Open Access Article
Open Access ArticleNovel TCNQ-stacking motifs in (12-crown-4)-complexes of alkali metal TCNQ salts†
Bingjia
Yan‡
 ,
Peter N.
Horton
,
Peter N.
Horton
 ,
Simon C.
Weston§
,
Simon C.
Weston§
 ,
Andrea E.
Russell
,
Andrea E.
Russell
 and
Martin C.
Grossel
and
Martin C.
Grossel
 *
*
School of Chemistry, University of Southampton, Highfield, Southampton, SO17 1BJ, UK. E-mail: M.C.Grossel@soton.ac.uk; Fax: +44 (0)23 8059 3781; Tel: +44 (0)23 8059 3153
First published on 31st August 2021
Abstract
An investigation of the solid-state behaviour of five 12-crown-4 alkali metal TCNQ complexes, (12-crown-4)2LiTCNQ (1), (12-crown-4)2NaTCNQ (2), (12-crown-4)2Li(TCNQ)2 (3), (12-crown-4)2Na(TCNQ)2 (4), and (12-crown-4)2K(TCNQ)2 (5), reveals an unusual “cross-stitch” packing motif with the extended face-to-face π-stacked TCNQ˙− columns present in complexes 1 and 2. The effect of the presence of additional neutral TCNQ0 has also been explored.
The ability of 7,7′,8,8′-tetracyanoquinodimethane (TCNQ) to act as a good one-electron acceptor results in charge-transfer complexes,1–7 which can exhibit interesting solid-state electrical and magnetic properties,8,9 arising from the presence of unpaired electrons.10–16 The TCNQ˙− moieties have a tendency to form face-to-face π-stacked dimers, which can assemble into infinite columns within a crystal.17–19 Furthermore, neutral TCNQ0 can be incorporated into these anion stacks resulting in a considerable change in the solid-state electronic properties of such materials.20 The combination of the potential packing arrangements of the TCNQ units and the size and nature of the counter-cation are the dominant factors controlling the solid-state architectures of such complexes.21
We have previously reported detailed studies of the solid-state behaviour of a range of ionophore-complexed alkali metal–TCNQ˙− salts including 15-crown-5 and 18-crown-6 complexes of Li+, Na+, K+, Tl+ and Rb+TCNQ˙−1,17,19,22–25 and 15-crown-5 complexes of Li+ and Na+ (TCNQ˙−)(TCNQ0).25 Recently, Akutagawa and co-workers26,27 have reported the crystal structure of Li+(12-crown-4)2(TCNQ)2 and Na+(12-crown-4)2(TCNQ)2, but there is limited information to the solid-state behaviour of other 12-crown-4 alkali metal TCNQ complexes.
In this paper, we report the synthesis and crystallographic studies of five 12-crown-4 alkali metal TCNQ complexes, two simple TCNQ salts: (12-crown-4)2LiTCNQ (1), and (12-crown-4)2NaTCNQ (2); and three related complex TCNQ salts: (12-crown-4)2Li(TCNQ)2 (3), (12-crown-4)2Na(TCNQ)2 (4), and (12-crown-4)2K(TCNQ)2 (5) (see Scheme 1).
