Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The cytochrome b5 CybE is regulated by iron availability and is crucial for azole resistance in A. fumigatus
Matthias
Misslinger
 a,
Fabio
Gsaller
a,
Fabio
Gsaller
 b,
Peter
Hortschansky
c,
Christoph
Müller
d,
Franz
Bracher
d,
Michael J.
Bromley
b and
Hubertus
Haas
b,
Peter
Hortschansky
c,
Christoph
Müller
d,
Franz
Bracher
d,
Michael J.
Bromley
b and
Hubertus
Haas
 *a
*a
aDivision of Molecular Biology, Biocenter, Medical University of Innsbruck, Innrain 80, 6020 Innsbruck, Austria. E-mail: hubertus.haas@i-med.ac.at
bManchester Fungal Infection Group, Institute of Inflammation and Repair, University of Manchester, Manchester, UK
cDepartment of Molecular and Applied Microbiology, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Jena, Germany
dDepartment of Pharmacy, Center for Drug Research, Ludwig-Maximilians-University Munich, Munich, Germany
First published on 26th October 2017
Abstract
Cytochrome P450 enzymes (P450) play essential roles in redox metabolism in all domains of life including detoxification reactions and sterol biosynthesis. The activity of P450s is fuelled by two electron-transferring mechanisms, heme-independent P450 reductase (CPR) and the heme-dependent cytochrome b5 (CYB5)/cytochrome b5 reductase (CB5R) system. In this study, we characterised the role and regulation of the cytochrome b5 CybE in the fungal pathogen Aspergillus fumigatus. Deletion of the CybE encoding gene (cybE) caused a severe growth defect in two different A. fumigatus isolates, emphasising the importance of the CB5R system in this pathogen, while the non-essentiality of cybE indicates the partial redundancy of the CPR and CB5R systems. Interestingly, the growth defect caused by the cybE loss of function was even more drastic in A. fumigatus strain AfS77 compared to strain A1160P+ indicating a strain-dependent degree of compensation, which is supported by azole resistance studies. In agreement with CybE being important for the assistance of the ergosterol biosynthetic P450 Cyp51, deletion of cybE decreased resistance to the Cyp51-targeting antifungal voriconazole and caused accumulation of the ergosterol pathway intermediate eburicol. Northern analysis indicated that CybE deficiency leads to the compensatory transcriptional upregulation of Cyp51-encoding cyp51A and CPR-encoding cprA. Overexpression of cybE did not affect azole resistance suggesting that CybE activity is not rate limiting. Expression of cybE was found to be repressed during iron starvation by the iron-regulatory transcription factor HapX demonstrating iron dependence of CybE not only at the level of enzyme activity but also at the level of gene expression.
Significance to metallomicsCytochrome P450 enzymes (P450) are involved in oxidative metabolism and biosynthesis in all kingdoms of life. The required electrons for this redox chemistry are provided by an iron-dependent and an iron-independent system. With a focus on the iron-dependent cytochrome b5, this study gives new insights about the fine-tuning of electron transfer systems dependent on iron availability. |
Introduction
Cytochrome P450 enzymes (P450) are found in all domains of life. They play important roles in xenobiotic metabolism/biotransformation,1 secondary metabolism2 and primary metabolism such as biosynthesis of fatty acids, vitamins3,4 and sterols.5 Although P450 amino acid sequences are extremely diverse between species, their structures are highly conserved. This structural conservation allows all P450s to bind the cofactor heme, which is essential for their function.6 In addition to the structural conservation, members of this family share the catalytic property to activate molecular oxygen to incorporate one molecule of oxygen into a substrate, while the other is reduced to water by NAD(P)H.4 This reductive activation is highly dependent on free electrons and on the co-factor heme of the respective cytochrome P450.6 The heme-bound iron is a crucial factor for this chemistry, as it can adopt different oxidation states and hence serves as an electron acceptor and donor.7 In many species including the yeast Saccharomyces cerevisiae, the two electrons, which are required for a P450 reaction, are provided by two partially redundant systems.8,9 In one system, electrons are passed from NADPH by a cytochrome P450 reductase (CPR) to the respective P450. In the second system, electrons are supplied to P450s by the cytochrome b5 (CYB5)/NADH cytochrome b5 reductase (CB5R) system. It is speculated that CYB5 optimises the kinetics of P450 reactions by allosteric enhancement and by acting as an electron store. In the latter mechanism, the second electron needed for P450 reactions is supplied by CYB5 immediately after the activation of oxygen.10 This facilitation of electron supply increases the formation of reaction products and decreases the release superoxide.CYB5 is also involved in P450-independent reactions as an electron shuttle in systems such as the electron transfer to microsomal desaturases, which are important for unsaturated fatty acid synthesis.11,12 CYB5 also contains heme as a prosthetic group, which is involved in shuttling electrons to the respective target.13,14
Ergosterol is the predominant sterol in fungal membranes.5 One step in ergosterol biosynthesis includes sterol-C14α demethylation, which is performed by the P450 sterol-C14α demethylase CYP51.13 This enzyme is the target of the azole class of antifungals, which bind to CYP51, and inhibit its function and thereby the biosynthesis of ergosterol, resulting in lethality.5 In two yeast species, S. cerevisiae and Candida albicans, individual inactivation of either CPR or CYB5 was not lethal and decreased but did not block ergosterol biosynthesis.14–16 In contrast, simultaneous inactivation of both CYB5 and CPR was lethal,14,17 which corroborates the partial redundancy of these systems in these yeast species. In addition, the CYB5/CB5R system was found to fully support yeast CYP51 in vitro.13 Due to decreased CYP51 activity, loss of CPR leads to increased azole susceptibility, and overexpression of the CYB5 encoding gene in such a setting increased azole resistance in contrast to the CPR knockout, which emphasises partial redundancy of the CYB5/CB5R and CPR systems in electron transport to CYP51 in yeast.14
The filamentous fungus Aspergillus fumigatus is an opportunistic human pathogen and is the major cause for life-threatening invasive aspergillosis.18 Immunocompromised patients are especially at high risk for being infected. One of the most common treatments of fungal infections including aspergillosis is the inhibition of the sterol-C14α-demethylase CYP51 (termed Erg11 in yeast species) with triazole-type antifungals.19–21 The A. fumigatus genome encodes two CYP51 enzymes termed Cyp51A and Cyp51B.22 Both are able to perform sterol-C14α demethylation with similar activity.23,24 However, most mechanisms described for azole resistance are linked to Cyp51A.21,25
The A. fumigatus genome, similar to other filamentous fungal genomes, comprises 77 genes encoding for P450s.2,26 This quantity is striking compared to the widely used model organism S. cerevisiae, which only possesses three P450s: the sterol-C14α-demethylase CYP51/Erg11 and the sterol-C22-desaturase CYP61/Erg5, which are both involved in ergosterol biosynthesis, and the dityrosine hydroxylase CYP56, which is important for yeast sporulation.13 In contrast to S. cerevisiae, mould species like A. fumigatus produce a wide range of secondary metabolites, the biosynthesis of which requires numerous P450-dependent oxygenation reactions.27 Consequently, the expansion of P450s in moulds compared to S. cerevisiae is most likely linked to their complex secondary metabolism. The A. fumigatus genome encodes one putative CYB5, termed CybE (AFUA_2G04710), and two putative CPRs, termed CprA (AFUA_6G10990) and CprB (AFUA_2G07940), which are potentially involved in the biosynthesis of ergosterol and secondary metabolites.
