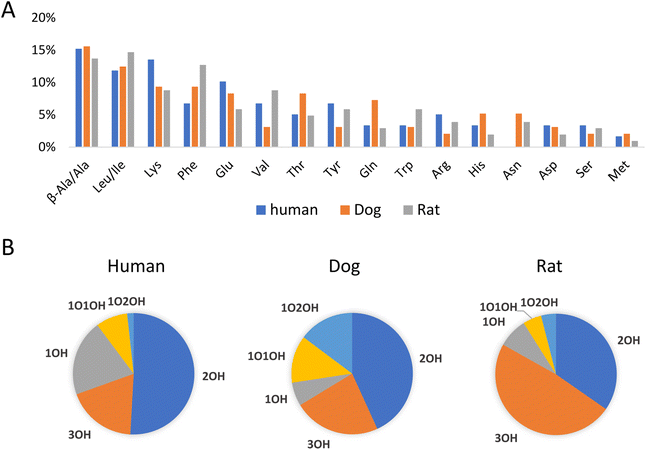 | ||
| Fig. 4 Distribution of (A) amino acids and (B) bile acids in amino acid-conjugated bile acids in human, dog, and rat feces. Statistics were based on the numbers of bile acids annotated. | ||
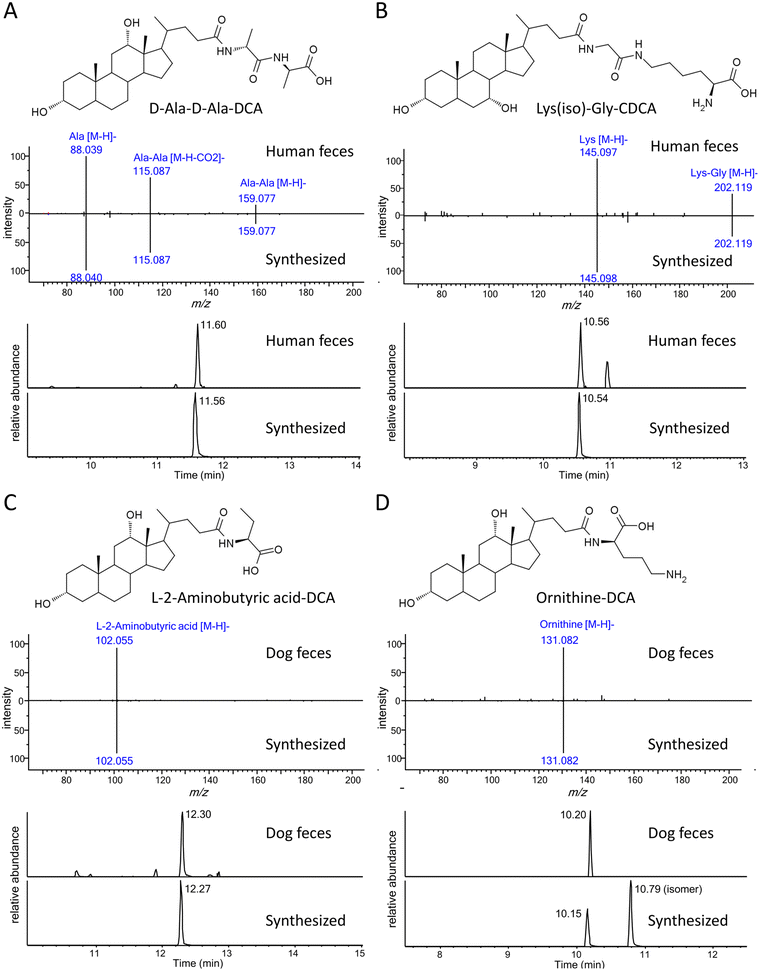
So far, the current approach was limited to only 18 common amino acid-conjugated bile acids. To discover potential new bile acid conjugates, we assumed that other broadly defined amino acids (i.e., small molecules with amino and carboxyl groups) might also conjugate with bile acids and their MS/MS fragmentation patterns would be similar to the common amino acid conjugates. Based on the above assumptions, a new feature for unknown conjugated bile acid annotation was added to BAFinder 2.0 software. When a list of customized amino acid masses was imported, BAFinder 2.0 in silico conjugated them with 10 general bile acid skeletons and calculated their theoretical precursor m/z in negative and positive mode. The unknown MS/MS spectra passing the precursor filter were screened with the characteristic product ions of [M − H]− and [M + H]+ of the amino acids in the negative and positive mode, respectively. In addition, the positive mode spectra were searched against the library of free bile acids, in a similar way as for the common amino acid conjugates. To showcase the usefulness of this feature, 7326 structures with both amino and carboxyl groups were extracted from the HMDB database,23 and 855 unique formulas were obtained with only common organic elements (C, H, O, N, P, S) and a molecular weight smaller than 500 Da. Their exact masses were imported into BAFinder 2.0, and the corresponding conjugated bile acids were searched in the fecal dataset. Two new conjugates, C6H12N2O3 and C8H17N3O3, were found in the human/rat fecal samples to be conjugated with 1OH, 2OH, and 3OH-BA. Examination of their MS/MS spectra (Fig. 5A and B) revealed fragment ions of alanine (m/z 88) and lysine (m/z 145), respectively. Therefore, they were tentatively annotated as Ala-Ala and Gly-Lys (or Lys-Gly). After comparison with the retention times of the synthesized compounds, they were finally identified as D-Ala-D-Ala-LCA, D-Ala-D-Ala-DCA, D-Ala-D-Ala-HDCA, Lys(iso)-Gly-CDCA, and Lys(iso)-Gly-CA (Table 2). Here, “iso” indicates that lysine was connected to glyco-BA via an isopeptide bond, which eluted later than its isomer with a common