Experimental procedure
General consideration
All experiments were conducted under a nitrogen atmosphere unless stated otherwise. Acetonitrile was dried over calcium hydride and distilled before use. Diethyl ether was dried over sodium wire and was freshly decanted before use. All chemicals were commercially available and used as received, unless otherwise stated.Physical measurements
Melting points were measured on an electrothermal melting point apparatus and are uncorrected. Elemental analyses were performed by Medac Ltd., Chobham Business Centre, Chertsey Road, Chobham, Surry, GU24 8JB. IR (KBr discs) spectra were obtained using a Golden Gate sampling attachment on a Mattson Satellite 3000 FTIR. Raman spectra were collected using a Renishaw In-Via system with a high powered near Infrared (HPNIR) 785 nm laser and microscope using a 50× objective. ESI mass spectra were obtained using a solariX (Bruker Daltonics, Bremen, Germany) mass spectrometer equipped with a 4.7 T magnet and FT-ICR cell.Single crystal X-ray diffraction data collection and processing
A suitable crystal was selected and measured following a standard method28 for each compound. For 1 on a Rigaku 007HF diffractometer equipped with Varimax confocal mirrors and an AFC11 goniometer and an enhanced sensitivity (HG) Saturn 944+ detector at 100 K. For 2 on a Rigaku SPIDER RAPID diffractomer at 120 K with an image plate detector. For 3, 4, and 5 on a Rigaku AFC12 goniometer equipped with HG Saturn724+ detector mounted at the window of a FR-E+ SuperBright molybdenum rotating anode generator with either VHF Varimax optics (70 μm focus) (3 and 4) or HF Varimax optics (100 μm focus) (5) at 100 K. Cell determination and data collection were carried out using CrystalClear.29 Data reduction, cell refinement, and absorption correction were carried out using either CrystalClear29 (2) or CrysAlisPro (1, 3, 4, 5).30 Structures were solved with Olex231 using the ShelXT32 and refined using ShelXL.33General procedure for preparation of simple TCNQ salts (1) and (2)
A solution of the alkali metal TCNQ salt (1 mol equiv.) and 12-crown-4 (2 mol equiv.) in anhydrous acetonitrile (50 ml) was boiled for 5 minutes, and filtered whilst hot. The mixture was then left to cool. The solvent was allowed to evaporate slowly over a period of several days during which a crystalline solid formed which was separated by filtration. This solid was washed with dry diethyl ether (50 ml), and was then dried in vacuum to give the resulting small crystals.Preparation of (12-crown-4)2LiTCNQ (1)
Reaction of LiTCNQ (0.424 g, 2 mmol) and 12-crown-4 (0.700 g, 4 mmol) achieved small crystals of 1 as black chunks (0.437 g, 78%). MS (solution) (MeCN) (ESI−) m/z: 204.1 (TCNQ˙−). (ESI+) m/z: 375.2 (2 × crown + Na+), 199.2 (crown + Na+). IR vmax/cm−1 2968, 2915, 2875 (saturated![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) stretch), 2173, 2151 (
stretch), 2173, 2151 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1589 (
stretch), 1589 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (CN)2 stretch), 1505 (
(CN)2 stretch), 1505 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1361 (
ring stretch), 1361 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bend), 1177 (
bend), 1177 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N and
N and ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 986 (C–C ring stretch), 819, 718 (
ring stretch), 986 (C–C ring stretch), 819, 718 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) out of plane bend). Raman vmax/cm−1 2214 (
out of plane bend). Raman vmax/cm−1 2214 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1604 (
stretch), 1604 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1384 (
ring stretch), 1384 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N stretch), 1208 (C
N stretch), 1208 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bending). M.p. 234 °C (dec.). Elemental analysis: calculated: C: 59.68%, H: 6.44%, N: 9.94%. Found: C: 59.80%, H: 6.15%, N: 10.36%.
bending). M.p. 234 °C (dec.). Elemental analysis: calculated: C: 59.68%, H: 6.44%, N: 9.94%. Found: C: 59.80%, H: 6.15%, N: 10.36%.
Preparation of (12-crown-4)2NaTCNQ (2)
Reaction of NaTCNQ (0.454 g, 2 mmol) and 12-crown-4 (0.700 g, 4 mmol) afforded 2 as small shiny black needles (0.269 g, 47%). MS (solution) (MeCN) (ESI−) m/z: 204.1 (TCNQ˙−). (ESI+) m/z: 375.2 (2 × crown + Na+), 199.2 (crown + Na+). IR vmax/cm−1 2962, 2916, 2875 (saturated![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) stretch), 2172, 2150 (
stretch), 2172, 2150 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch) (lit.34 2177, 2156), 1567 (
stretch) (lit.34 2177, 2156), 1567 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (CN)2 stretch), 1505 (
(CN)2 stretch), 1505 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1361 (
ring stretch), 1361 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bend), 1177 (
bend), 1177 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N and
N and ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 986 (C–C ring stretch), 820, 718 (
ring stretch), 986 (C–C ring stretch), 820, 718 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) out of plane bend). Raman vmax/cm−1 2257 (
out of plane bend). Raman vmax/cm−1 2257 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1609 (
stretch), 1609 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1389 (
ring stretch), 1389 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N stretch), 1206 (C
N stretch), 1206 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bending). M.p. 226 °C (dec.) (lit.34 185–187 °C). Elemental analysis: calculated: C: 58.03%, H: 6.22%, N: 9.67%. Found: C: 57.64%, H: 6.16%, N: 9.48%.