Iron is essential for almost all organisms, including the heme function in P450s and CYB5. Iron acquisition and the expression of iron-consuming genes has to be tightly regulated to keep the balance between iron toxicity and iron shortage.28 In A. fumigatus, this includes gene regulation by the iron-responsive transcription factor HapX, which represses iron-consuming pathways during iron starvation in order to spare iron and activates the respective genes during iron excess.29 Adaptation to iron starvation including high-affinity acquisition via siderophores and HapX-mediated transcriptional reprogramming has been demonstrated to be crucial for the virulence of A. fumigatus.30 This iron-responsive reprogramming also involves the SREBP (sterol regulatory element-binding protein)-type regulator SrbA, which links the control of iron metabolism, ergosterol biosynthesis and adaptation to low oxygen conditions.31
In this study, we characterised the function of CybE in A. fumigatus via gene inactivation and overexpression, including its role in ergosterol biosynthesis, demonstrating its regulation by iron availability via the transcription factor HapX.
Experimental
Growth conditions
In liquid cultures, strains were grown at 37 °C and 200 rpm in Aspergillus minimal medium (MM),32 containing 1% glucose (carbon source), 20 mM glutamine (nitrogen source) and 30 μM FeSO4 (if not otherwise indicated). For plate assays, MM agar or agarose was used (agarose for −Fe conditions to avoid iron contaminations). Plates were grown for 48 h at 37 °C. For low oxygen experiments, strains were grown for 48 h at 37 °C with 1% O2 and 5% CO2.A. fumigatus strains, generation of cybE deletion mutants, cybE inducible strains and reconstitution of ΔcybE
The coding sequence of cybE (AFUA_2G04710) was replaced by a hygromycin (hph) resistance conferring cassette in the AfS77 and A1160P+ strains. The deletion construct was generated by fusion PCR following an approach previously described.33 In the first step, around 1 kb of the cybE 5′-flanking region and 1 kb of the 3′-flanking region were amplified by PCR using primers the AACCCATCCTGTCTGACCGA & TAGTTCTGTTACCGAGCCGGCGGTTTATCTGGATTGGTGACG and GCTCTGAACGATATGCTCCCGCAAGGGGAAAGGAGGTCTG & GGCGTCATCCCTTCCATTGA. Subsequently, fragments were linked to a hygromycin resistance cassette (amplified with the primers CCGGCTCGGTAACAGAACTAACGGCGTAACCAAAAGTCAC and GGGAGCATATCGTTCAGAGCTCTTGACGACCGTTGATCTG) by PCR fusion using the primers CCCTGAAGATGTGCTGCTGA and GTTGTCGATCACAGCGATGC.To reconstitute ΔcybE, plasmid pcybEREC, comprising a pyrithiamine resistance cassette, was generated. The backbone of the pyrithiamine resistance cassette carrying plasmid pSK275 (syn. pME302434) was amplified using the primers ACCGGTCAAAGCTAAAGAGG and ACTAGTTCTAGAGCGGCCG.
The cybE coding sequences including 1.5 kb 5′- and 1.9 kb 3′-flanking regions were amplified with the primers CGGCCGCTCTAGAACTAGTCTGCTCTGTGGCCTGTGATA and CCTCTTTAGCTTTGACCGGTCGCGTGCAGATAAAGATCAA. Both fragments were connected via Gibson Assembly®.
To generate a strain in which cybE is inducible by the addition of xylose to the growth medium (cybExylP), the promoter of cybE was separated from the coding sequence by a hygromycin cassette and a xylose inducible xylP promoter.35 The construct was generated using the NEBuilder® kit (New England Biolabs), with the primers MM118 (AATTCGAGCTCGGTACAGCGGAAGACCATTC), MM119 (TACCTAGGTGTGTAGAAATTATTCGGTTT), MM120 (TCTACACACCTAGGTACAGAAGTCC), MM121 (GCGGACATGGTTGGTTCTTCGAGT), MM122 (AACCAACCATGTCCGCCTCCAAG), MM123 (GCCAAGCTTGCATGCCCACTCCACTGTTGAGAG), and linear pUC19 (amplified with the primers GGCATGCAAGCTTGGCGT and GTACCGAGCTCGAATTCACTG) as a plasmid backbone. MM118/119, MM120/121 and MM122/123 were used to amplify the 5′-homologous region, a hygromycin-xylP cassette and the 3′-homologous region, respectively.
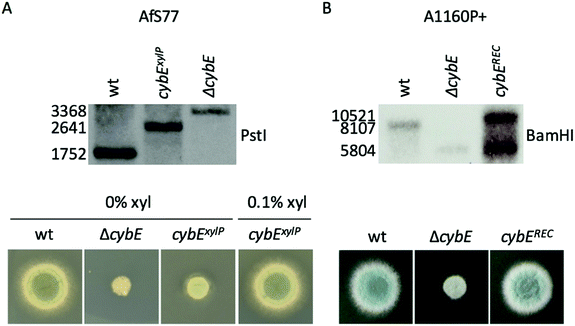
Linear knockout- and promoter exchange constructs, as well as pcybEREC, were transformed into A. fumigatus protoplasts and integrated into the genome by homologous recombination. Integration was proven by Southern blot analysis (Fig. 1). pcybEREC restored the wildtype phenotype in the ΔcybE strain, just as cybE induction did in the cybExylP strain (Fig. 1). Two different A. fumigatus isolates were used for this study, AfS7736 and A1160P+.33 The strains used in this study are listed in Table 1.