peptide bond. These dipeptide-conjugated bile acids were reported for the first time in biological samples. Two other novel conjugates, L-2-aminobutyric acid and ornithine, were annotated in similar ways (Fig. 5C and D). Similar to lysine, ornithine contained two amino groups and thus could generate two conjugate isomers. The one with earlier retention times was detected in dog and rat feces. The structure was tentatively identified as the peptide bond form based on the experience of lysine conjugates.
| Conjugate | Bile acid | [M − H]− | RT (min) | Human | Dog | Rat |
|---|---|---|---|---|---|---|
| D-Ala-D-Ala | LCA | 517.365 | 13.39 | ✓ | ||
| D-Ala-D-Ala | DCA | 533.360 | 11.60 | ✓ | ||
| D-Ala-D-Ala | HDCA | 533.361 | 9.16 | ✓ | ||
| Lys(iso)-Gly | CDCA | 576.402 | 10.56 | ✓ | ||
| Lys(iso)-Gly | CA | 592.397 | 8.13 | ✓ | ||
| L-2-Aminobutyric acid | 12o-DCA | 474.323 | 11.00 | ✓ | ||
| L-2-Aminobutyric acid | DCA | 476.338 | 12.30 | ✓ | ✓ | |
| L-2-Aminobutyric acid | ω-MCA | 492.333 | 7.02 | ✓ | ||
| L-2-Aminobutyric acid | CA | 492.333 | 10.10 | ✓ | ||
| Ornithine | DCA | 505.366 | 10.20 | ✓ | ||
| Ornithine | CA | 521.360 | 7.20 | ✓ | ✓ | |
| Ornithine | α-MCA | 521.360 | 3.74 | ✓ | ||
| Ornithine | β-MCA | 521.361 | 3.66 | ✓ | ||
| Ornithine | ω-MCA | 521.362 | 3.52 | ✓ |
The sources and functions of these novel bile acid conjugates remain to be discovered. A microbial origin was very likely by analogy with the common amino acid conjugates. A previous study found 27 species of gut bacteria, most in the Bifidobacteriaceae, Lachnospiraceae, and Bacteroidaceae families, were capable of conjugating one or more of 16 common amino acids to CDCA, DCA, or CA.30 Similar screening experiments could find the species responsible for the production of the new metabolites discovered in this study. Conjugations of amino acids such as D-Ala, Lys, and Gly increased the hydrophilicities of bile acids, while dipeptide conjugates (D-Ala-D-Ala and Lys-Gly) were even more hydrophilic, according to their retention times. This may then affect their emulsifying, signaling, and antimicrobial properties.3 Some amino acid-conjugated bile acids, namely Phe-CA, Tyr-CA, and Leu-CA, were found at higher levels in patients with inflammatory bowel disease and cystic fibrosis.7 Further investigations are required to reveal the associations between these conjugated bile acids and health or the disease state.