bending). M.p. 226 °C (dec.) (lit.34 185–187 °C). Elemental analysis: calculated: C: 58.03%, H: 6.22%, N: 9.67%. Found: C: 57.64%, H: 6.16%, N: 9.48%.
Preparation of (12-crown-4)2Li(TCNQ)2 (3)27
Reaction of LiTCNQ (0.0211 g, 0.1 mmol), neutral TCNQ0 (0.0204 g, 0.1 mmol) and 12-crown-4 (0.0352 g, 0.2 mmol) gave 3 as dark green crystalline solid (0.02122 g, 28%). MS (solution) (MeCN) (ESI−) m/z: 204.7 (TCNQ˙−). (ESI+) m/z: 375.6 (2 × crown + Na+), 199.4 (crown + Na+). IR vmax/cm−1 2970, 2940 (saturated![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) stretch), 2215, 2188 (
stretch), 2215, 2188 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1558 (
stretch), 1558 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (CN)2 stretch), 1507 (
(CN)2 stretch), 1507 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1327 (
ring stretch), 1327 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bend), 1134 (
bend), 1134 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N and
N and ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 953 (C–C ring stretch), 861, 694 (
ring stretch), 953 (C–C ring stretch), 861, 694 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) out of plane bend). Raman vmax/cm−1 2216 (
out of plane bend). Raman vmax/cm−1 2216 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1606 (
stretch), 1606 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1385 (
ring stretch), 1385 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N stretch), 1207 (C
N stretch), 1207 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bending). M.p. 312 °C (dec.).
bending). M.p. 312 °C (dec.).
Preparation of (12-crown-4)2Na(TCNQ)2 (4)26
Reaction of NaTCNQ (0.0227 g, 0.1 mmol), neutral TCNQ0 (0.0204 g, 0.1 mmol) and 12-crown-4 (0.0352 g, 0.2 mmol) gave 4 as black crystalline solid (0.01085 g, 14%). MS (solution) (MeCN) (ESI−) m/z: 204.5 (TCNQ˙−). (ESI+) m/z: 375.7 (2 × crown + Na+), 199.5 (crown + Na+). IR vmax/cm−1 2969, 2935, 2881 (saturated![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) stretch), 2223, 2201, 2177 (
stretch), 2223, 2201, 2177 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1560 (
stretch), 1560 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (CN)2 stretch), 1506 (
(CN)2 stretch), 1506 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1302 (
ring stretch), 1302 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bend), 1136 (
bend), 1136 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N and
N and ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 951 (C–C ring stretch), 848, 689 (
ring stretch), 951 (C–C ring stretch), 848, 689 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) out of plane bend). Raman vmax/cm−1 2212 (
out of plane bend). Raman vmax/cm−1 2212 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1602 (
stretch), 1602 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1382 (
ring stretch), 1382 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N stretch), 1206 (C
N stretch), 1206 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bending). M.p. 310 °C (dec.)
bending). M.p. 310 °C (dec.)
Preparation of (12-crown-4)2K(TCNQ)2 (5)
Reaction of KTCNQ (0.0972 g, 0.4 mmol), neutral TCNQ0 (0.0816 g, 0.4 mmol) and 12-crown-4 (0.1408 g, 0.8 mmol) afforded 5 as a black crystalline solid (0.01942 g, 24%). MS (solution) (MeCN) (ESI−) m/z: 204.4 (TCNQ˙−). (ESI+) m/z: 375.6 (2 × crown + Na+), 199.4 (crown + Na+). IR vmax/cm−1 2965, 2926, 2877 (saturated![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) stretch), 2195, 2179, 2167 (
stretch), 2195, 2179, 2167 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1560 (
stretch), 1560 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (CN)2 stretch), 1508 (
(CN)2 stretch), 1508 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1363 (
ring stretch), 1363 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bend), 1181 (
bend), 1181 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N and
N and ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 956 (C–C ring stretch), 828, 700 (
ring stretch), 956 (C–C ring stretch), 828, 700 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) out of plane bend). Raman vmax/cm−1 2217 (
out of plane bend). Raman vmax/cm−1 2217 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) stretch), 1608 (
stretch), 1608 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ring stretch), 1390 (
ring stretch), 1390 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) N stretch), 1204 (C
N stretch), 1204 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –
–![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bending). M.p. >300 °C (dec.). Elemental analysis: calculated: C: 60.06%, H: 5.04%, N: 14.00%. Found: C: 59.82%, H: 4.44%, N: 15.74%.