| Strain | Genotype | Ref. |
|---|---|---|
| wt, A1160P+ | A1160, ΔakuB::pyrG+ | 33 |
| ΔcybE, A1160P+ | A1160P+, ΔcybE::hph | This study |
| cybE REC | A1160P+, ΔcybE::hph, pcybEREC, ptrA | This study |
| wt, AfS77 | ATCC46645, ΔakuA::loxP | 36 |
| ΔcybE, AfS77 | AfS77, ΔcybE::hph | This study |
| ΔcybExylP | AfS77, pcybE::hph-pxylP | This study |
| ΔsrbA | AfS77, ΔsrbA::hph | This study |
RNA isolation and northern blot analysis
RNA was isolated using TRI Reagent® (Sigma) according to the manufacturer's manual. 10 μg of RNA was used for electrophoresis on 2.2 M formaldehyde agarose gels and subsequently blotted onto Hybond N membranes (Amersham). DIG labelled probes were amplified by PCR.Southern blot analysis
DNA was isolated by PCI extraction and isopropanol precipitation. To confirm the gene-specific restriction pattern of the genetic manipulations, the DNA was digested with PstI (AfS77 strains) or BamHI (A1160P+ strains). The resulting restriction fragments were separated on an agarose gel and transferred to a Hybond N membrane (Amersham) by capillary blotting with NaOH. The deletion of cybE caused the deletion of a PstI site in the CDS and the insertion of a BamHI site. The xylP-promoter insertion elongated the space between the PstI sites. Integration of pcybEREC generated a second 5′-region of cybE. These differences in fragment size were detected with a DIG-labelled probe binding to the 5′-region of cybE. A successful genetic manipulation leads to the following fragments: wt PstI 1752 bp, ΔcybE PstI 3368 bp, cybExylP PstI 2641 bp, wt BamHI 8107 bp, ΔcybE BamHI 5804 bp, cybEREC BamHI 5804 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 521 bp.
521 bp.
Susceptibility tests
For diffusion tests, conidia were mixed with 25 mL of MM to a final concentration of 106 conidia per mL. These plates with 25 mL of MM were poured, and a paper disc was placed in the middle or a hole was pricked out, respectively. For oxidative stress tests, the hole was filled with 100 μL H2O2 (34 mg mL−1). For antifungal susceptibility tests, the paper disc was drenched with voriconazole (10 μL of a 320 μg mL−1 solution in DMSO) or with terbinafine (10 μL of a 100 μg mL−1 solution in DMSO).Sterol measurements with gas chromatography ion trap mass spectrometry (GC-IT-MS)
Sterols were analysed as their corresponding trimethylsilyl (TMS) ethers. The sterols were identified by their relative retention time and mass spectra.37–39 Strains were grown in MM for 24 h at 37 °C and 200 rpm. Mycelia were harvested, freeze-dried, ground and dissolved in 2 M NaOH. Subsequent procedures were performed according to Müller et al.38 The content for each sterol was calculated according to Müller and Bracher.40 The samples were measured in biological triplicates.Results and discussion
Deletion of cybE causes severe growth defects in A. fumigatus
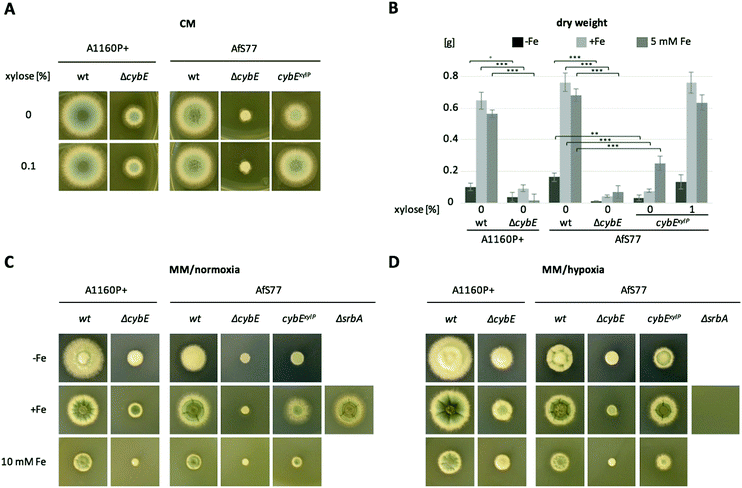
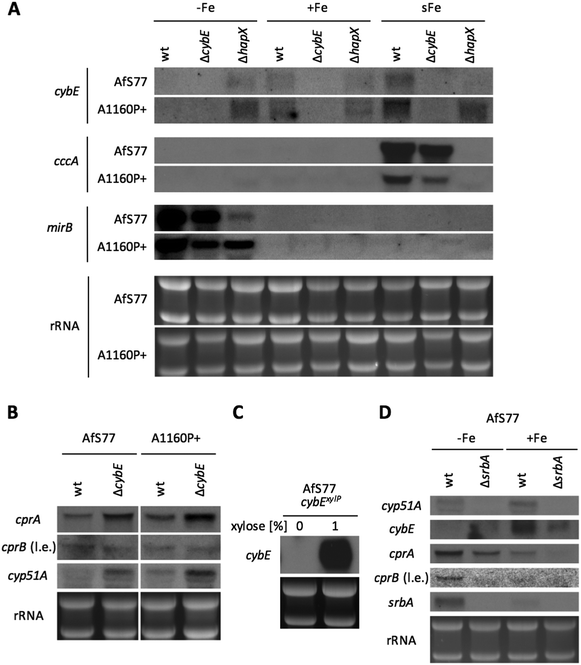
To analyse the function of cytochrome b5, mutant strains were generated lacking CybE (AFUA_2G04710), here referred to as ΔcybE, by replacing the encoding gene cybE with a hygromycin resistance cassette, as described in the Experimental section. This gene deletion was performed in the A. fumigatus isolate A1160P+,33 which is an akuB::pyrG derivative of CEA10 (termed wt A1160P+). To confirm the cybE specificity of this genetic manipulation, the cybE gene was reinserted in the ΔcybE mutant strain in single copy at the cybE locus yielding cybEREC (see the Experimental section). Correct integration of the deletion construct and the reconstitution construct, respectively, was proven by PCR analysis (not shown) and Southern blot analysis (Fig. 1). In addition, a ΔcybE strain was generated in AfS77,36 which is an akuA::loxP derivative of ATCC46645 (termed wt AfS77). Gene deletion in both genetic backgrounds caused a clear reduced growth phenotype. Reinsertion of cybE in ΔcybE in the A1160P+ genetic background (strain cybEREC) restored the wt phenotype, which proves that the growth defect is cybE deletion specific and not an off-target effect (Fig. 1). To study the effects of cybE overexpression, the gene was expressed in AfS77 under the control of the xylose-inducible xylP-promoter.35 Growth of this strain, termed strain cybExylP, under non-inducing conditions (0% xylose) resulted in decreased growth, albeit less extreme compared to cybE deletion, indicating decreased cybE expression compared to the wt strain (Fig. 1). Induction of cybE gene expression (0.1% xylose) again fully restored the wt phenotype.In both genetic backgrounds (A1160P+ and AfS77), the deletion of cybE led to severe growth defects on plates, as well as in liquid cultures in both minimal (MM) and complex (CM) growth media (Fig. 2A–C). Interestingly, the growth defect caused by cybE deletion was more severe in the AfS77 genetic background compared to A1160P+.