Conclusions
To annotate unknown amino acid-conjugated bile acids, a semi-empirical MS/MS library of 180 common amino acid-conjugated bile acids were developed in negative ESI mode. This library was combined with characteristic fragments in positive ESI mode to further increase the confidence of annotation. A software named BAFinder 2.0 was provided to perform the annotation workflow automatically. As a proof of concept, fecal extracts of human, beagle dog, and rat were analyzed, and 171 amino acid-conjugated bile acids were annotated. Among them, 105 were validated with the retention times of synthesized compounds. Most common amino acids, except for proline and cysteine, and two uncommon amino acids (β-alanine and D-glutamic acid) were detected as bile acid conjugates. Furthermore, four novel conjugates of bile acids, D-Ala-D-Ala, Lys(iso)-Gly, L-2-aminobutyric acid, and ornithine, were discovered, and confirmed with synthesized compounds. This new tool facilitates the analysis of microbially conjugated bile acids, which may promote further studies on their biological roles and effects on the host. The semi-empirical library and BAFinder 2.0 software are freely available at https://github.com/BAFinder/bafinder.github.io/tree/BAFinder-2.0.Author contributions
Yan Ma: conceptualization, methodology, software, validation, writing – original draft, writing – review & editing. Yang Cao: investigation, data curation. Xiaocui Song: investigation, data curation. Weichen Xu: resources. Zichen Luo: resources. Jinjun Shan: resources. Jingjie Zhou: resources.Ethics statement
All procedures were approved by the medical ethics committee of the Affiliated Jiangyin Hospital of Nanjing University of Chinese Medicine and followed the tenets of the Declaration of Helsinki (SR202201701).Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by the National Key Research and Development Program of China (grant no. 2020YFF01014505). The authors would acknowledge Qingcui Wu and Dr Xiangbing Qi for their helpful suggestions on the synthesis of amino acid-conjugated bile acids.References
- S. L. Collins, J. G. Stine, J. E. Bisanz, C. D. Okafor and A. D. Patterson, Nat. Rev. Microbiol., 2023, 21, 236–247 CrossRef CAS.
- H. Dai, J. Han, T. Wang, W.-B. Yin, Y. Chen and H. Liu, Nat. Prod. Rep., 2023, 40, 1078–1093 RSC.
- D. V. Guzior and R. A. Quinn, Microbiome, 2021, 9, 1–13 CrossRef.
- Y. Kiriyama and H. Nochi, Microorganisms, 2021, 10, 68 CrossRef PubMed.
- A. B. Larabi, H. L. Masson and A. J. Bäumler, Gut Microbes, 2023, 15, 2172671 CrossRef PubMed.
- C. N. Falany, M. R. Johnson, S. Barnes and R. B. Diasio, J. Biol. Chem., 1994, 269, 19375–19379 CrossRef CAS PubMed.
- R. A. Quinn, A. V. Melnik, A. Vrbanac, T. Fu, K. A. Patras, M. P. Christy, Z. Bodai, P. Belda-Ferre, A. Tripathi and L. K. Chung, Nature, 2020, 579, 123–129 CrossRef CAS.
- L. Lucas, K. Barrett, R. Kerby, Q. Zhang, L. Cattaneo, D. Stevenson, F. Rey and D. Amador-Noguez, Msystems, 2021, 6, e00805–e00821 CrossRef CAS PubMed.
- C. J. Garcia, V. Kosek, D. Beltrán, F. A. Tomás-Barberán and J. Hajslova, Biomolecules, 2022, 12, 687 CrossRef CAS PubMed.
- M. A. Hoffmann, L.-F. Nothias, M. Ludwig, M. Fleischauer, E. C. Gentry, M. Witting, P. C. Dorrestein, K. Dührkop and S. Böcker, Nat. Biotechnol., 2022, 40, 411–421 CrossRef CAS PubMed.
- Q.-F. Zhu, Y.-Z. Wang, N. An, J.-D. Hao, P.-C. Mei, Y.-L. Bai, Y.-N. Hu, P.-R. Bai and Y.-Q. Feng, Anal. Chem., 2022, 94, 2655–2664 CrossRef CAS PubMed.
- Y.-Z. Wang, P.-C. Mei, P.-R. Bai, N. An, J.-G. He, J. Wang, Q.-F. Zhu and Y.-Q. Feng, Anal. Chim. Acta, 2023, 1239, 340691 CrossRef CAS PubMed.
- T. Kind, K.-H. Liu, D. Y. Lee, B. DeFelice, J. K. Meissen and O. Fiehn, Nat. Methods, 2013, 10, 755–758 CrossRef CAS PubMed.
- U. Keshet, T. Kind, X. Lu, S. Devi and O. Fiehn, Anal. Chem., 2022, 94, 2732–2739 CrossRef CAS PubMed.
- Y. Ma, Y. Cao, X. Song, Y. Zhang, J. Li, Y. Wang, X. Wu and X. Qi, Anal. Chem., 2022, 94, 6242–6250 CrossRef CAS PubMed.