bending). M.p. >300 °C (dec.). Elemental analysis: calculated: C: 60.06%, H: 5.04%, N: 14.00%. Found: C: 59.82%, H: 4.44%, N: 15.74%.
Result and discussion
Complexes involving “simple” TCNQ˙− salts (1 and 2)
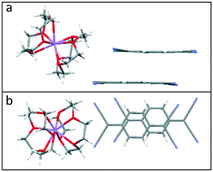
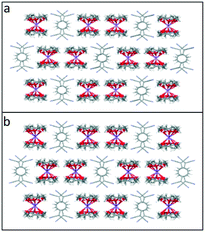
Crystallisation of Li or NaTCNQ from anhydrous acetonitrile in the presence of two equivalents of 12-crown 4 afforded black single crystals of the bis-crown ether complexes (1) and (2) suitable for X-ray structural study. It should be noted that crystallisation of LiTCNQ with 1 equivalent of 12-crown-4 also only produced that bis-crown ether complex (1). Attempts to isolate crystalline samples of (12-crown-4)2KTCNQ also failed, however the salt (12-crown-4)2K(TCNQ)2 (5) was successfully prepared instead.For (12-crown-4)2LiTCNQ (1) and (12-crown-4)2NaTCNQ (2), some difficulties were encountered during the X-ray crystal structure solution as the 12-crown-4 moieties are disordered. The atomic occupancy of the disordered 12-crown-4 units in 1 is 0.504(2): 0.496(2) and 0.5: 0.5 in 2, respectively. Complexes 1 and 2 are iso-structural. In each case, the alkali metal cation (Li+ and Na+) is sandwiched between two 12-crown-4 units and is thus completely screened from the TCNQ˙− counter anion (see Fig. 1). This situation is similar to that seen previously for the (15-crown-5)2KTCNQ (6),1 in which one K+ cation is sandwiched between two disordered 15-crown-5 ether ligands at room temperature, the presence of the ionophore sandwich preventing direct cation–anion contacts.1
 | ||
| Fig. 1 Unit cell of TCNQ complexes (a). (12-crown-4)2LiTCNQ (1) and (b). (12-crown-4)2NaTCNQ (2). In each case, the 12-crown-4 ether ligands are disordered. | ||
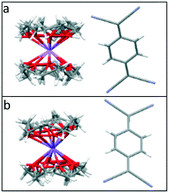
The TCNQ˙− anions are face-to-face π-stacked in both 1 and 2 (see Fig. 2), in which neighbouring anion planes are tilted in respect to each other (6.27° in 1 and 6.89° in 2) forming a novel “cross-stitch”-like stack (see Fig. 2a and c). A top view of adjacent TCNQ˙− units reveals that neighbouring TCNQ˙− anions are significantly long-axis slipped and slightly short-axis slipped (see Table 1 and Fig. 3 for the key structural details). Furthermore, adjacent TCNQ˙− anions are mutually twisted in respect to each other (26.02° in 1 and 26.00° in 2) (see Fig. 2b and d). To our knowledge, this “cross-stitch”-like twisted packing motif of neighbouring TCNQ˙− units is a novel packing motif for alkali metal TCNQ complexes.