CybE has a heme-binding domain and is supposed to be a hemoprotein like other cytochrome b5 proteins.9 Consequently, it requires iron for proper function. Therefore, we compared the growth of wt, ΔcybE and cybExylP strains under conditions of different iron availabilities. Despite the growth defect, both the ΔcybE and uninduced cybExylP strains responded to starvation and excess of iron similar to the wt strain (Fig. 2B–D). This underpins an important role of CybE independent of iron availability.
Taken together, the severe growth defect caused by CybE deficiency in two different A. fumigatus isolates emphasises the importance of the CB5R system in this species while the non-essentiality of cybE indicates the partial redundancy of the CPR and CB5R systems. Moreover, the difference in the degree of growth impairment in the two A. fumigatus strains demonstrates the strain-dependent degree of compensation.
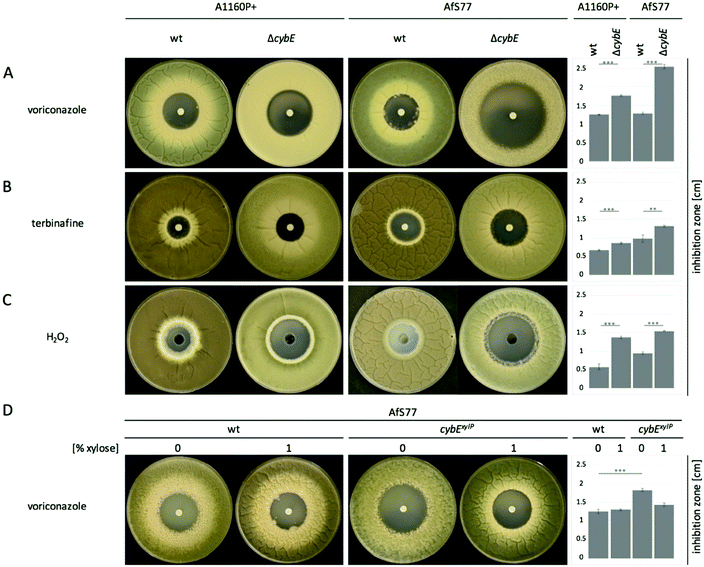
CybE deficiency increases susceptibility to voriconazole, terbinafine, oxidative stress, and cellular accumulation of the ergosterol biosynthetic pathway intermediate eburicol
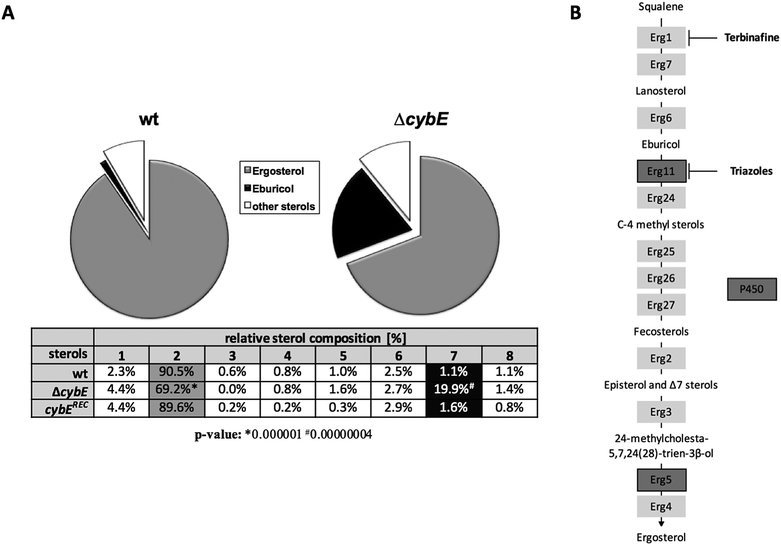
The ergosterol biosynthetic P450 enzyme sterol-C14α-demethylase, termed Cyp51, is the target for triazole-type antifungals such as voriconazole, and previous studies have indicated a link between the roles of CybE and Cyp51.5,13 To investigate a potential involvement of CybE in ergosterol biosynthesis in A. fumigatus, we compared the voriconazole susceptibility of wt, ΔcybE and cybExylP strains in paper disc diffusion assays (Fig. 3A and D). Both wt strains displayed about the same susceptibility to voriconazole. CybE deficiency significantly increased the susceptibility to this antifungal in both genetic backgrounds. However, the increase in susceptibility was more dramatic in AfS77 compared to A1160P+ (Fig. 3A) underlining the strain-specific effects already seen at the level of growth (see above). In line with the fact that CybE deficiency was more detrimental at the level of growth in AfS77 compared to A1160P+, the voriconazole susceptibility data demonstrate that mechanisms that compensate for CybE deficiency are more efficient in the A1160P+ strain compared to AfS77. The cybExylP strain displayed an increased voriconazole susceptibility under non-inducing conditions (without xylose, Fig. 3D), although the increase was lower compared to cybE gene deletion indicating leaky expression below the wt level (Fig. 3A and D), which is in line with the growth pattern of this strain (see above). Inducing conditions (1% xylose), which causes huge overexpression of cybE as shown in Fig. 5C, did not increase voriconazole resistance of the cybExylP strain compared to the wt strain (Fig. 3D), which indicates that CybE activity is not a limiting factor for the Cyp51 activity in A. fumigatus, even under triazole stress. These data also indicate that mutations leading to gain of function of CybE are not expected to play a role in the development of resistance to triazoles in clinical settings.The increase in the voriconazole susceptibility caused by loss of CybE indicates the role of this enzyme in sterol biosynthesis, perhaps via Cyp51. This is corroborated by the analysis of the cellular sterol content of wt and ΔcybE strains (Fig. 4). In line with CybE assisting Cyp51 enzyme activity, CybE deficiency caused a 0.76-fold decrease in the cellular ergosterol content and an 18-fold increased accumulation of eburicol, the substrate of Cyp51. Such a change of the cellular sterol profile is also caused by deletion of one of the two Cyp51-encoding genes or by triazole treatment.5 Reintegration of cybE into ΔcybE (strain cybEREC) restored the wt sterol profile (Fig. 4), proving that the changes observed are indeed caused by gene deletion.