- F. Venturoni, A. Gioiello, R. Sardella, B. Natalini and R. Pellicciari, Org. Biomol. Chem., 2012, 10, 4109–4115 RSC.
- M. C. Chambers, B. Maclean, R. Burke, D. Amodei, D. L. Ruderman, S. Neumann, L. Gatto, B. Fischer, B. Pratt and J. Egertson, Nat. Biotechnol., 2012, 30, 918–920 CrossRef CAS.
- X. Zhang, X. Liu, J. Yang, F. Ren and Y. Li, Metabolites, 2022, 12, 633 CrossRef CAS.
- S. Yin, M. Su, G. Xie, X. Li, R. Wei, C. Liu, K. Lan and W. Jia, Anal. Bioanal. Chem., 2017, 409, 5533–5545 CrossRef CAS.
- Y. Alnouti, I. L. Csanaky and C. D. Klaassen, J. Chromatogr. B: Biomed. Sci. Appl., 2008, 873, 209–217 CrossRef CAS.
- C. A. Smith, E. J. Want, G. O'Maille, R. Abagyan and G. Siuzdak, Anal. Chem., 2006, 78, 779–787 CrossRef CAS PubMed.
- L. He, J. Diedrich, Y.-Y. Chu and J. R. Yates III, Anal. Chem., 2015, 87, 11361–11367 CrossRef CAS PubMed.
- D. S. Wishart, A. Guo, E. Oler, F. Wang, A. Anjum, H. Peters, R. Dizon, Z. Sayeeda, S. Tian and B. L. Lee, Nucleic Acids Res., 2022, 50, D622–D631 CrossRef CAS PubMed.
- M. Maekawa, M. Shimada, T. Iida, J. Goto and N. Mano, Steroids, 2014, 80, 80–91 CrossRef CAS PubMed.
- H. Tsugawa, T. Cajka, T. Kind, Y. Ma, B. Higgins, K. Ikeda, M. Kanazawa, J. VanderGheynst, O. Fiehn and M. Arita, Nat. Methods, 2015, 12, 523–526 CrossRef CAS PubMed.
- X. Qiao, M. Ye, C.-f. Liu, W.-z. Yang, W.-j. Miao, J. Dong and D.-a. Guo, Steroids, 2012, 77, 204–211 CrossRef CAS PubMed.
- P. Zhang, W. Chan, I. L. Ang, R. Wei, M. M. Lam, K. M. Lei and T. C. Poon, Sci. Rep., 2019, 9, 1–10 CrossRef PubMed.
- M. Matsumoto, A. Kunisawa, T. Hattori, S. Kawana, Y. Kitada, H. Tamada, S. Kawano, Y. Hayakawa, J. Iida and E. Fukusaki, Sci. Rep., 2018, 8, 17915 CrossRef CAS PubMed.
- R. Thakare, J. A. Alamoudi, N. Gautam, A. D. Rodrigues and Y. Alnouti, J. Appl. Toxicol., 2018, 38, 1323–1335 CrossRef CAS PubMed.
- L. Lucas, K. Barrett, R. Kerby, Q. Zhang, L. Cattaneo, D. Stevenson, F. Rey and D. Amador-Noguez, MSystems, 2021, 6, 00805–00821, DOI:10.1128/msystems.
Footnote |
| † Electronic supplementary information (ESI) available: Characteristic product ions of amino acid-conjugated bile acids in negative and positive ESI mode (Tables S1 and 2), identification of synthesized compounds using the semi-empirical library (Table S3), amino acid-conjugated bile acids detected in human, dog and rat fecal samples (Table S4), comparison of MS intensities of amino acid-conjugated LCA, DCA, and CA in the negative and positive mode (Fig. S1), MS/MS spectra of cysteine-conjugated LCA, DCA, and CA in negative mode (Fig. S2), workflow of library development (Fig. S3), MS/MS spectra of amino acid-conjugated CA in the positive mode (Fig. S4). See DOI: https://doi.org/10.1039/d3an01237a |
| This journal is © The Royal Society of Chemistry 2023 |

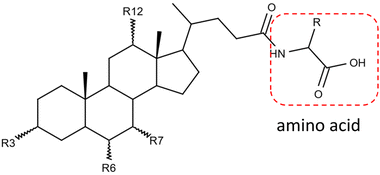
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. R represents the side chain specific to each amino acid. Conjugates are actually not limited to the α-amino acids.
O. R represents the side chain specific to each amino acid. Conjugates are actually not limited to the α-amino acids.