 | ||
| Fig. 2 Side (a) and top (b) views of the TCNQ˙− dimer in (12-crown-4)2LiTCNQ (1) in respect to the views (c and d) of (12-crown-4)2NaTCNQ (2). | ||
| Complex | π–π separation/Å | Long-axis slippage/Å | Short-axis slippage/Å | Ref. |
|---|---|---|---|---|
| 1. (at 100 K) | — | 2.40 | 0.26 | This work |
| 2. (at 100 K) | — | 2.34 | 0.23 | This work |
| 3. (at 100 K) | 3.153 | 1.99 | 0.48 | This work |
| 4. (at 100 K) | 3.123 | 2.04 | 0.33 | This work |
| 5. (at 100 K) | 3.125 | 2.03 | 0.30 | This work |
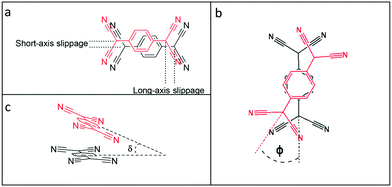 | ||
| Fig. 3 Definition of short and long-axis slippage (a), twist angle φ (b), and tilt angle δ (c) in this study (see Table 1). | ||
In the solid state, both 1 and 2 the TCNQ˙− moiety and the cation-crown ether complex (12-crown-4)2M+ (M+ = Li+ and Na+) form mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 alternating sheets, in a manner similar to that previously reported for 61 (see Fig. 4a and b).
2 alternating sheets, in a manner similar to that previously reported for 61 (see Fig. 4a and b).
 | ||
| Fig. 4 A top view of the solid-state structure of (12-crown-4)2LiTCNQ (1, a) and (12-crown-4)2NaTCNQ (2, b). | ||
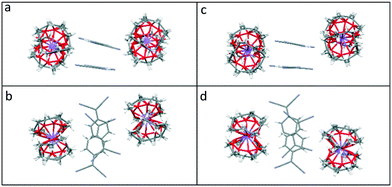
Viewed from the side, both the TCNQ˙− dimers and the cation barrels formed by the (12-crown-4)2M+ (M+ = Li+ and Na+) moieties in 1 and 2 are assembled further into infinite extended columns (see Fig. 5). The centroid–centroid distance between adjacent TCNQ˙− units within the column is 4.155 Å in 1 and 4.154 Å in 2. This packing motif in both 1 and 2 is clearly different from that seen for 6. In the latter case, TCNQ˙− anion dimers are isolated from each other, being surrounded by (15-crown-5)2K+ cation barrels.1
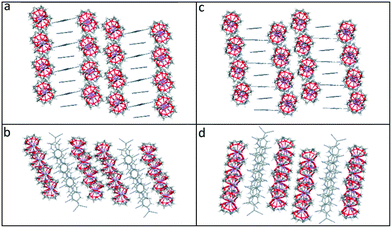 | ||
| Fig. 5 Comparison of the side and top views of the extended columns formed by (12-crown-4)2LiTCNQ (1), (a) and (b) respectively, and by (12-crown-4)2NaTCNQ (2) (c) and (d) respectively. | ||
Complex TCNQ salts (3, 4, and 5)
Crystallisation of Li+, Na+, and K+TCNQ˙− from anhydrous acetonitrile in the presence of 12-crown-4 and one molar equivalent of neutral TCNQ0 afforded crystals of the complex salts of (12-crown-4)2M(TCNQ˙−)(TCNQ0) (M+ = Li+, Na+, or K+) 3, 4, and 5, respectively. These three complex TCNQ salts (3, 4, and 5) are iso-structural.In the solid-state, the alkali metal cation (Li+, Na+, or K+) in each of the complex TCNQ salts (3–5) is coordinated to and sandwiched between two 12-crown-4 units. The resulting cation “barrels” of (12-crown-4)2M+ (M+ = Li+, Na+, and K+) inhibit any direct interaction between alkali metal cations and the TCNQ units (M+–NC), with the cation complexes sitting in channels between the infinite columns of face-to-face π-stacked (TCNQ)2˙− moieties (see Fig. 6 for example).
In 3–5, neighbouring (TCNQ)2˙− units form extended face-to-face π-stacked columns. Within the (TCNQ)2˙− units, the TCNQ moieties are significantly long-axis slipped (see Table 1 and Fig. 3 for the key structural details) with a vertical π–π separation of 3.153 Å in 3, 3.123 Å in 4, and 3.125 Å in 5, respectively. Additionally, individual TCNQ units adopt a shallow boat conformation, as previously reported by Akutagawa for Na+([12]-crown-4)2(TCNQ)2 at 100 K26 (see Fig. 7).