The viability and ergosterol production of CybE-deficient strains indicate that alternative systems are able to compensate for this defect. In agreement, CprA has been shown to be able to assist the activity of both A. fumigatus Cyp51A and Cyp51B in vitro.41 Our data provide the first proof of involvement of CybE in A. fumigatus Cyp51 enzyme activity.
The role of CybE in sterol biosynthesis is further exemplified in susceptibility tests to the allylamine-type antifungal terbinafine, which inhibits squalene epoxidase (Erg1), involved in an early step in ergosterol biosynthesis (Fig. 4B), and is used therapeutically against dermatophytes.42 The two wt isolates demonstrated a distinct susceptibility to terbinafine, with AfS77 being more susceptible compared to A1160P+ (Fig. 3B), again illustrating differences between these two strains. CybE deficiency slightly increased the susceptibility to terbinafine in both genetic backgrounds. Similarly, deletion of the cybE homolog in C. albicans increased the susceptibility to both triazoles and terbinafine.16 It has been shown that squalene epoxidase, although not a member of the P450 family, accomplishes its function in a CPR-dependent manner, i.e. CPR provides electrons to squalene epoxidase using NADPH.43 As the CybE homolog appears to be able to compensate for CPR deficiency in yeast species with azole susceptibility as the read out,15,17 the modest increase in terbinafine susceptibility caused by CybE deficiency in C. albicans16 and here in A. fumigatus might indicate a role of CybE in assisting squalene epoxidase activity.
AfS77 was also found to be less resistant to hydrogen peroxide, and in both genetic backgrounds CybE deficiency increased the susceptibility to this oxidative stressor (Fig. 3C). A. fumigatus must cope with oxidative stress in various environments. Detoxification of oxidative stress is particularly important during host infection, as reactive oxygen species are produced by phagocytes during the immune defense.44 A huge part of intrinsic oxidative stress is caused by respiration and by P450 reactions, which form reactive oxygen species as by-products that harm cell components.45 During P450 reactions, two electrons are required for substrate oxidation. In P450 reactions, unwanted superoxide is formed due to the premature release of the superoxide anion before the second electron can be acquired for proper catalysis.10 CybE is believed to support the donation of the second electron and thereby reduces uncoupling of superoxide anions. As A. fumigatus possesses 77 P450s2,26 including Cyp51, the oxidative-stress susceptibility caused by CybE deficiency might indicate that CybE is important to avoid the generation of intrinsic oxidative stress.
Expression of cybE is HapX-dependent regulated by iron availability
In A. fumigatus as in other fungal species,46,47 transcription of numerous genes that encode iron-dependent proteins or genes that are involved in iron-dependent pathways is downregulated by the transcription factor HapX in cooperation with the CCAAT-binding complex (CBC). Moreover, transcription of this gene set is induced by the HapX–CBC complex within 30 minutes in a shift from iron starvation to iron sufficiency.29 Northern blot analysis demonstrated that cybE expression is also responsive to iron availability (Fig. 5A). Compared to iron sufficiency (+Fe), the cybE transcript level was decreased to undetectability during iron starvation (−Fe) in both wt genetic backgrounds, while the repression of cybE transcription was impaired by HapX deficiency in both genetic backgrounds. These results demonstrate that cybE expression is repressed by HapX during iron starvation. As a control for iron starvation conditions, the expression of the iron-repressed, siderophore transporter-encoding mirB gene was monitored.48 The transcript levels of mirB are lower in hapX-deficient backgrounds because HapX is also involved in the activation of iron acquisition during iron starvation.38,39 In both genetic wt backgrounds, cybE transcription was induced within 30 minutes after the addition of iron to a final concentration of 30 μM (iron shift, sFe), while this induction was abrogated by HapX deficiency in the AfS77 background suggesting that the short-term activation of cybE transcription is mediated by HapX. In contrast, HapX deficiency did not abrogate cybE induction in the A1160P+ genetic background. As a control for the HapX-mediated short-term activation of transcription in the iron shift, we monitored transcript levels of cccA, which encodes a vacuolar iron transporter that is essential for iron detoxification.49The transcription of cccA was induced in the iron shift in both genetic backgrounds in a HapX-dependent manner, thus demonstrating the general functionality of this regulatory circuit in both genetic backgrounds. The difference in the short-term response of cybE transcript levels in the two HapX-deficient genetic backgrounds reveals that cybE expression is subject to different regulatory circuits in the different strains, which might mask the HapX-dependency in A1160P+. The undetectability of cybE transcripts in ΔcybE strains in both genetic backgrounds confirms the successful gene deletion.
Taken together, cybE expression was found to be transcriptionally repressed during iron starvation in a HapX-dependent manner in both genetic backgrounds, while the short-term iron-induction was dependent on HapX in the AfS77, but not the A1160P+ background, revealing an interesting strain-specificity of this regulation.
CybE deficiency causes transcriptional upregulation of the CPR system and cyp51A
Northern blot analysis revealed that CybE deficiency leads to upregulation of cprA transcript levels in both genetic backgrounds, indicating that compensation for loss of CybE involves the upregulation of the alternative CPR system (Fig. 5B). Remarkably, the second CPR encoding gene, cprB, was expressed at a significantly lower level (detectable only after long exposure of the blot) and not affected by CybE deficiency (Fig. 5B).Moreover, CybE deficiency resulted in the transcriptional upregulation of cyp51A in both genetic backgrounds (Fig. 5B). This compensatory transcriptional upregulation of cyp51A emphasises the link between CybE, ergosterol biosynthesis and Cyp51. This transcriptional response is reminiscent of the increase of cyp51A transcription level upon treatment with triazoles,50 which also impairs Cyp51 activity.
Taken together, these data emphasise the links of CybE with CPR and Cyp51A via transcriptional responses caused by the CybE defect and demonstrate the cells' effort to compensate for defects by transcriptional upregulation of a target gene and the CybE alternative system.
The transcription factor SrbA regulates not only cyp51 expression but also Cyp51 associated genes
SrbA is a SREBP-type transcriptional activator that plays an important role in the regulation of sterol biosynthesis, and adaptation to hypoxia and iron starvation in A. fumigatus.51,52 One of the SrbA target genes is cyp51A and, consequently, SrbA deficiency leads to a dramatically increased susceptibility to triazole-type antifungals.53,54 In agreement, northern blot analysis confirmed the transcriptional downregulation of cyp51A in response to SrbA deficiency in the AfS77 background during both iron sufficiency and starvation (Fig. 5D). Furthermore, SrbA deficiency was found to result in the downregulation of transcript levels of cprA during iron starvation and sufficiency, cybE during iron sufficiency, and cprB during iron starvation (Fig. 5D). These data indicate that these Cyp51-assisting protein-encoding genes belong to the SrbA regulon in order to support the coordination of sterol biosynthesis. In accordance with previous findings,31srbA was transcriptionally upregulated during iron starvation (Fig. 5D). Interestingly, also cprA and cprB were found to be transcriptionally upregulated during iron starvation, while cybE was downregulated under this condition as already discussed above (Fig. 5D). The inverse regulation of cybE and cprA/B genes in response to iron availability indicates the cellular optimisation of partially alternative iron-dependent and iron-independent systems to iron availability.CybE is not essential under low oxygen conditions
A. fumigatus can cope with extremely low oxygen conditions, which is important to adapt to certain niches, including inflamed or necrotic tissue during host infection.52 During hypoxia, several enzymes involved in ergosterol biosynthesis are transcriptionally elevated.55 This might be due to the fact that oxygen is required in several steps of ergosterol biosynthesis including Cyp51 enzyme activity.56 Decreased availability of the substrate oxygen is expected to decelerate the reaction speed of the involved enzymes. Increasing the amount of the enzymes would compensate for this to maintain the rate of ergosterol production when oxygen is rare. As CybE supports the Cyp51/Erg11 reaction kinetics,10 we addressed the question of whether CybE is also involved in this adaptation to hypoxia. We tested whether limited oxygen availability worsens the growth of ΔcybE strains with plate growth assays performed at low oxygen conditions (1% O2, 5% CO2). As a control under low oxygen conditions, we included a mutant strain (ΔsrbA) lacking SrbA, a transcription factor that is essential for adaptation to low oxygen conditions including the transcriptional activation of the ergosterol biosynthetic pathway.55 In comparison to normoxic conditions, low oxygen conditions did not decrease the growth of the cybE-deficient strains, in contrast to ΔsrbA (Fig. 2C and D). These data demonstrate that CybE is not essential during low oxygen conditions and that alternative electron-shuttling systems are also able to partially compensate for the loss of CybE under this condition.Conclusion
This study illustrates that CybE assists Cyp51 enzyme activity in A. fumigatus and is consequently important for resistance to ergosterol biosynthesis pathway targeting antifungals. However, overexpression of cybE does not lead to increased resistance to these antifungals, making this enzyme an unlikely cause for resistance in clinical settings. Expression of cybE was found to be regulated by iron availability mediated by the transcription factor HapX. Recently, the CBC–HapX complex has been identified as a repressor of cyp51A/erg11 gene expression in A. fumigatus.54 Consequently, our data demonstrating the HapX-dependent iron regulation of cybE expression expand and underline the link between ergosterol biosynthesis, HapX and iron. Northern blot analysis revealed the compensation of CybE deficiency by the transcriptional upregulation of the alternative CPR system and the target gene cyp51A. Moreover, this study revealed remarkable differences between the two analysed A. fumigatus isolates, which highlights the extent to which different isolates of A. fumigatus can vary with respect to sterol homeostasis. This phenomenon has recently been observed regarding virulence and photoresponse in other A. fumigatus strains.57–59Conflict of interests
The authors declare no conflict of interest.Acknowledgements
This work was supported by the joint D-A-CH program ‘Novel molecular mechanisms of iron sensing and homeostasis in filamentous fungi’ (Deutsche Forschungsgemeinschaft (DFG) DFG_HO 2596/1-1 to PH and Austrian Science Fund (FWF) I1346-B22 to HH). The funders had no role in study design, data collection and analysis; decision to publish; or preparation of the manuscript.References
- V. Burkina, M. K. Rasmussen, N. Pilipenko and G. Zamaratskaia, Comparison of xenobiotic-metabolising human, porcine, rodent, and piscine cytochrome P450, Toxicology, 2017, 375, 10–27 CrossRef CAS PubMed.
- P. Durairaj, J.-S. Hur and H. Yun, Versatile biocatalysis of fungal cytochrome P450 monooxygenases, Microb. Cell Fact., 2016, 15, 125 CrossRef PubMed.
- R. Bernhardt, Cytochromes P450 as versatile biocatalysts, J. Biotechnol., 2006, 124, 128–145 CrossRef CAS PubMed.
- I. G. Denisov, T. M. Makris, S. G. Sligar and I. Schlichting, Structure and Chemistry of Cytochrome P450, Chem. Rev., 2004, 105, 2253–2277 CrossRef PubMed.
- L. Alcazar-Fuoli and E. Mellado, Ergosterol biosynthesis in Aspergillus fumigatus: Its relevance as an antifungal target and role in antifungal drug resistance, Front. Microbiol., 2012, 3, 439 Search PubMed.
- D. Werck-Reichhart and R. Feyereisen, Protein family review Cytochromes P450: a success story, Genome Biol., 2000, 1, 1–9 CrossRef PubMed.
- K. J. McLean, M. Sabri, K. R. Marshall, R. J. Lawson, D. G. Lewis, D. Clift, P. R. Balding, A. J. Dunford, A. J. Warman, J. P. McVey, A.-M. Quinn, M. J. Sutcliffe, N. S. Scrutton and A. W. Munro, Biodiversity of cytochrome P450 redox systems, Biochem. Soc. Trans., 2005, 33, 796–801 CrossRef CAS PubMed.
- H. J. M. van den Brink, R. F. M. van Gorcom, C. A. M. J. J. van den Hondel and P. J. Punt, Cytochrome P450 Enzyme Systems in Fungi, Fungal Genet. Biol., 1998, 23, 1–17 CrossRef CAS PubMed.
- T. D. Porter, The roles of cytochrome b5 in cytochrome P450 reactions, J. Biochem. Mol. Toxicol., 2002, 16, 311–316 CrossRef CAS PubMed.
- J. B. Schenkman and I. Jansson, The many roles of cytochrome b5, Pharmacol. Ther., 2003, 97, 139–152 CrossRef CAS PubMed.
- P. Strittmatter, L. Spatz, D. Corcoran, M. J. Rogers, B. Setlow and R. Redline, Purification and properties of rat liver microsomal stearyl coenzyme A desaturase, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 4565–4569 CrossRef CAS.
- G. Vergères and L. Waskell, Cytochrome b5, its functions, structure and membrane topology, Biochimie, 1995, 77, 604–620 CrossRef.
- D. C. Lamb, D. E. Kelly, N. J. Manning, M. A. Kaderbhai and S. L. Kelly, Biodiversity of the P450 catalytic cycle: Yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction, FEBS Lett., 1999, 462, 283–288 CrossRef CAS PubMed.