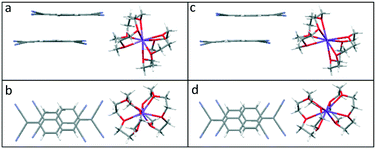 | ||
| Fig. 7 Side and top views of the (TCNQ)2˙− dimer formed in (12-crown-4)2Na(TCNQ)2 (4) (a and b) and in (12-crown-4)2K(TCNQ)2 (5) (c and d). | ||
Furthermore, in 3–5, two pairs of TCNQ dimer units assemble into extended tetramers forming parallel sheets, which are separated by the corresponding cation barrels of (12-crown-4)2M+ (M+ = Li+, Na+, and K+) (see Fig. 8). This observation of tetramer packing motif is consistent with that reported by Akutagawa and co-workers for Li+([12]-crown4)2(TCNQ)2, and Na+([12]-crown4)2(TCNQ)2, in which two kinds of TCNQ units (A and B) form (A⋯B) face-to-face π-stacked dimers at 100 K with additional interdimer interactions ⋯(A⋯B)⋯(B′⋯A')⋯.26,27Fig. 8 illustrates this behaviour for 3 (please refer to ESI† (Fig. S1 and S2) for the corresponding information in 4 and 5.).
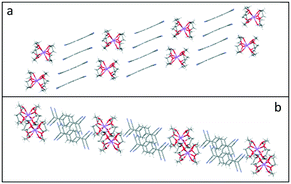 | ||
| Fig. 8 Side (a) and top (b) views of the sheets of face-to-face π-stacked columns of TCNQ dimers formed in (12-crown-4)2Li(TCNQ)2 (3), revealing the long-axis slipped character of the TCNQ dimer. | ||
This repetitive pattern of tetramers further assembles into infinite TCNQ columns (see Fig. 9). The similarity of the bond lengths within the TCNQ columns makes it difficult to distinguish between the TCNQ˙− and neutral TCNQ0 components [please refer to ESI† (Fig. S3 and S4) for the corresponding figures for 4 and 5.].
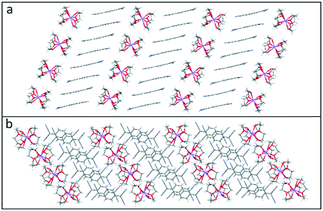 | ||
| Fig. 9 Side (a) and top (b) views of the packing motif of the alternating infinite columns of TCNQ units and cation barrels in (12-crown-4)2Li(TCNQ)2 (3). | ||
Vibrational spectra of ionophore-encapsulated alkali metal TCNQ complexes (1–5)
Vibrational spectroscopy provides a convenient probe into the chemical nature of TCNQ species.35Table 2 presents key data for TCNQ complexes 1–5 together with literature data for comparison. In the infrared (IR) region, two bands are seen which are associated with the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretches characteristic of the TCNQ˙− anion in 1 and 2. An extra band (at ∼2200 cm−1) indicates the presence of neutral TCNQ0 in 3–5. In the spectra, the C
N stretches characteristic of the TCNQ˙− anion in 1 and 2. An extra band (at ∼2200 cm−1) indicates the presence of neutral TCNQ0 in 3–5. In the spectra, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretching bands of TCNQ units will generate obvious Raman peaks because the π electrons are strongly polarisable.37 For the TCNQ complexes (1–5), the peaks in the region 2217 and 2206 cm−1 are ascribed to stretching bands for cyano groups in TCNQ molecule.37 The peak between 1390 and 1380 cm−1 reflects electron transfer between neutral TCNQ0 and TCNQ˙−.37,38
N stretching bands of TCNQ units will generate obvious Raman peaks because the π electrons are strongly polarisable.37 For the TCNQ complexes (1–5), the peaks in the region 2217 and 2206 cm−1 are ascribed to stretching bands for cyano groups in TCNQ molecule.37 The peak between 1390 and 1380 cm−1 reflects electron transfer between neutral TCNQ0 and TCNQ˙−.37,38
| Compound | Infrared data/cm−1 | Raman data/cm−1 | Ref. | |||