- G. Truan, J. C. Epinat, C. Rougeulle, C. Cullin and D. Pompon, Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain, Gene, 1994, 142, 123–127 CrossRef CAS PubMed.
- K. Venkateswarlu, D. C. Lamb, D. E. Kelly, N. J. Manning and S. L. Kelly, The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity, J. Biol. Chem., 1998, 273, 4492–4496 CrossRef CAS PubMed.
- K. M. Rogers, C. A. Pierson, N. T. Culbertson, C. Mo, A. M. Sturm, J. Eckstein, R. Barbuch, N. D. Lees and M. Bard, Disruption of the Candida albicans CYB5 gene results in increased azole sensitivity, Antimicrob. Agents Chemother., 2004, 48, 3425–3435 CrossRef CAS PubMed.
- T. R. Sutter and J. C. Loper, Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P450 reductase causes increased sensitivity to ketoconazole, Biochem. Biophys. Res. Commun., 1989, 160, 1257–1266 CrossRef CAS PubMed.
- K. J. Kwon-Chung and J. A. Sugui, Aspergillus fumigatus-What Makes the Species a Ubiquitous Human Fungal Pathogen?, PLoS Pathog., 2013, 9, e1003743 Search PubMed.
- T. F. Patterson, G. R. Thompson, D. W. Denning, J. A. Fishman, S. Hadley, R. Herbrecht, D. P. Kontoyiannis, K. A. Marr, V. A. Morrison, M. H. Nguyen, B. H. Segal, W. J. Steinbach, D. A. Stevens, T. J. Walsh, J. R. Wingard, J.-A. H. Young and J. E. Bennett, Executive Summary: Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America, Clin. Infect. Dis., 2016, 63, 433–442 CrossRef PubMed.
- T. J. Walsh, E. J. Anaissie, D. W. Denning, R. Herbrecht, D. P. Kontoyiannis, K. A. Marr, V. A. Morrison, B. H. Segal, W. J. Steinbach, D. A. Stevens, J. van Burik, J. R. Wingard and T. F. Patterson, Treatment of Aspergillosis: Clinical Practice Guidelines of the Infectious Diseases Society of America, Clin. Infect. Dis., 2008, 46, 327–360 CrossRef CAS PubMed.
- R. Garcia-Rubio, M. Cuenca-Estrella and E. Mellado, Triazole Resistance in Aspergillus Species: An Emerging Problem, Drugs, 2017, 77, 599–613 CrossRef CAS PubMed.
- E. Mellado, T. M. Diaz-Guerra, M. Cuenca-Estrella and J. L. Rodriguez-Tudela, Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species, J. Clin. Microbiol., 2001, 39, 2431–2438 CrossRef CAS PubMed.
- E. Mellado, G. Garcia-Effron, M. J. Buitrago, L. Alcazar-Fuoli, M. Cuenca-Estrella and J. L. Rodriguez-Tudela, Targeted Gene Disruption of the 14-Sterol Demethylase (cyp51A) in Aspergillus fumigatus and Its Role in Azole Drug Susceptibility, Antimicrob. Agents Chemother., 2005, 49, 2536–2538 CrossRef CAS PubMed.
- W. Hu, S. Sillaots, S. Lemieux, J. Davison, S. Kauffman, A. Breton, A. Linteau, C. Xin, J. Bowman, J. Becker, B. Jiang and T. Roemer, Essential Gene Identification and Drug Target Prioritization in Aspergillus fumigatus, PLoS Pathog., 2007, 3, e24 Search PubMed.
- L. Alcazar-Fuoli and E. Mellado, Current status of antifungal resistance and its impact on clinical practice, Br. J. Haematol., 2014, 166, 471–484 CrossRef PubMed.
- J. Park, S. Lee, J. Choi, K. Ahn, B. Park, J. Park, S. Kang and Y.-H. Lee, Fungal cytochrome P450 database, BMC Genomics, 2008, 9, 402 CrossRef PubMed.
- L. M. Podust, D. H. Sherman, F. H. Arnold, B. Tudzynski, R. A. E. Butchko, M. Kimura, B. Youn, B. S. Kim, J. S. Ahn, D. H. Sherman, Y. J. Yoon, M. J. Sutcliffe, N. S. Scrutton, A. W. Munro, J. R. Falck, T. Shimada, M. R. Waterman, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O’Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill and D. A. Hopwood, Diversity of P450 enzymes in the biosynthesis of natural products, Nat. Prod. Rep., 2012, 29, 1251 RSC.
- H. Haas, Iron – A Key Nexus in the Virulence of Aspergillus fumigatus., Front. Microbiol., 2012, 3, 28 CAS.
- F. Gsaller, P. Hortschansky, S. R. Beattie, V. Klammer, K. Tuppatsch, B. E. Lechner, N. Rietzschel, E. R. Werner, A. A. Vogan, D. Chung, M. Kato, R. A. Cramer, A. A. Brakhage and H. Haas, The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess, EMBO J., 2014, 33, 2261–2276 CrossRef CAS PubMed.
- M. Schrettl and H. Haas, Iron homeostasis-Achilles’ heel of Aspergillus fumigatus?, Curr. Opin. Microbiol., 2011, 14, 400–405 CrossRef CAS PubMed.
- M. Blatzer, B. M. Barker, S. D. Willger, N. Beckmann, S. J. Blosser, E. J. Cornish, A. Mazurie, N. Grahl, H. Haas and R. a Cramer, SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus, PLoS Genet., 2011, 7, e1002374 CAS.
- G. Pontecorvo, J. A. Roper, L. M. Hemmons, K. D. Macdonald and A. W. J. Bufton, The genetics of Aspergillus nidulans, Adv. Genet., 1953, 5, 141–238 CAS.
- M. G. Fraczek, M. Bromley, A. Buied, C. B. Moore, R. Rajendran, R. Rautemaa, G. Ramage, D. W. Denning and P. Bowyer, The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus, J. Antimicrob. Chemother., 2013, 68, 1486–1496 CrossRef CAS PubMed.
- S. Krappmann, N. Jung, B. Medic, S. Busch, R. A. Prade and G. H. Braus, The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis, Mol. Microbiol., 2006, 61, 76–88 CrossRef CAS PubMed.
- I. Zadra, B. Abt, W. Parson and H. Haas, xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE, Appl. Environ. Microbiol., 2000, 66, 4810–4816 CrossRef CAS PubMed.
- T. Hartmann, M. Dümig, B. M. Jaber, E. Szewczyk, P. Olbermann, J. Morschhäuser and S. Krappmann, Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system, Appl. Environ. Microbiol., 2010, 76, 6313–6317 CrossRef CAS PubMed.