|---|---|---|---|---|---|---|
C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretch N stretch |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch C stretch |
C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretch N stretch |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch C stretch |
C–C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretch N stretch |
||
| TCNQ0 | 2228, 2225 | 1545 | 2225 | 1600 | 1450 | 36 |
| TCNQ0 | 2224, 2220 | 1545 | 2230 | 1603 | 1454 | This work |
| LiTCNQ | 2201, 2171 | 1501 | This work | |||
| NaTCNQ | 2197, 2184, 2160 | 1505 | 2204 | 1602 | 1384 | This work |
| KTCNQ | 2215, 2152 | 1505 | This work | |||
| 1 | 2173, 2151 | 1505 | 2214 | 1604 | 1384 | This work |
| 2 | 2172, 2150 | 1505 | 2206 | 1609 | 1389 | This work |
| 3 | 2222, 2198, 2165 | 1507 | 2216 | 1606 | 1385 | This work |
| 4 | 2223, 2201, 2177 | 1506 | 2212 | 1602 | 1382 | This work |
| 5 | 2195, 2179, 2167 | 1508 | 2217 | 1608 | 1390 | This work |
Conclusions
In this paper, we have explored the solid-state architecture of five ionophore-encapsulated alkali metal TCNQ complexes (1–5). The “simple” TCNQ˙− salts 1 and 2 are iso-structural. The 12-crown-4 moiety in both complexes is disordered with one alkali metal cation sandwiched between two crown ethers. Neighbouring TCNQ˙− anions are twisted with respect to each other and form face-to-face π-stacks with further assembling into infinite columns instead of isolated dimers as observed in (15-crown-5)2K+ analogue.1 In the solid-state, the three complex TCNQ salts (3, 4, and 5) consist of (TCNQ)2˙− units, which are packed into repetitive tetramers throughout the crystal structures and separated by cation barrels of (12-crown-4)2M+ moieties.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the EPSRC UK National Crystallography Service at the University of Southampton for the collection of the crystallographic data.28References
- M. C. Grossel and S. C. Weston, Chem. Mater., 1996, 8, 977–980 CrossRef CAS.
- M. C. Grossel and S. C. Weston, Contemp. Org. Synth., 1994, 1, 367–386 RSC.
- V. F. Kaminskii, R. P. Shibaeva and L. O. Atovmyan, J. Struct. Chem., 1974, 14, 1014–1019 CrossRef.
- V. F. Kaminskii, R. P. Shibaeva and L. O. Atovmyan, J. Struct. Chem., 1974, 14, 646–650 CrossRef.
- R. P. Shibaeva and L. O. Atovmyan, J. Struct. Chem., 1972, 13, 514–531 CrossRef.
- R. Shibaeva, L. Atovmyan, V. Ponomarjev, O. Philipenko and L. Rozenberg, Tetrahedron Lett., 1973, 14, 185–188 CrossRef.
- M. C. Grossel, P. B. Hitchcock, K. R. Seddon, T. Welton and S. C. Weston, Chem. Mater., 1994, 6, 1106–1108 CrossRef CAS.
- L. R. Melby, R. J. Harder, W. R. Hertler, W. Mahler, R. E. Benson and W. E. Mochel, J. Am. Chem. Soc., 1962, 84, 3374–3387 CrossRef CAS.
- J. G. Vegter, T. Hibma and J. Kommandeur, Chem. Phys. Lett., 1969, 3, 427–429 CrossRef CAS.
- D. Chesnut and W. Phillips, J. Chem. Phys., 1961, 35, 1002–1012 CrossRef CAS.
- D. Chesnut and P. Arthur, J. Chem. Phys., 1962, 36, 2969–2975 CrossRef CAS.
- P. Nordio, Z. Soos and H. McConnell, Annu. Rev. Phys. Chem., 1966, 17, 237–260 CrossRef CAS.
- T. Hibma, P. Dupuis and J. Kommandeur, Chem. Phys. Lett., 1972, 15, 17–20 CrossRef CAS.
- T. Hibma and J. Kommandeur, Phys. Rev. B: Solid State, 1975, 12, 2608 CrossRef CAS.
- A. Silverstein and Z. Soos, Chem. Phys. Lett., 1976, 39, 525–530 CrossRef CAS.
- J. R. Morton, K. F. Preston, M. D. Ward and P. J. Fagan, J. Chem. Phys., 1989, 90, 2148–2153 CrossRef CAS.
- M. C. Grossel and S. C. Weston, J. Phys. Org. Chem., 1992, 5, 533–539 CrossRef CAS.
- M. C. Grossel, F. A. Evans, J. A. Hriljac, K. Prout and S. C. Weston, J. Chem. Soc., Chem. Commun., 1990, 1494–1495 RSC.
- R. C. Hynes, J. R. Morton, K. F. Preston, A. J. Williams, F. Evans, M. C. Grossel, L. H. Sutcliffe and S. C. Weston, J. Chem. Soc., Faraday Trans., 1991, 87, 2229–2233 RSC.
- J. Lu, X. Qu, G. Peleckis, J. F. Boas, A. M. Bond and L. L. Martin, J. Org. Chem., 2011, 76, 10078–10082 CrossRef CAS PubMed.
- J. Huang, S. Kingsbury and M. Kertesz, Phys. Chem. Chem. Phys., 2008, 10, 2625–2635 RSC.
- M. C. Grossel, F. A. Evans, J. A. Hriljac, J. R. Morton, Y. LePage, K. F. Preston, L. H. Sutcliffe and A. J. Williams, J. Chem. Soc., Chem. Commun., 1990, 439–441 RSC.
- M. C. Grossel, F. A. Evans, J. A. Hriljac, K. Prout and S. C. Weston, J. Chem. Soc., Chem. Commun., 1990, 1494–1495 RSC.
- M. C. Grossel and S. C. Weston, J. Chem. Soc., Chem. Commun., 1992, 1510–1512 RSC.
- B. Yan, P. N. Horton, A. E. Russell, C. J. Wedge, S. C. Weston and M. C. Grossel, CrystEngComm, 2019, 21, 3273–3279 RSC.
- K. Sambe, N. Hoshino, T. Takeda, T. Nakamura and T. Akutagawa, Cryst. Growth Des., 2020, 20, 3625–3634 CrossRef CAS.
- K. Sambe, N. Hoshino, T. Takeda, T. Nakamura and T. Akutagawa, J. Phys. Chem. C, 2020, 124, 13560–13571 CrossRef CAS.
- S. J. Coles and P. A. Gale, Chem. Sci., 2012, 3, 683–689 RSC.
- CrystalClear, Rigaku Corporation, The Woodlands, Texas, U.S.A., 2008-2014 Search PubMed.
- CrysAlisPro Software System, Rigaku Oxford Diffraction, 2021 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- S. C. Weston, PhD thesis, University of London, Royal Holloway and Bedford New College, 1992 Search PubMed.
- R. Ramanathan, S. Walia, A. E. Kandjani, S. Balendran, M. Mohammadtaheri, S. K. Bhargava, K. Kalantar-Zadeh and V. Bansal, Langmuir, 2014, 31, 1581–1587 CrossRef PubMed.
- T. Takenaka, Bull. Inst. Chem. Res., Kyoto Univ., 1969, 47, 387–400 CAS.
- E. Faulques, A. Leblanc, P. Molinié, M. Decoster, F. Conan, J. E. Guerchais and J. Sala-Pala, Spectrochim. Acta, Part A, 1995, 51, 805–819 CrossRef.
- C. Ye, G. Cao, F. Fang, H. Xu, X. Xing, D. Sun and G. Chen, Micron, 2005, 36, 461–464 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Including additional figures and key crystallographic information. CCDC 2101940–2101944. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1ce01075a |
| ‡ Current address: Leibniz-Forschungsinstitut für Molekulare Pharmakologie, 13125 Berlin, Germany. |
| § Current address: Corporate Strategic Research, ExxonMobil Research & Engineering Company, Annandale, NJ 08801, USA. |
| This journal is © The Royal Society of Chemistry 2021 |