- L. Alcazar-Fuoli, E. Mellado, G. Garcia-Effron, J. F. Lopez, J. O. Grimalt, J. M. Cuenca-Estrella and J. L. Rodriguez-Tudela, Ergosterol biosynthesis pathway in Aspergillus fumigatus, Steroids, 2008, 73, 339–347 CrossRef CAS PubMed.
- C. Müller, V. Staudacher, J. Krauss, M. Giera and F. Bracher, A convenient cellular assay for the identification of the molecular target of ergosterol biosynthesis inhibitors and quantification of their effects on total ergosterol biosynthesis, Steroids, 2013, 78, 483–493 CrossRef PubMed.
- P. Keller, C. Müller, I. Engelhardt, E. Hiller, K. Lemuth, H. Eickhoff, K.-H. Wiesmüller, A. Burger-Kentischer, F. Bracher and S. Rupp, An Antifungal Benzimidazole Derivative Inhibits Ergosterol Biosynthesis and Reveals Novel Sterols, Antimicrob. Agents Chemother., 2015, 59, 6296–6307 CrossRef CAS PubMed.
- C. Müller and F. Bracher, Determination by GC-IT/MS of phytosterols in herbal medicinal products for the treatment of lower urinary tract symptoms and food products marketed in Europe, Planta Med., 2015, 81, 613–620 CrossRef PubMed.
- A. G. S. Warrilow, J. E. Parker, C. L. Price, W. D. Nes, S. L. Kelly and D. E. Kelly, In Vitro Biochemical Study of CYP51-Mediated Azole Resistance in Aspergillus fumigatus, Antimicrob. Agents Chemother., 2015, 59, 7771–7778 CrossRef CAS PubMed.
- A. K. Gupta, M. Paquet and F. C. Simpson, Therapies for the treatment of onychomycosis, Clin. Dermatol., 2013, 31, 544–554 CrossRef PubMed.
- T. D. Porter, Electron Transfer Pathways in Cholesterol Synthesis, Lipids, 2015, 50, 927–936 CrossRef CAS PubMed.
- K. B. Boyle, L. R. Stephens and P. T. Hawkins, Activation of the neutrophil NADPH oxidase by Aspergillus fumigatus, Ann. N. Y. Acad. Sci., 2012, 1273, 68–73 CrossRef CAS PubMed.
- T. Finkel and N. J. Holbrook, Oxidants, oxidative stress and the biology of ageing, Nature, 2000, 408, 239–247 CrossRef CAS PubMed.
- M. Schrettl, N. Beckmann, J. Varga, T. Heinekamp, I. D. Jacobsen, C. Jöchl, T. a Moussa, S. Wang, F. Gsaller, M. Blatzer, E. R. Werner, W. C. Niermann, A. a Brakhage and H. Haas, HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus, PLoS Pathog., 2010, 6, e1001124 Search PubMed.
- P. Hortschansky, M. Eisendle, Q. Al-Abdallah, A. D. Schmidt, S. Bergmann, M. Thön, O. Kniemeyer, B. Abt, B. Seeber, E. R. Werner, M. Kato, A. a Brakhage and H. Haas, Interaction of HapX with the CCAAT-binding complex-a novel mechanism of gene regulation by iron, EMBO J., 2007, 26, 3157–3168 CrossRef CAS PubMed.
- H. Haas, M. Schoeser, E. Lesuisse, J. F. Ernst, W. Parson, B. Abt, G. Winkelmann and H. Oberegger, Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C, Biochem. J., 2003, 371, 505–513 CrossRef CAS PubMed.
- F. Gsaller, M. Eisendle, B. E. Lechner, M. Schrettl, H. Lindner, D. Müller, S. Geley and H. Haas, The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus, Metallomics, 2012, 4, 1262–1270 RSC.
- A. Buied, C. B. Moore, D. W. Denning and P. Bowyer, High-level expression of cyp51B in azole-resistant clinical Aspergillus fumigatus isolates, J. Antimicrob. Chemother., 2013, 68, 512–514 CrossRef CAS PubMed.
- C. M. Bien and P. J. Espenshade, Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis, Eukaryotic Cell, 2010, 9, 352–359 CrossRef CAS PubMed.
- N. Grahl, K. M. Shepardson, D. Chung and R. A. Cramer, Hypoxia and fungal pathogenesis: to air or not to air?, Eukaryotic Cell, 2012, 11, 560–570 CrossRef CAS PubMed.
- S. J. Blosser and R. A. Cramer, SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A), Antimicrob. Agents Chemother., 2012, 56, 248–257 CrossRef CAS PubMed.
- F. Gsaller, P. Hortschansky, T. Furukawa, P. D. Carr, B. Rash, J. Capilla, C. Müller, F. Bracher, P. Bowyer, H. Haas, A. A. Brakhage and M. J. Bromley, Sterol Biosynthesis and Azole Tolerance Is Governed by the Opposing Actions of SrbA and the CCAAT Binding Complex, PLoS Pathog., 2016, 12, e1005775 Search PubMed.
- D. Chung, B. M. Barker, C. C. Carey, B. Merriman, E. R. Werner, B. E. Lechner, S. Dhingra, C. Cheng, W. Xu, S. J. Blosser, K. Morohashi, A. Mazurie, T. K. Mitchell, H. Haas, A. P. Mitchell and R. A. Cramer, ChIP-seq and In Vivo Transcriptome Analyses of the Aspergillus fumigatus SREBP SrbA Reveals a New Regulator of the Fungal Hypoxia Response and Virulence, PLoS Pathog., 2014, 10, e1004487 Search PubMed.
- A. M. Galea and A. J. Brown, Special relationship between sterols and oxygen: Were sterols an adaptation to aerobic life?, Free Radical Biol. Med., 2009, 47, 880–889 CrossRef CAS PubMed.
- N. P. Keller, Heterogeneity Confounds Establishment of ‘a’ Model Microbial Strain, mBio, 2017, 8, e00135 CrossRef PubMed.
- C. H. Kowalski, S. R. Beattie, K. K. Fuller, E. A. McGurk, Y.-W. Tang, T. M. Hohl, J. J. Obar and R. A. Cramer, Heterogeneity among Isolates Reveals that Fitness in Low Oxygen Correlates with Aspergillus fumigatus Virulence, mBio, 2016, 7, e01515 CrossRef PubMed.
- K. K. Fuller, R. A. Cramer, M. E. Zegans, J. C. Dunlap and J. J. Loros, Aspergillus fumigatus Photobiology Illuminates the Marked Heterogeneity between Isolates, mBio, 2016, 7, e01517 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2017 |